Introduction
Lower back pain is a common health problem
worldwide. Chronic pain makes life difficult, and in some cases, it
can even develop into disability: Statistics have shown that lower
back pain has been one of the three leading causes of disability in
the past two decades (1), directly
leading to long-term heavy burdens for both the immediate family
and in society at large. Intervertebral disc degeneration (IDD) is
generally regarded as the predominant causative factor for lower
back pain (2). The process of IDD
is spontaneous with age, and both lack of vascularization and
limited cell presence in the disc make self-repair impossible
(3). When degeneration occurs, the
homeostatic environment is disturbed and the balance between
anabolism and catabolism is disrupted, leading to an increased
apoptosis of nucleus pulposus (NP) cells accompanied by the loss of
the extracellular matrix (ECM) (3,4). NP
cells are the major cells in NP tissue and have an important role
in resisting mechanical loading by synthesizing ECM (4). The apoptosis of NP cells is
considered crucial in the degeneration process (5). Thus, the search for new molecules
able to prevent apoptosis is fundamental in the search to provide
better therapeutic interventions against IDD.
Syringic acid (SyrA), a natural phenolic compound,
exists in various plant species and possesses many biologically
important properties. A recent review by Srinivasulu et al
(6) details how SyrA has been
shown to fulfill various roles as an anti-oxidant,
anti-inflammatory, chemo-protective and anti-microbial compound.
The chemo-protective activity of SyrA is effective on multiple
cells with different functions. For example, pre-treatment with
SyrA has been shown to reverse neuronal injury as well as the
apoptosis of hippocampal neuronal cells, processes that were
induced by oxygen-glucose deprivation/reperfusion, and the
neuroprotective effect of SyrA was shown to be dose-dependent
(7). In addition,
H2O2-induced apoptosis of retinal ganglion
cells, apoptosis of cardiomyocytes following hypoxia/reoxygenation
treatment and apoptosis in rat renal ischemia-reperfusion injury
have all been shown to be inhibited by pre-treatment or treatment
with SyrA (8–10). However, to date, the effect of SyrA
on NP cells and their apoptosis is unknown, although these
aforementioned studies could suggest that this molecule might
reverse the apoptosis of NP cells.
The present study investigated the role of SyrA in
the lipopolysaccharide (LPS)-induced apoptosis of NP cells, and
following SyrA treatment, the differentially expressed genes (DEGs)
were analyzed using RNA-Seq analysis. Reverse
transcription-quantitative PCR (RT-qPCR) and bioinformatics
analyses were also performed to study the potential mechanisms
involved. The results of the present study may provide fresh
avenues for further exploring the reversion of apoptotic NP cells
and the therapeutic intervention of IDD.
Materials and methods
Cells and cell culture
Human NP cells which had previously been isolated
from NP tissues of patients with lumbar fracture (AO
classification, B1) were kindly provided by Dr. Guo (Shanghai
Changzheng Hospital, Shanghai, China) (11). NP cells were cultured in DMEM/F12
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and
1% penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.)
at 37°C in an incubator in an atmosphere of 5% CO2.
Cell viability
Cell Counting kit-8 (CCK-8; Beyotime Institute of
Biotechnology) was used to evaluate cell survival. Briefly, NP
cells were seeded in a 96-well plate (5×103 cells/well)
and incubated overnight. Subsequently, LPS (Sigma-Aldrich; Merck
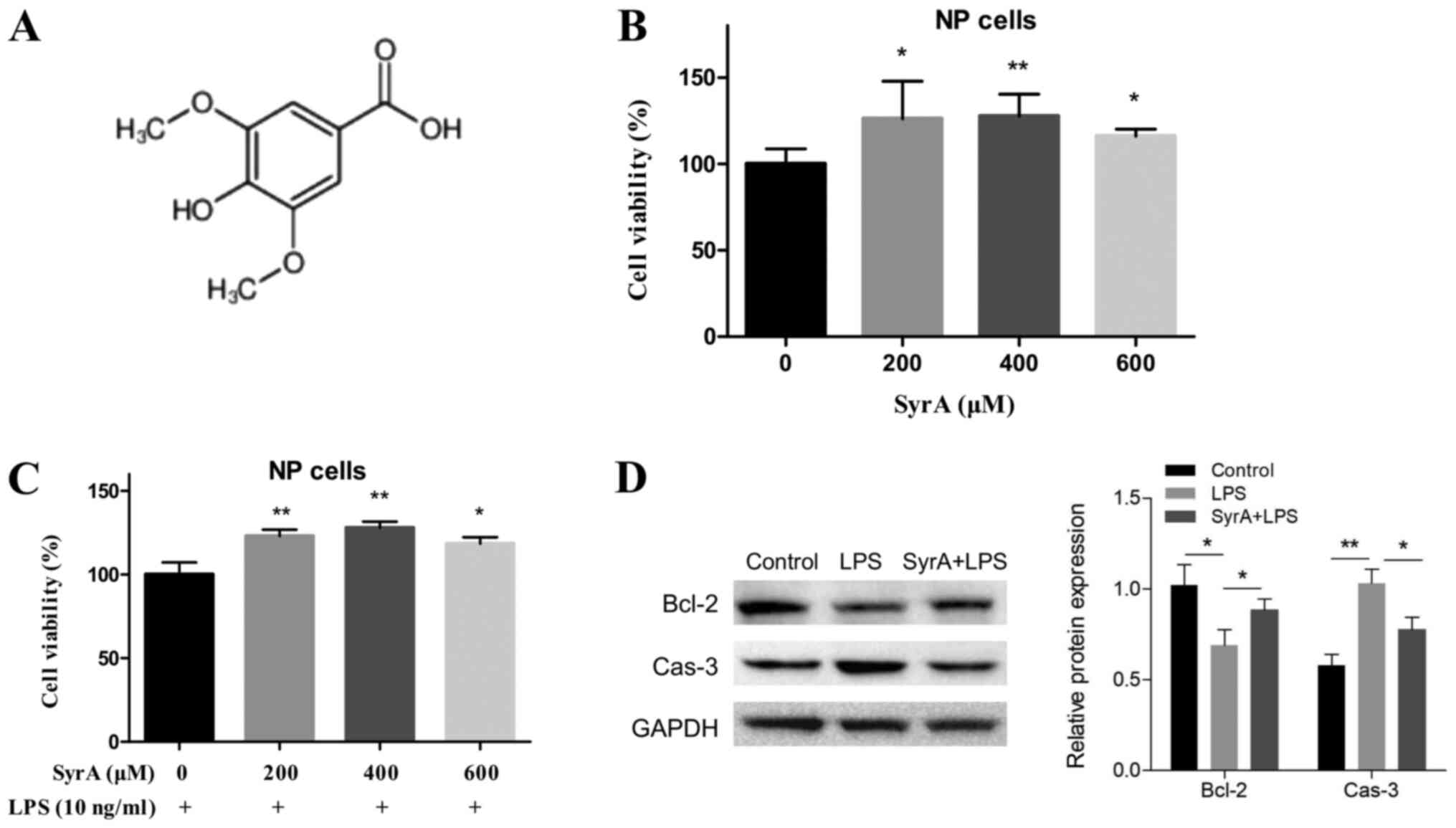
KGaA) or SyrA (Sigma-Aldrich; Merck KGaA) (Fig. 2A) were added at a final
concentration of 0, 0.001, 0.01, 0.1, 1, 10 and 100 µg/ml for LPS
(12,13), and 0, 200, 400 and 600 µM/ml for
SyrA. After 3 days, 10 µl CCK-8 reagent was added to each well and
cultured for a further 2 h. Subsequently, the plate was transferred
into a microplate reader (BioTek Instruments, Inc.), and the
absorbance at 450 nm was detected.
Western blot analysis
NP cells were seeded in a 6-well plate
(4×105 cells/well) and treated with LPS (0.01 µg/ml), or
with combined SyrA (400 µM) and LPS (0.01 µg/ml) for 48 h. Cell
lysates were then prepared using radioimmunoprecipitation assay
(RIPA) buffer (Sigma-Aldrich; Merck KGaA) and total protein
concentration was determined using a BCA protein assay kit
(Biosharp Life Sciences). A total of 10 µg protein was separated by
SDS-PAGE (12% gels) and transferred to polyvinylidene fluoride
membranes. Subsequently, these membranes were blocked in blocking
buffer (Beyotime Institute of Biotechnology) at room temperature
for 60 min and separately incubated overnight at 4°C with
antibodies against caspase-3 (cat. no. 9662; 1:1,000), Bcl-2 (cat.
no. 15071; 1:1,000), and GAPDH (cat. no. 5174; 1:1,000). The next
day, the membranes were incubated with horseradish
peroxidase-conjugated secondary antibody (cat. no. 7076; 1:1,000)
at room temperature for 2 h. All the antibodies were purchased from
Cell Signaling Technology, Inc., and protein expression was
normalized against GAPDH. Band densitometry was conducted and
semi-quantified using ImageJ software version 1.8.0 (National
Institutes of Health), and the relative gray value was used to
represent the relative protein expression.
RNA-Seq analysis
NP cells were treated with LPS (0.01 µg/ml) or
combined LPS (0.01 µg/ml)/SyrA (400 µM) for 48 h; control cells
were given no treatment. Total RNA from the three groups was
isolated using an RNeasy Mini kit (Qiagen GmbH) and poly
A-containing mRNA molecules were purified using poly T
oligo-attached magnetic beads. Next, the mRNA was fragmented into
small pieces, and these fragments were reverse-transcribed into
first-strand cDNA using RNA Seq First Strand Master Mix (Agilent
Technologies, Inc.) following manufacturer's instructions.
Subsequently, second-strand cDNA was synthesized using DNA
polymerase I and RNase H. After an end-repair process, consisting
of the addition of a single adenosine base, the products were
purified and enriched with PCR to create the final cDNA library.
Purified libraries were quantified using a Qubit® 2.0
Fluorometer (Thermo Fisher Scientific, Inc.) and validated using an
Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.) to confirm
the size of the insert and calculate the molar concentration.
Clusters were generated using the Illumina software cBot, with the
library diluted to 10 pM and then sequenced on an Illumina NovaSeq
6000 instrument (Illumina, Inc.). All gene level files were
imported into the Agilent GeneSpring GX software version 11.5
(Agilent Technologies, Inc.) for further analysis. Only genes that
fulfilled the two criteria [log2fold change (FC) (abs,
absolute value)≥1 and P<0.05], were filtered as DEGs.
RT-qPCR
Total RNA from NP cells of each group was obtained
using the RNAsimple Total RNA Extraction kit (Tiangen Biotech Co.,
Ltd.). Subsequently, 1 µg total RNA was used to synthesize
complementary DNA (cDNA) with a RT PCR kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.). Subsequently, 0.5 µl of the
resulting cDNA was added to a reaction system according to the
instructions of the SYBR Premix Ex Taq™ II kit (Takara
Biotechnology Co., Ltd.). Thermocycling conditions were as follows:
95°C for 60 sec, followed by 40 cycles at 95°C for 15 sec and 60°C
for 60 sec. Primers with the sequences listed in Table I were provided by Sangon Biotech
Co., Ltd. Gene expression data were processed using the
2−ΔΔCq method (14) and
normalized to GAPDH.
 | Table I.Primer sequences for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences for reverse
transcription-quantitative PCR.
| Primer name | Primer sequence
(5′-3′) |
|---|
| UPK3BL1 | F:
TTCAGCAGCCACAACATCTC |
| UPK3BL1 | R:
GCAGCCAATTTTTGTTGGTT |
| CACNA2D1 | F:
ACGCAGCAGTCCATATTCCTA |
| CACNA2D1 | R:
GCCACAATAATGAAGGGTCTTCC |
| PPP1R9A | F:
ATCGAACTGAGTTTCAGGCAC |
| PPP1R9A | R:
GGTTCCATACCCATCTGCATAAA |
| WRN | F:
CACAGCAGCGGAAATGTCCT |
| WRN | R:
GAGCAATCACTAGCATCGTAACT |
| PLK4 | F:
AAGCTCGACACTTCATGCACC |
| PLK4 | R:
GCATTTTCAGTTGAGTTGCCAG |
| SLC9A6 | F:
TCACCCTCACCATTCTCACA |
| SLC9A6 | R:
CACTTCACAGCTCAGGGTCA |
| NR3C1 | F:
ACAGCATCCCTTTCTCAACAG |
| NR3C1 | R:
AGATCCTTGGCACCTATTCCAAT |
| GAPDH | F:
GGAGCGAGATCCCTCCAAAAT |
| GAPDH | R:
GGCTGTTGTCATACTTCTCATGG |
Bioinformatics analyses
Gene Ontology (GO) (15) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) (16) signaling
pathway analyses were performed to analyze the biological functions
and the potential relevant signaling pathways of the DEGs using
clusterProfiler, a R/Bioconductor implementation. P<0.05 was
used as a threshold to screen relevant GO terms and KEGG pathways,
and the top 30 pathways were selected for drawing.
Statistical analyses
Statistical analyses were performed using SPSS 19.0
software (IBM Corp.). The results are presented as the mean ±
standard deviation (SD). For comparisons between groups, one-way
analysis of variance was used, followed by Tukey's multiple
comparison tests. All the experiments, except RNA-Seq, were
performed at least three times. P<0.05 was considered to
indicate a statistically significant difference.
Results
LPS induces apoptosis of NP cells
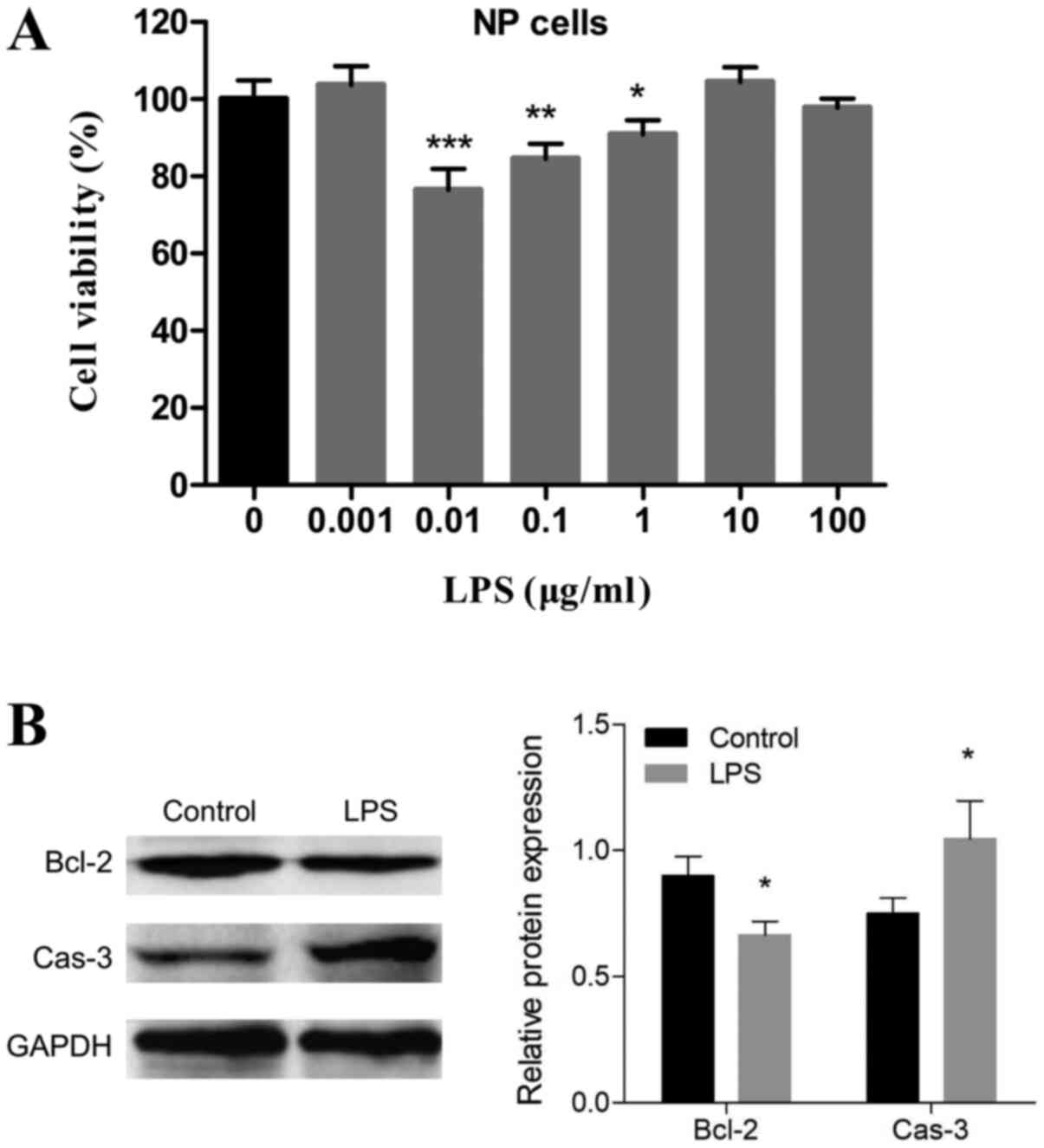
To confirm the appropriate concentration of LPS, NP
cells were treated with 0, 0.001, 0.01, 0.1, 1, 10 and 100 µg/ml
LPS. As shown in Fig. 1A,
significant cell inhibition of LPS was observed at 0.01, 0.1 and 1,
and 0.01 µg/ml LPS showed the greatest effect on the primary NP
cells' viability. Hence, the 0.01 µg/ml concentration was selected
for further studies. Western blot analysis showed that the protein
level of caspase-3 significantly increased and that Bcl-2
expression levels significantly decreased (Fig. 1B), which suggested that there was
an onset of apoptosis after LPS stimulation.
SyrA reverses LPS-induced apoptosis in
NP cells
Before studying how SyrA (structure shown in
Fig. 2A) affected LPS-induced
apoptosis, its cytotoxicity on NP cells was evaluated. The results
of the CCK-8 assay indicated that there were no adverse cytotoxic
effects on NP cells at any of the three concentrations; however,
there was a significant proliferative effect (Fig. 2B). Subsequently, the effects of the
three aforementioned concentrations of SyrA on LPS-induced NP cell
apoptosis were studied; the highest rate of reversion of cell
apoptosis was induced by 400 µM SyrA (Fig. 2C). The protein levels of caspase-3
and Bcl-2 were subsequently detected, and the results indicated
that their expression in NP cells was reversed following treatment
with 400 µM SyrA (Fig. 2D).
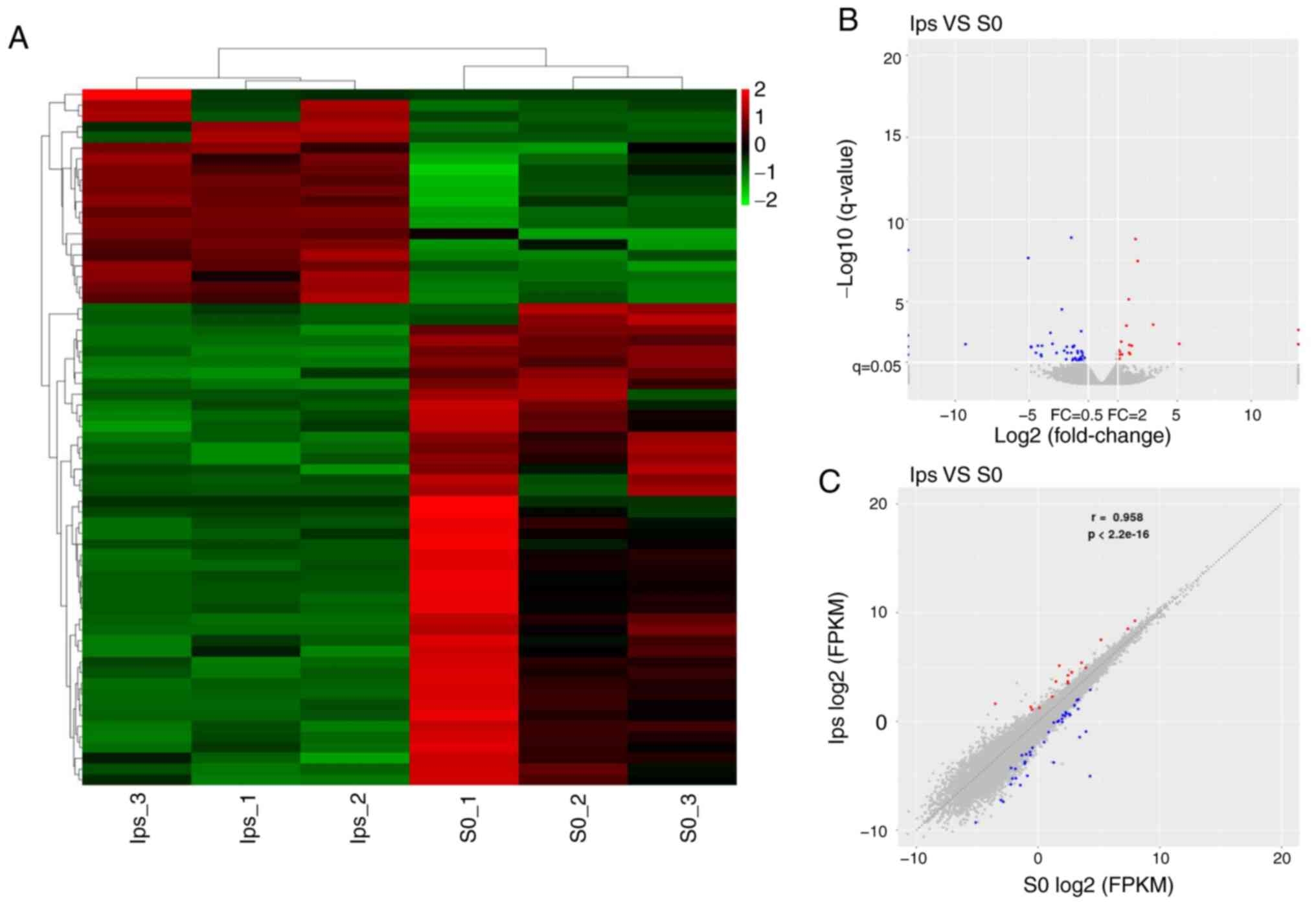
RNA-Seq analysis of NP cells treated
with LPS
To develop a better understanding of the apoptosis
reversal induced by SyrA, RNA-Seq of NP cells from LPS groups (LPS
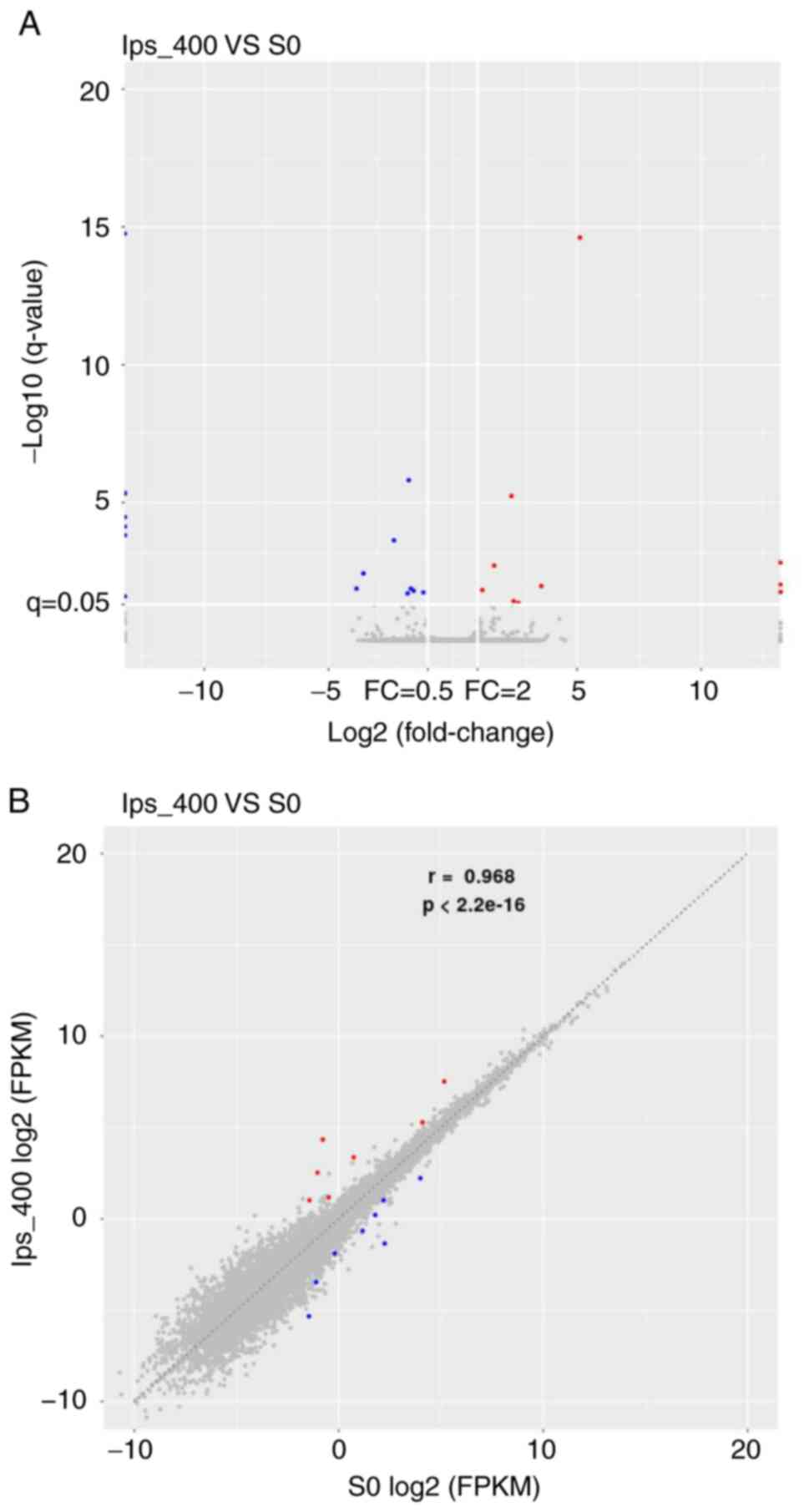
group vs. control group) was performed. As shown in Fig. 3A, 65 DEGs were screened, and the
majority of them were downregulated following LPS treatment.
Volcano plots and scatter plots (Fig.
3B and C) indicated that there were 20 upregulated (red dots)
and 45 downregulated (blue dots) DEGs. Among the DEGs, five were
previously unknown genes (data not shown), and the top 15 known
DEGs are listed in Table II. The
log2FC (abs) of the top 15 DEGs varied from 2.4 to 5.2,
and most of the top 15 DEGs were downregulated.
 | Table II.Top 15 differentially expressed genes
from lipopolysaccharide-stimulated groups. |
Table II.
Top 15 differentially expressed genes
from lipopolysaccharide-stimulated groups.
| No. | Gene ID | Gene name | log2FC | FC | P-value |
Up/downregulated |
|---|
| 1 |
ENSG00000267368 | UPK3BL1 | 5.136 | 35.159 |
2.52×10−6 | Up |
| 2 |
ENSG00000197603 | CPLANE1 | −4.547 | 0.043 |
2.25×10−5 | Down |
| 3 |
ENSG00000151967 | SCHIP1 | −4.411 | 0.047 |
4.55×10−6 | Down |
| 4 |
ENSG00000153956 | CACNA2D1 | −4.206 | 0.054 |
3.40×10−5 | Down |
| 5 |
ENSG00000158528 | PPP1R9A | −4.179 | 0.055 |
4.67×10−5 | Down |
| 6 |
ENSG00000228526 | MIR34AHG | −4.131 | 0.057 |
5.00×10−6 | Down |
| 7 |
ENSG00000133110 | POSTN | −3.563 | 0.085 |
4.15×10−7 | Down |
| 8 |
ENSG00000214827 | MTCP1 | −3.425 | 0.093 |
2.55×10−6 | Down |
| 9 |
ENSG00000210140 | MT-TC | 3.382 | 10.423 |
8.75×10−8 | Up |
| 10 |
ENSG00000120262 | CCDC170 | −3.147 | 0.113 |
2.42×10−5 | Down |
| 11 |
ENSG00000171466 | ZNF562 | −2.795 | 0.144 |
8.90×10−9 | Down |
| 12 |
ENSG00000165392 | WRN | −2.636 | 0.161 |
1.35×10−5 | Down |
| 13 |
ENSG00000142731 | PLK4 | −2.500 | 0.177 |
8.87×10−5 | Down |
| 14 |
ENSG00000198689 | SLC9A6 | −2.459 | 0.182 |
6.73×10−6 | Down |
| 15 |
ENSG00000113580 | NR3C1 | −2.419 | 0.187 |
2.23×10−5 | Down |
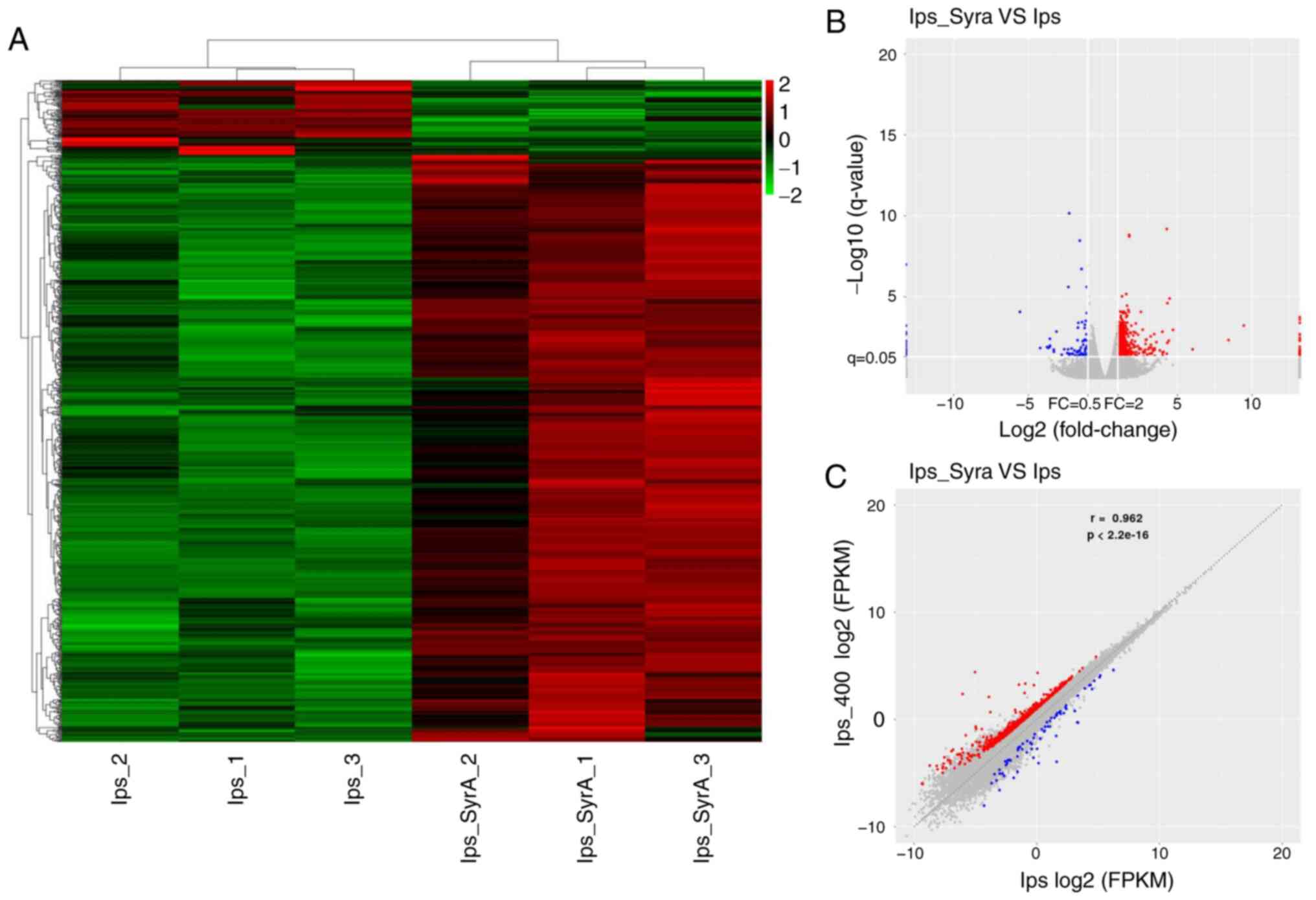
RNA-Seq analysis of LPS-stimulated NP
cells treated with SyrA
The effect of SyrA on LPS-stimulated NP cells was
much more comprehensive compared with that of LPS on NP cells. A
total of 819 DEGs from SyrA-reversed groups (SyrA plus LPS group
vs. LPS group) were screened, and these are presented in the
heat-map (Fig. 4A). Among the
DEGs, 727 were upregulated (red dots) and 92 were down-regulated
(blue dots); the resulting volcano plots and scatter plots
(Fig. 4B and C) showed that the
log2FC (abs) of the DEGs was 2–5. The unknown DEGs
(named with alphanameric code, lacking a specific gene name) in the
two groups reached 57 DEGs (6.96%; data not shown), and the top 15
known DEGs are presented in Table
III. Notably, the upregulation of uroplakin 3B-like 1 (UPK3BL1)
in LPS-stimulated NP cells (log2FC=5.136) was reversed
by SyrA, resulting in its downregulation (log2FC=−5.558)
(Tables II and III). A similar trend could also be
observed in the expression of the gene for voltage-dependent
calcium channel subunit α-2/δ-1 (CACNA2D1), with the
log2FC changing from −4.206 to 3.468 (Tables II and III). These changes suggested that there
might be a broad reversion of SyrA on LPS-resulted DEGs in NP
cells. Thus, DEGs between the SyrA plus LPS group and the control
group were also analyzed. As shown in Fig. 5A and B, the total DEGs (red and
blue dots) number was only 25, which indicated that SyrA could
reverse most of the 65 DEGs resulting from LPS stimulation.
 | Table III.Top 15 differentially expressed genes
from the syringic acid-reversed groups. |
Table III.
Top 15 differentially expressed genes
from the syringic acid-reversed groups.
| No. | Gene ID | Gene name | log2FC | FC | P-value |
Up/downregulated |
|---|
| 1 |
ENSG00000267368 | UPK3BL1 | −5.558 | 0.021 |
8.55×10−8 | Down |
| 2 |
ENSG00000114790 | ARHGEF26 | 4.423 | 21.446 |
6.89×10−5 | Up |
| 3 |
ENSG00000231292 | IGKV1OR2-108 | −4.206 | 0.054 |
4.18×10−4 | Down |
| 4 |
ENSG00000237248 | LINC00987 | 4.013 | 16.142 |
2.54×10−3 | Up |
| 5 |
ENSG00000164669 | INTS4P1 | 3.989 | 15.877 |
7.96×10−6 | Up |
| 6 |
ENSG00000197978 | GOLGA6L9 | 3.822 | 14.147 |
7.75×10−4 | Up |
| 7 |
ENSG00000086696 | HSD17B2 | −3.739 | 0.075 |
3.34×10−4 | Down |
| 8 |
ENSG00000284118 | MIR4707 | −3.653 | 0.079 |
4.09×10−4 | Down |
| 9 |
ENSG00000237380 | HOXD-AS2 | −3.557 | 0.085 |
3.13×10−04 | Down |
| 10 |
ENSG00000005471 | ABCB4 | 3.476 | 11.123 |
3.02×10−3 | Up |
| 11 |
ENSG00000153956 | CACNA2D1 | 3.468 | 11.069 |
4.39×10−4 | Up |
| 12 |
ENSG00000187790 | FANCM | 3.443 | 10.874 |
4.97×10−4 | Up |
| 13 |
ENSG00000283824 | MIR22 | 3.435 | 10.817 |
1.41×10−5 | Up |
| 14 |
ENSG00000021645 | NRXN3 | 3.379 | 10.400 |
8.66×10−4 | Up |
| 15 |
ENSG00000198088 | NUP62CL | 3.356 | 10.239 |
7.47×10−4 | Up |
RT-qPCR validation of the RNA-Seq
analysis
The FC of the 65 DEGs from the LPS-stimulated groups
was compared with that of the 819 DEGs from the SyrA-reversed
groups to screen the genes that were regulated by both LPS and
SyrA, which therefore could have an important role in preventing
apoptosis. As a result, 33 DEGs were screened, i.e., nearly half of
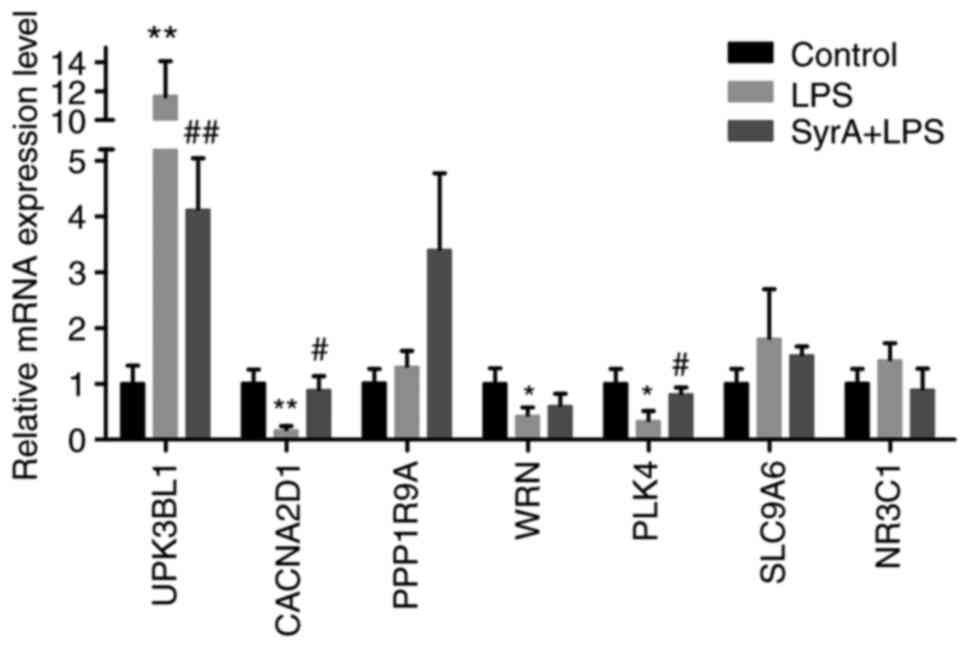
the total DEGs of the LPS-stimulated group (Table SI). Among the top 15 most
significantly dysregulated DEGs, the expression of seven was
completely reversed following SyrA treatment (Table IV), and RT-qPCR analysis indicated
that the expression of UPK3BL1 was significantly upregulated after
LPS stimulation, and subsequently downregulated by SyrA (Fig. 6). In addition, the altered
expression patterns of CACNA2D1 and polo-like kinase 4 (PLK4) were
consistent with the RNA-Seq results (Fig. 6).
 | Table IV.The FC of seven DEGs out of the top
15 DEGs in LPS-stimulated groups were reversed after SyrA
treatment. |
Table IV.
The FC of seven DEGs out of the top
15 DEGs in LPS-stimulated groups were reversed after SyrA
treatment.
|
|
| LPS-stimulated
groups | SyrA-reversed
groups |
|---|
|
|
|
|
|
|---|
| No. | Gene name | log2FC | FC |
Up/downregulated | log2FC | FC |
Up/downregulated |
|---|
| 1 | UPK3BL1 | 5.136 | 35.159 | Up | −5.558 | 0.021 | Down |
| 2 | CACNA2D1 | −4.206 | 0.054 | Down | 3.468 | 11.069 | Up |
| 3 | PPP1R9A | −4.179 | 0.055 | Down | 3.279 | 9.707 | Up |
| 4 | WRN | −2.636 | 0.161 | Down | 2.207 | 4.618 | Up |
| 5 | PLK4 | −2.500 | 0.177 | Down | 1.578 | 2.985 | Up |
| 6 | SLC9A6 | −2.459 | 0.182 | Down | 1.751 | 3.367 | Up |
| 7 | NR3C1 | −2.419 | 0.187 | Down | 1.711 | 3.275 | Up |
Bioinformatics analyses
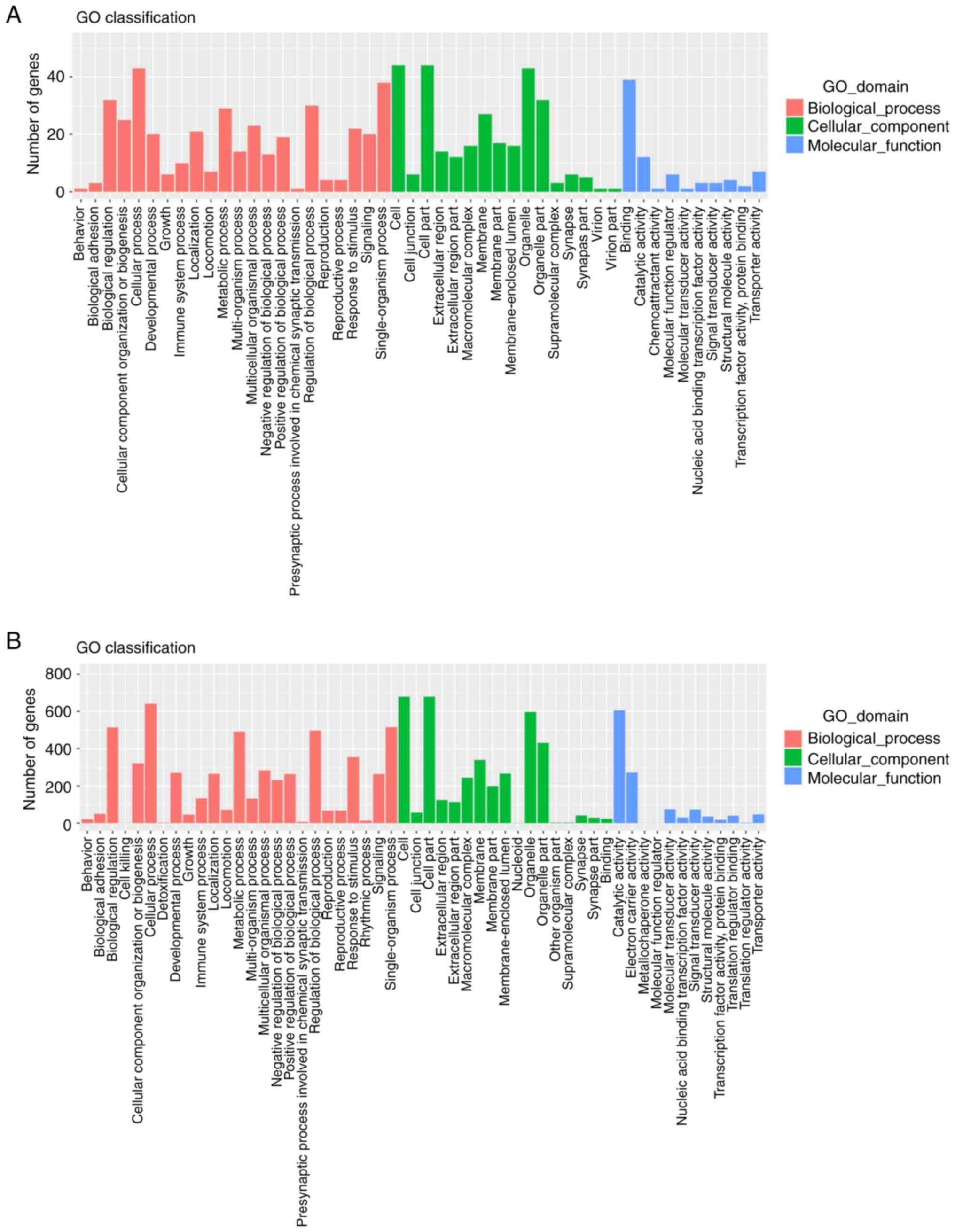
For an improved understanding of these DEGs, GO
analysis was conducted. As shown in Fig. 7A and B, there was a high
correspondence of DEGs from LPS-stimulated groups and SyrA-reversed
groups in terms of the GO classifications. The 65 DEGs were mostly
annotated for ‘biological regulation’, ‘cellular process’ and
‘single-organism process’ within the ‘Biological Process’; ‘cell’,
‘cell part’ and ‘organelle’ within ‘Cellular Component’; and
‘binding’ followed by ‘catalytic activity’ within ‘Molecular
Function’ (Fig. 7A). Similar
distribution trends could also be found among the 819 DEGs in the
SyrA-reversed groups (Fig. 7B).
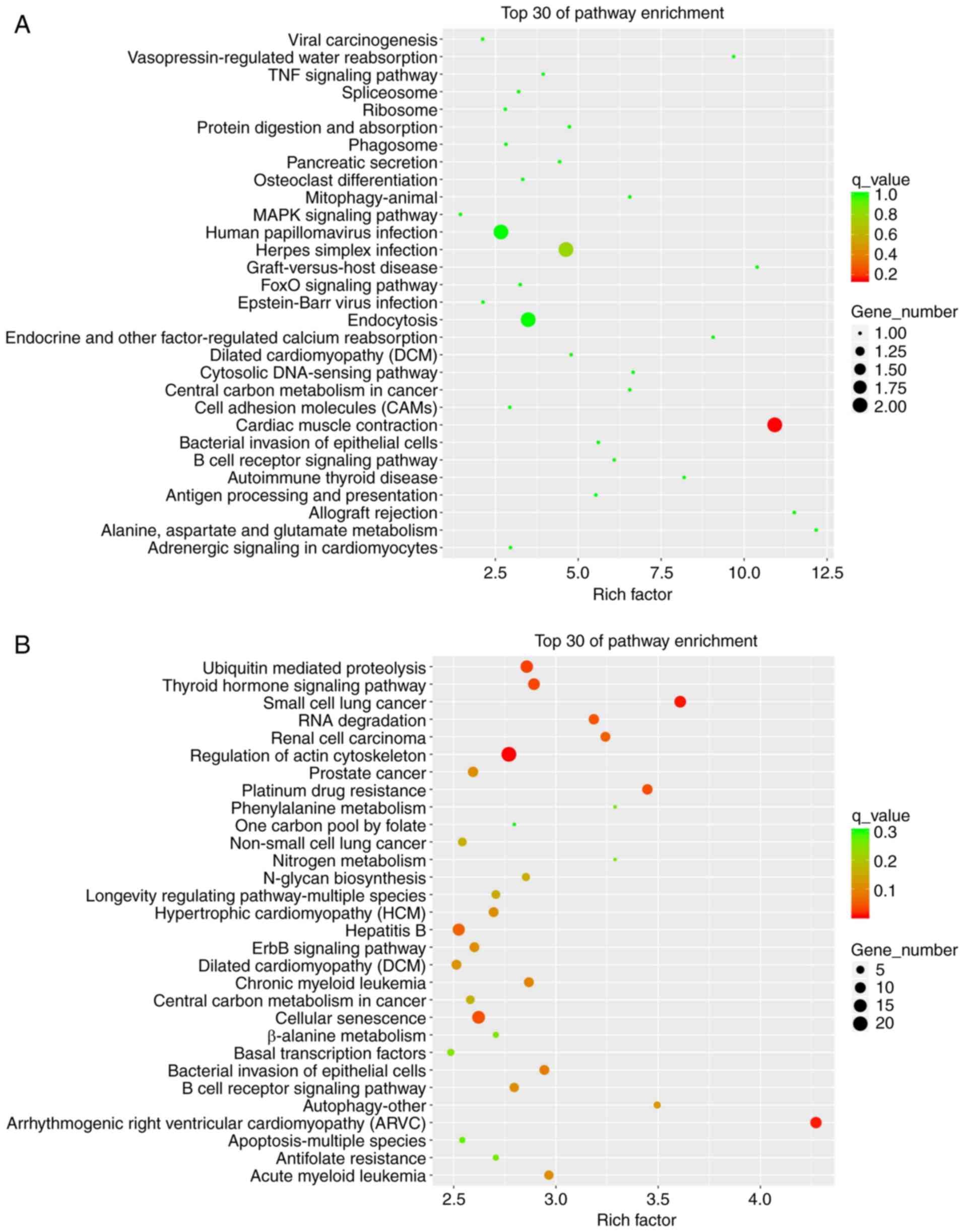
KEGG classification analysis was also conducted: The top 30
pathways were screened, and are presented in Fig. 8. As shown in Fig. 8A, DEGs from LPS-stimulated groups
were mainly enriched in ‘Cardiac muscle contraction’, ‘Endocytosis’
and the ‘MAPK signaling pathway’. In the SyrA-reversed group, DEGs
were involved in the ‘Regulation of the actin cytoskeleton’,
‘Ubiquitin-mediated proteolysis’, ‘ErbB signaling pathway’,
‘Autophagy-other’, ‘Cellular senescence’ and ‘Apoptosis-multiple
species’ (Fig. 8B).
Discussion
To date, studies have primarily focused on the
mechanisms of apoptosis of NP cells based on RNAs and long
non-coding RNAs (17–19). However, significant obstacles still
remain in terms of their clinical application, such as poor uptake,
low potency at target sites, and off-target effects (20). Consequently, seeking novel
molecules to prevent the apoptosis of NP cells and further slow IDD
might be a more feasible option for clinical implementation. The
present study identified that SyrA was able to reverse both
apoptosis and the abnormal expression of certain mRNAs in the NP
cells that were induced by LPS.
LPS is a stimulant that has been used to induce
cellular apoptosis, including apoptosis of NP cells (12,13,21).
The stimulatory action of LPS on NP cells not only involves the
initiation of apoptosis, but also promotes the expression of
ECM-degrading enzymes, thereby accelerating IDD (22,23).
However, the experimental concentration of LPS that was used to
induce NP cellular apoptosis in these studies varied widely,
ranging between 10 ng/ml and 10 µg/ml (12,13).
Therefore, in the present study, different concentrations of LPS
were used with NP cells in culture, and the results showed that
0.01 µg/ml (10 ng/ml) LPS induced apoptosis the most efficiently,
consistent with the results of Liu et al (13). Notably, the LPS concentration and
its apoptosis induction rate were found to be negatively related,
in contrast with other studies (12,24).
A previous study showed that no significant survival inhibition was
observed after nasal epithelial cells were treated with a high dose
of LPS, which was demonstrated to be the autophagy activator
(25). Hence, it was hypothesized
that the negative relationship between LPS concentration and
survival inhibition could have been caused by autophagy initiated
by a higher dose of LPS.
Another study, NP cells pretreated with SyrA then
treated with LPS, was also conducted and the results indicated that
SyrA could exert a protection effect compared to cells treated with
LPS alone, which is similar to the results in the present study
(cells treated with SryA and LPS together). Then, the easier
treatment way, treated with SryA and LPS together, was selected for
further study.
Usually, RNA-Seq is used to analyze the DEGs between
two groups; in the present study, the mRNA expression in three
groups were detected, and DEGs were identified between the pairing
of groups. The major objective of the present study was to search
for the main target genes that are involved in both LPS stimulation
and SyrA reversion, even though the total effect of SyrA
interference on LPS-stimulated NP cells compared with the control
NP cells was also observed. In the LPS-stimulated groups, a total
of 65 DEGs were identified from 819 DEGs following treatment with
SyrA. The effect of SyrA on LPS-stimulated NP cells was greater
compared with that of LPS on NP cells. This suggested that there
would be a greater influence of SyrA plus LPS on the transcriptome
of NP cells. In contrast to this, the results demonstrated that the
alterations in the expression of the 65 DEGs were reversed by
treatment with SyrA, which made the abnormally expressed
transcriptome of LPS-stimulated NP cells return to a relatively
normal phenotype.
The comparison between the top 15 DEGs from the
LPS-stimulated groups and the 819 DEGs from the SyrA-reversed
groups showed that seven were involved in LPS stimulation and SyrA
reversion. The results of RT-qPCR validation screened three DEGs
(CACNA2D1, PLK4 and UPK3BL1), which were in line with the trend of
each FC. CACNA2D1 is a subunit of the Ca2+-channel
complex, and to the best of the authors' knowledge, no research has
previously reported the role of this gene in apoptosis. However,
the protein CACNA2D1 shares a similar secondary and tertiary
structure with CACNA2D2, which is reported to be able to directly
regulate the apoptosis of non-small cell lung cancer cells, and its
regulation was associated with disruption of the mitochondrial
membrane integrity (26). These
data suggest that there might be some as yet unknown relationship
between CACNA2D1 and LPS-induced apoptosis of NP cells. Studies
involved in the regulation of apoptosis by PLK4, a unique member of
the PLK family, are relatively abundant. For instance, the
depletion of PLK4 significantly promoted apoptosis of
hepatocellular carcinoma cells and suppressed cell proliferation
and tissue invasion (27). In
neuroblastoma cells, downregulation of PLK4 could also dramatically
increase the rate of apoptosis and lead to an inhibition of cell
migratory and invasive abilities (28). Additionally, apoptotic death
occurred in lung cancer cells after treatment with CFI-400945, a
PLK4 inhibitor (29). In the
present study, the expression of PLK4 was downregulated in
LPS-induced apoptosis and reversed by SyrA treatment. The
relationship between the expression level of PLK4 and apoptosis
inducement/inhibition in NP cells identified in the present study
was consistent with the aforementioned three studies. This
indicated that PLK4 possibly exerts important roles in both
LPS-induced and SyrA-reversed apoptosis of the NP cells. Until now,
to the best of the authors' knowledge, no studies have been
published on the role of UPK3BL1: Its potentially dominant effect
on the apoptosis of NP cells is an hypothesis that requires further
experimental validation.
Compared with GO classification analysis, KEGG
pathway enrichment analysis appeared to have a higher level of
precision and helped the study in enabling us to look for some
potential genes involved in the apoptosis of NP cells, as well as
other mechanisms responsible for survival induced by SyrA. The KEGG
pathway analysis revealed that DEGs from LPS-stimulated groups were
enriched in the MAPK signaling pathway. The role of the MAPK
pathway in the apoptosis of NP cells has previously been reported
(30,31), and the MAPK pathway might be
involved in the apoptosis observed in the present study. A DEG
identified that is known to be involved with the MAPK pathway was
CACNA2D1, which could also indicate the potential role of CACNA2D1
in the apoptosis of NP cells.
Following treatment with SyrA, DEGs were found to be
enriched in pathways of interest to the present study, such as the
‘Autophagy-other’ and ‘Apoptosis-multiple species’ pathways. The
number of DEGs in the ‘Autophagy-other’ pathway was 14 (consisting
of BIRC3, CUL5, CUL2, HERC3, WWP1, NEDD4, UBR5, MID1, UBA6, ITCH,
UBE4A, UBE2N, CUL4B and HERC1). Autophagy is known to be an
effective and important way for cells to survive in an unfavorable
environment (32). The viability
of the damaged NP cells reversed by SyrA was not limited to the
inhibition of apoptosis, and autophagy might also be initiated
after SyrA treatment. Among the 14 DEGs, the majority have
important roles in autophagy (33–35);
however, their effects on the autophagy of NP cells are still
unknown and require further research. Notably, the level of cullin
4B (CUL4B) was found to be significantly upregulated in IDD samples
compared with the controls, and this was positively correlated with
the severity of IDD (36). In
addition, the expression of CUL4B was enhanced after treatment with
the inflammatory factors interleukin (IL)-6 and tumour necrosis
factor (TNF)-α, and reversed by IL-6 and TNF-α inhibitors in
primary NP cells (37). The
previously reported close association between CUL4B and
inflammation suggested that there might an anti-inflammatory effect
exerted by SyrA in the survival of damaged NP cells. Indeed, the
onset of inflammation induced by LPS in NP cells and the
anti-inflammatory effect of SyrA has been confirmed in many prior
reports (37–41). DEGs in the ‘Apoptosis-multiple
species’ pathway include APAF1, BIRC3 and BCL2. The role of Bcl-2
in apoptotic NP cells has been demonstrated in previous studies
(18,41), including in the work presented
here. The other two DEGs might also have a role in the apoptosis of
NP cells, but further studies are still needed to confirm the roles
of these proteins.
Collectively, the present study has shown that SyrA
can reverse the LPS-induced apoptosis of NP cells, which was
attributed to an extensive reversion of the dysregulated mRNA
levels to near-normal expression levels. The regulation of the
CACNA2D1 and PLK4 genes may be responsible for the reversal of
apoptosis. Additionally, the complete survival reversion induced by
SyrA on damaged NP cells might be due to the onset of autophagy,
which prevents apoptosis and promotes cell viability. All these
hypotheses require further study to help elucidate the potential
mechanisms for apoptosis/survival reversion induced by SyrA in NP
cells.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by Research Project of
Inheritance and Innovation of Traditional Chinese Medicine and
Ethnic Medicine in Health Department of Guangxi Zhuang Autonomous
Region (grant no. GZPT13-41).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CZ and QF designed and supervised the project. HZ
and HQ performed the major research and wrote the manuscript. YY,
JS and YT analyzed and interpreted the data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
GBD 2017 Disease, Injury Incidence and
Prevalence Collaborators: Global, regional, and national incidence,
prevalence, and years lived with disability for 354 diseases and
injuries for 195 countries and territories, 1990–2017: A systematic
analysis for the Global Burden of Disease Study 2017. Lancet.
392:1789–1858. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vadala G, Russo F, Di Martino A and Denaro
V: Intervertebral disc regeneration: From the degenerative cascade
to molecular therapy and tissue engineering. J Tissue Eng Regen
Med. 9:679–690. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sakai D and Grad S: Advancing the cellular
and molecular therapy for intervertebral disc disease. Adv Drug
Deliv Rev. 84:159–171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sampara P, Banala RR, Vemuri SK, Av GR and
Gpv S: Understanding the molecular biology of intervertebral disc
degeneration and potential gene therapy strategies for
regeneration: A review. Gene Ther. 25:67–82. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ding F, Shao ZW and Xiong LM: Cell death
in intervertebral disc degeneration. Apoptosis. 18:777–785. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Srinivasulu C, Ramgopal M, Ramanjaneyulu
G, Anuradha CM and Suresh Kumar C: Syringic acid (SA) a review of
its occurrence, biosynthesis, pharmacological and industrial
importance. Biomed Pharmacother. 108:547–557. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cao Y, Zhang L, Sun S, Yi Z, Jiang X and
Jia D: Neuroprotective effects of syringic acid against
OGD/R-induced injury in cultured hippocampal neuronal cells. Int J
Mol Med. 38:567–573. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Song M, Du Z, Lu G, Li P and Wang L:
Syringic acid protects retinal ganglion cells against
H2O2-induced apoptosis through the activation
of PI3K/Akt signaling pathway. Cell Mol Biol (Noisy-le-grand).
62:50–54. 2016.PubMed/NCBI
|
|
9
|
Ding SK, Wang LX, Guo LS, Luo P, Du JJ,
Zhao ZL and Wang GG: Syringic acid inhibits apoptosis pathways via
downregulation of p38MAPK and JNK signaling pathways in H9c2
cardiomyocytes following hypoxia/reoxygenation injury. Mol Med Rep.
16:2290–2294. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sancak EB, Akbas A, Silan C, Cakir DU,
Turkon H and Ozkanli SS: Protective effect of syringic acid on
kidney ischemia-reperfusion injury. Ren Fail. 38:629–635. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang S, Sun J, Yang H, Zou W, Zheng B,
Chen Y, Guo Y and Shi J: Profiling and bioinformatics analysis of
differentially expressed circular RNAs in human intervertebral disc
degeneration. Acta Biochim Biophys Sin (Shanghai). 51:571–579.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chai X, Si H, Song J, Chong Y, Wang J and
Zhao G: miR-486-5p inhibits inflammatory response, matrix
degradation and apoptosis of nucleus pulposus Cells through
directly targeting FOXO1 in intervertebral disc degeneration. Cell
Physiol Biochem. 52:109–118. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu J, Jiang T, He M, Fang D, Shen C, Le
Y, He M, Zhao J and Zheng L: Andrographolide prevents human nucleus
pulposus cells against degeneration by inhibiting the NF-kappaB
pathway. J Cell Physiol. 234:9631–9639. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
The Gene Ontology Consortium: The Gene
Ontology Resource: 20 years and still Going strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47:D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang Q, Guo XP, Cheng YL and Wang Y:
MicroRNA-143-5p targeting eEF2 gene mediates intervertebral disc
degeneration through the AMPK signaling pathway. Arthritis Res
Ther. 21:972019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang R, Wen B and Sun D: miR-573 regulates
cell proliferation and apoptosis by targeting Bax in nucleus
pulposus cells. Cell Mol Biol Lett. 24:22019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu Y, Zhang X, Li Z, Kong L and Huang Y:
LncRNA HOTAIR suppresses TNF-alpha induced apoptosis of nucleus
pulposus cells by regulating miR-34a/Bcl-2 axis. Biomed
Pharmacother. 107:729–737. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Barata P, Sood AK and Hong DS:
RNA-targeted therapeutics in cancer clinical trials: Current status
and future directions. Cancer Treat Rev. 50:35–47. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Yang J, Zhou X, Wang N, Li Z,
Zhou Y, Feng J, Shen D and Zhao W: Knockdown of miR-222 inhibits
inflammation and the apoptosis of LPS-stimulated human
intervertebral disc nucleus pulposus cells. Int J Mol Med.
44:1357–1365. 2019.PubMed/NCBI
|
|
22
|
Liu H, Pan H, Yang H, Wang J, Zhang K, Li
X, Wang H, Ding W, Li B and Zheng Z: LIM mineralization protein-1
suppresses TNF-alpha induced intervertebral disc degeneration by
maintaining nucleus pulposus extracellular matrix production and
inhibiting matrix metalloproteinases expression. J Orthop Res.
33:294–303. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhongyi S, Sai Z, Chao L and Jiwei T:
Effects of nuclear factor kappa B signaling pathway in human
intervertebral disc degeneration. Spine (Phila Pa 1976).
40:224–232. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li B, Zhang H, Zeng M, He W, Li M, Huang
X, Deng DY and Wu J: Bone marrow mesenchymal stem cells protect
alveolar macrophages from lipopolysaccharide-induced apoptosis
partially by inhibiting the Wnt/β-catenin pathway. Cell Biol Int.
39:192–200. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang XH, Zhang ZH, Cai XL, Ye P, Feng X,
Liu TT and Li XZ: Lipopolysaccharide induces autophagy by targeting
the AMPK-mTOR pathway in human nasal epithelial cells. Biomed
Pharmacother. 96:899–904. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Carboni GL, Gao B, Nishizaki M, Xu K,
Minna JD, Roth JA and Ji L: CACNA2D2-mediated apoptosis in NSCLC
cells is associated with alterations of the intracellular calcium
signaling and disruption of mitochondria membrane integrity.
Oncogene. 22:615–626. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bao J, Yu Y, Chen J, He Y, Chen X, Ren Z,
Xue C, Liu L, Hu Q, Li J, et al: MiR-126 negatively regulates PLK-4
to impact the development of hepatocellular carcinoma via ATR/CHEK1
pathway. Cell Death Dis. 9:10452018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tian X, Zhou D, Chen L, Tian Y, Zhong B,
Cao Y, Dong Q, Zhou M, Yan J, Wang Y, et al: Polo-like kinase 4
mediates epithelial-mesenchymal transition in neuroblastoma via
PI3K/Akt signaling pathway. Cell Death Dis. 9:542018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kawakami M, Mustachio LM, Zheng L, Chen Y,
Rodriguez- Canales J, Mino B, Kurie JM, Roszik J, Villalobos PA,
Thu KL, et al: Polo-like kinase 4 inhibition produces polyploidy
and apoptotic death of lung cancers. Proc Natl Acad Sci USA.
115:1913–1918. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang T, Wang CJ, Tian S and Song HB:
Overexpressed IGFBP5 promotes cell proliferation and inhibits
apoptosis of nucleus pulposus derived from rats with disc
degeneration through inactivating the ERK/MAPK axis. J Cell
Biochem. 120:18782–18792. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu Q, Fang H, Zhao L, Zhang C, Zhang L and
Tian B: Mechano growth factor attenuates mechanical
overload-induced nucleus pulposus cell apoptosis through inhibiting
the p38 MAPK pathway. Biosci Rep. 39:BSR201824622019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jiang L, Yuan F, Yin X and Dong J:
Responses and adaptations of intervertebral disc cells to
microenvironmental stress: A possible central role of autophagy in
the adaptive mechanism. Connect Tissue Res. 55:311–321. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Antonioli M, Albiero F, Nazio F, Vescovo
T, Perdomo AB, Corazzari M, Marsella C, Piselli P, Gretzmeier C,
Dengjel J, et al: AMBRA1 interplay with cullin E3 ubiquitin ligases
regulates autophagy dynamics. Dev Cell. 31:734–746. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xie W, Jin S and Cui J: The NEDD4-USP13
axis facilitates autophagy via deubiquitinating PIK3C3. Autophagy.
16:1150–1151. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rossi M, Rotblat B, Ansell K, Amelio I,
Caraglia M, Misso G, Bernassola F, Cavasotto CN, Knight RA,
Ciechanover A and Melino G: High throughput screening for
inhibitors of the HECT ubiquitin E3 ligase ITCH identifies
antidepressant drugs as regulators of autophagy. Cell Death Dis.
5:e12032014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen Z, Han Y, Deng C, Chen W, Jin L, Chen
H, Wang K, Shen H and Qian L: Inflammation-dependent downregulation
of miR-194-5p contributes to human intervertebral disc degeneration
by targeting CUL4A and CUL4B. J Cell Physiol. 234:19977–19989.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang H, Hao P, Zhang H, Xu C and Zhao J:
MicroRNA-223 inhibits lipopolysaccharide-induced inflammatory
response by directly targeting Irak1 in the nucleus pulposus cells
of intervertebral disc. IUBMB Life. 70:479–490. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Guo F, Zou Y and Zheng Y: Moracin M
inhibits lipopolysaccharide-induced inflammatory responses in
nucleus pulposus cells via regulating PI3K/Akt/mTOR
phosphorylation. Int Immunopharmacol. 58:80–86. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ham JR, Lee HI, Choi RY, Sim MO, Seo KI
and Lee MK: Anti-steatotic and anti-inflammatory roles of syringic
acid in high-fat diet-induced obese mice. Food Funct. 7:689–697.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li Y, Zhang L, Wang X, Wu W and Qin R:
Effect of Syringic acid on antioxidant biomarkers and associated
inflammatory markers in mice model of asthma. Drug Dev Res.
80:253–261. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhu H, Sun B and Shen Q: TNF-α induces
apoptosis of human nucleus pulposus cells via activating the
TRIM14/NF-κB signalling pathway. Artif Cells Nanomed Biotechnol.
47:3004–3012. 2019. View Article : Google Scholar : PubMed/NCBI
|