Introduction
Patients undergoing major surgery, for example
abdominal cancer surgery, experience a clinically well-known
phenomenon of postoperative inflammation (1). This inflammation, together with
haemodynamic and metabolic instability, contribute to postoperative
morbidity (2).
Endogenous glucocorticoids (GCs) are an essential
part of the stress response in mammals and serve important roles in
immunological and haemodynamic homeostasis and metabolism (3). Deficient, or excessive, GC-signalling
leads to debilitating disease, especially under physiological
stress (4,5). In states of hyperinflammation, such as
autoimmune disease, synthetic GCs are used therapeutically.
However, although widely used and comprehensively studied, the
molecular mechanisms of GCs have not been fully explored (6). The response to GCs is also highly
variable between individuals and the cause of this diversity
remains unknown (7).
In our previous study it was demonstrated that
responsiveness to GCs may be related to a patient's ability to
recover from surgically induced inflammatory stress. In patients
undergoing major abdominal surgery, a negative correlation between
the regulation of the glucocorticoid receptor (GR)α gene nuclear
receptor subfamily 3 group C member 1 (NR3C1), in peripheral
blood leukocytes, and the length of stay in the intensive care unit
was observed (8). The GC signalling
pathway is intricate. Several GR isoforms exist and have been
investigated in relation to numerous conditions. These
investigations have revealed a diverse regulation of downstream
target genes (9–11). The gene NR3C1, encoding the
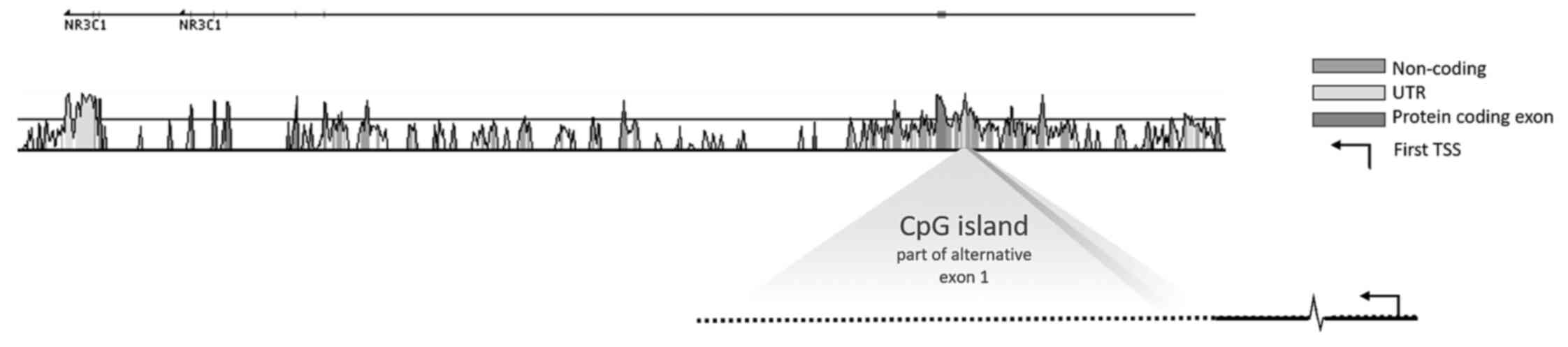
GR, consists of nine exons and is evolutionarily conserved. Exon 1
comprises nine different variants that are transcribed, but not
translated (12). Of these nine
non-coding exon 1 variants, seven are situated closely together and
contain a cytosine-phosphate-guanine (CpG) island spanning 3 kb
(Fig. 1), an established site for
differential regulation.
Methylation patterns in exon 1 differ between
individuals (13). Differences in
methylation in this region have been demonstrated to influence
various forms of stress responses in humans and mice (12,14–16).
The methylation pattern is manifested early in life and later
affects susceptibility to stressors in neuropsychological, as well
as other, conditions (17–20). DNA methylation of NR3C1 has
not been explored in settings of postoperative inflammatory stress.
A translational approach is warranted, allowing for the
investigation of inter-individual differences in the molecular
biology of the GR gene in relation to a clinically well-described
state of inflammation.
We therefore hypothesise that there is an
association between GR gene NR3C1 exon 1 methylation status
and postoperative complications following major abdominal surgery.
In the present study, the methylation status and genotype in the
CpG island at NR3C1 exon 1 in the peripheral blood of
patients who subsequently underwent major abdominal surgery was
explored. Correlations with postoperative complications were
further investigated.
Materials and methods
Patients
Ethical approval for the present study was granted
by the Stockholm Regional Ethical Review Board (Stockholm, Sweden;
approval no. 2009/366-31). Patients eligible for inclusion were
those >18 years, accepted and planned for surgery involving
resections of the pancreas or liver, pre-operatively estimated to
last >4 h. Patients requiring long-term GC treatment
pre-operatively or with any known disease of the glucocortid system
were excluded. The patients recruited for the present study were
enrolled after informed consent was obtained, orally and in
writing, in accordance with The Declaration of Helsinki. Whole
blood (4 ml) was collected from 24 patients who were to have major
abdominal (pancreatic or liver) surgery. Samples were drawn during
a pre-operative visit at the Department of Surgery, Karolinska
University Hospital Huddinge (Stockholm, Sweden) in 2014. The
present study did not allow for a power calculation of the required
number of participants, as the distribution of SNPs in the patient
population was not known.
Outcomes
Complications were graded according to Clavien-Dindo
classification (21), which is a
well-established grading system in the surgical community. It is an
ordinal scale, ranging from 1 to 5, with 1 denoting only a minor
deviance, from the expected postoperative course, i.e., additional
antiemetic treatment. Grade 2 denotes need for pharmacological
therapy not considered routine or standard post-operative
treatment. Grade 4 denotes organ failure in any organ system and
grade 5 denotes mortality of the patient due to any cause. Grade 3
signifies complications, such as anastomic leakage or wound
dehiscence, necessitating re-intervention and is divided into two
sub-classes depending on whether this re-intervention is performed
without (3a) or with (3b) general anaesthesia. Complications graded
≥3b are regarded as major.
Sampling and DNA extraction
Pre-operative samples of whole blood were collected
in EDTA tubes (BD Vacutainer; BD Diagnostics) and the erythrocytes
were immediately treated with cold (4–6°C) ammonium lysis buffer
(153 mM NH4Cl, 10 mM KHCO3 and 0.01 mM EDTA
adjusted to pH 7.4-7.5) until tranluscent. After lysis and washing
in PBS, leukocytes were resuspended in 350 µl of RLT Buffer from
the Qiagen RNEasy kit (Qiagen AB) and were subsequently frozen. DNA
was extracted using the Qiagen RNEasy kit in a QIAcube (Qiagen AB)
according to the manufacturer's protocol. Concentration and purity
assessments were performed using a NanoDrop 1000 (Thermo Fisher
Scientific, Inc.). The 260 nm/280 nm absorbance ratios were in the
range of 1.75-1.85.
Targeted bisulfite (BiS)
sequencing
The CpG island is located at chromosome
(chr)5:143402505-143405805 [Human Dec 2013 (GRCh38/hg38) Assembly],
i.e., GRCh38/hg38, Human Genome Reference Consortium, human genome
build 38 (human genome 38) (12).
Targeted BiS sequencing was performed by Zymo Research Corp.
Samples containing approximately 400–1400 µg DNA were diluted and
BiS-converted using the EZ DNA Methylation-Lightning kit (Zymo
Research Corp.) at 98°C for 8 min and 54°C for 60 min.
BiS-converted DNA was then PCR-amplified using ZymoTaq PreMix (Zymo
Research Corp.) for library construction, using a microfluidics
system (Fluidigm Access Array; Fludigm Corporation), the specific
cycling conditions and final concentrations are proprietary to Zymo
Research Corp. The primer sequences used are presented in Table SI. BiS-converted, amplified DNA
underwent next generation sequencing using the Illumina MiSeq
platform (Illumina, Inc.) and corresponding kits by Zymo Research
Corp. A paired-end 300 bp configuration was used and libraries were
loaded at 8 pM concentrations. The raw data was aligned for CpG
methylation calling. Sequence reads were identified using standard
Illumina base-calling software. Sequence alignment was made if the
coverage exceeded 10 reads. Furthermore, in addition to the CpG
methylation analysis, the full sequences were analysed for SNPs to
the extent allowed for following BiS conversion using Bioconductor
software package R (version 3.7, http://bioconductor.org/news/bioc_3_7_release/) with
the packages Rqc (version 1.10.2), Rsamtools (version 1.35.2), msa
version 1.12.0 and GenomicRanges (1.32.0).
Multifactorial analysis
Multifactorial analysis was done in R (version
3.5.0) (22). Consensus sequences
from each patient were extracted from the sequence data obtained
via BiS-sequencing, which used a threshold of 0.5 as minimum
probability threshold in the multiple sequence alignment package
(23). All biological parameters
were assessed using multiple factor analysis in Factominer (version
2.4) (24). This multifactorial
approach was selected as it enabled the analysis of individual and
combined SNP effects, methylation patterns, patient characteristics
and clinical data. No patient outliers skewed the model. No
individual factors, except for the Clavien-Dindo classification,
were able to explain the significant portion of the variance
observed. Therefore, all other factors were excluded from further
analysis.
Parameters with minimal risk of non-biological bias
were used to construct the statistical model, including liver or
pancreas surgery, age, sex, and CRP levels. Factors omitted at this
stage were those including some degree of subjective assessment,
for example ASA-classification or pre-operative chemotherapy. Those
variables were instead subsequently fitted into the model as
supplementary variables. Patients with postoperative complications
(defined as Clavien-Dindo ≥1; n=14) were included for partial least
squares (PLS) analysis which was performed using the PLS package.
PLS was chosen because it can accept ordinal variables, including
Clavien-Dindo classification. Moreover, Clavien-Dindo classifies
postoperative complications and therefore the non-differentiation
between patients without complications presented a lower boundary
of measurement. This truncation of the outcome variable,
complications (graded according to the Clavien-Dindo
classification), produced a region with zero variance that
disqualified it from appropriate factor analysis. Therefore, it was
only possible to analyse the severity of complications and patients
without complications were excluded from further analysis.
The Clavien-Dindo classification system was used as
the response variable and all investigated nucleotide positions
with more than zero variance were used as the predictor matrix.
‘Leave one out’ cross-validation in Factominer (version 2.4) was
used to avoid overfitting and the number of components with the
smallest mean square error of prediction was retained (24). Two components explaining 71% of the
variance were retained. Scores were used as an estimate of
contribution of each SNP. P<0.001 was considered to indicate a
statistically significant difference and was used as the cut-off
value.
Statistical analysis
Data are presented as medians with ranges or
interquartile ranges, as appropriate. Analysis of differential
allele comparisons was performed using an independent-samples
Mann-Whitney U Test (SPSS version 26; IBM Corp.). P<0.05 was
considered to indicate a statistically significant difference.
Bioinformatics
Genomic positions determined to be significant were
acquired from the statistical analysis and were investigated using
the University of California, Santa Cruz (UCSC) Genome Browser
Human Dec 2013 (GRCh38/hg38) Assembly (12). A VISTA plot was used to visualise
the gene (25). Allele frequencies
(MAFs) were examined in the 1000 Genomes Phase 3, ESP and gnomAD
(v2.1) cohorts (26–28)
Results
Patients
Patient characteristics and clinical data are
presented in Table I. The median
age distribution was 65.5 (range 42–81) years and sex distribution
was 11/13 male/female. Overall, 14/24 patients had a postoperative
complication. The specimen histopathology demonstrated different
clinical conditions of the liver or pancreas, as listed in Table I.
 | Table I.Patient characteristics. |
Table I.
Patient characteristics.
| No. | Age | Sexa | H (cm) | W (kg) | WBC billion
(cells/l) | Op. type | ASAb | Op.
dur.b (min) | Chemob | CDb | HDU
(days)b | ICUb | CRP D1 (mg/l) | CRP Max D1-3
(mg/l) | Histo.a |
|---|
| 1 | 60 | F | 177 | 91.6 | 6.1 | Panc | 1 | 197 | No |
| 5 | No | 46 | 146 | NET |
| 2 | 65 | F | 164 | 56.6 | 8.4 | Panc | 2 | 383 | No | 3b | 5 | No | 39 | 39 | IPMN |
| 3 | 76 | F | 172 | 67.1 | 5.8 | Panc | 3 | 352 | No | 4 | 3 | No | 55 | 127 | NET |
| 4 | 70 | F | 167 | 78.6 | 5.0 | Panc | 2 | 423 | No | 2 | 6 | No | 45 | 139 | PDAC |
| 5 | 59 | M | 176 | 104.4 | 5.2 | Panc | 3 | 290 | No |
| 4 | No | 48 | 192 | NET |
| 6 | 76 | M | 175 | 79.5 | 7.4 | Panc | 2 | 197 | No | 2 | 6 | No | 29 | 92 | IPMN |
| 7 | 78 | M | 180 | 70.0 | 6.0 | Panc | 2 | 315 | No | 3 | 6 | No | 50 | 90 | IPMN |
| 8 | 45 | F | 168 | 82.0 | 6.7 | Panc | 1 | 176 | No |
| 2 | No | 28 | 183 | IPMN |
| 9 | 70 | F | 176 | 68.2 | 6.9 | Panc | 2 | 314 | No |
| 5 | No | 54 | 98 | PDAC |
| 10 | 66 | M | 176 | 79.5 | 8.7 | Panc | 2 | 366 | No |
| 3 | No | 49 | 102 | PDAC |
| 11 | 81 | F | 169 | 67.0 | 5.6 | Panc | 3 | 283 | No |
| 2 | No | 57 | 82 | PDAC |
| 12 | 63 | F | 170 | 101.7 | 8.3 | Panc | 3 | 205 | No | 4,5 | 2 | Yes | 35 | 66 | PDAC |
| 13 | 72 | M | 182 | 94.6 | 4.7 | Hep | 3 | 250 | No | 3b | 2 | No | 25 | 177 | CRCM |
| 14 | 65 | M | 176 | 96.2 | 7.5 | Hep | 2 | 414 | No | 2 | 8 | Yes | 17 | 80 | HCC |
| 15 | 70 | M | 169 | 74.8 | 6.7 | Hep | 3 | 290 | Yes | 2 | 4 | No | 19 | 80 | CRCM |
| 16 | 62 | F | 173 | 76.7 | 7.8 | Hep | 2 | 105 | No |
| 2 | No | 17 | 111 | Benign |
| 17 | 57 | M | 191 | 88.8 | 10.2 | Hep | 2 | 280 | Yes | 3b | 5 | No | 84 | 129 | CRCM |
| 18 | 59 | M | 173 | 72.9 | 7.5 | Hep | 2 | 269 | No | 4a | 2 | Yes | 30 | 179 | GIST |
| 19 | 68 | M | 176 | 89.9 | 3.9 | Hep | 2 | 213 | Yes |
| 2 | No | N/A | 47 | CRCM |
| 20 | 47 | F | 170 | 81.0 | 7.4 | Hep | 2 | 288 | No | 1 | 0 | No | 32 | 48 | Benign |
| 21 | 70 | F | 165 | 62.0 | 7.4 | Hep | 3 | 210 | No |
| 2 | No | 23 | 88 | HCC |
| 22 | 58 | F | 173 | 74.6 | 4.8 | Hep | 2 | 275 | No | 3,5 | 2 | No | 19 | 134 | Benign |
| 23 | 69 | M | 187 | 92.0 | 9.5 | Hep | 2 | 227 | No |
| 2 | No | 67 | 146 | Chr. infl. |
| 24 | 42 | F | 160 | 49.0 | 5.3 | Hep | 1 | 343 | Yes | 5 | 2 | Yes | 33 | 96 | CRCM |
Of the parameters available for factor analysis, sex
and benign disease were statistically significant (P<0.05) in
dimension one and two with 7.1 and 6.7% of the variance,
respectively. These effects were not, however, assessed to skew the
model and consequently not considered in subsequent analyses.
Hence, a multifactorial PLS regression-based approach was employed
to assess the association of methylation with postoperative
complications, using the full BiS-converted sequencing data and the
Clavien-Dindo classification system only.
DNA methylation and postoperative
complications
When examining all 319 CpG islands in the
BiS-converted sequence, a homogeneous demethylated pattern was
demonstrated. The whole region was demethylated, except for the
last CpG island (position, chr5:143405794). There was no
association between CpG methylation at this site and postoperative
complications (Fig. 2).
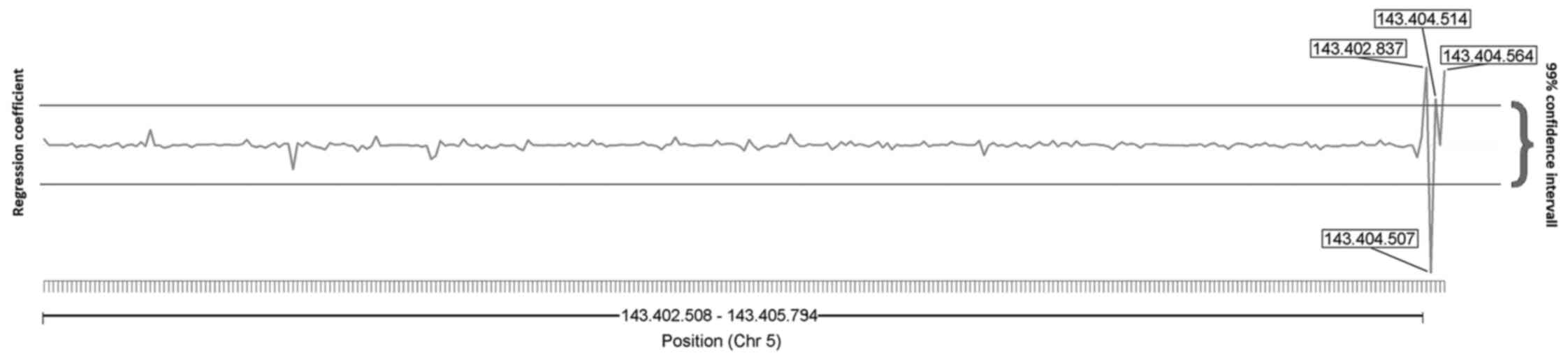
SNPs associated with postoperative
complications
The combined effect of multiple contributing
nucleotide sites was assessed, taking the clinically relevant
heterogeneity into account. The results demonstrated that 4
positions were significantly associated with postoperative
complications (Fig. 2). These four
sites were previously reported to be SNPs. The properties of these
SNPs are presented in Table II,
along with the summarised data from these aforementioned studies
[obtained using the UCSC genome browser (Fig. S1)]. For ease, the SNPs are numbered
1–4. Of the four SNPs identified in our analysis, two were
previously identified common SNPs; NCBI (National Center for
Biotechnology Information) dbSNP (data base for Single Nucleotide
Polymophisms) accession number rs10482614 (chr5:143402837; SNP1)
and NCBI dbSNP accession number rs3806855 (chr5:143404564; SNP4).
GRCh38/hg38 demonstrated that the highest population minor allele
frequencies (MAFs; observed in any population, including the 1000
Genomes Phase 3, ESP and gnomAD cohorts (26–28)
were 0.26 and 0.25, respectively. SNP1 consists of a minor allele
(A), which deletes a possible methylation site (CpG) (16). SNP4 has previously been described to
affect promotor activity (16). The
same nucleotide as in SNP4 can also be part of a three-nucleotide
deletion, (NCBI dbSNP accession number rs796817133), highest
population MAF <0.01), which was not encountered in our cohort
(29). NCBI dbSNP accession number
rs1039242888 (chr5:143404507; SNP2) has not been thoroughly
investigated but has previously been reported to have been
encountered (highest population MAF 0.02). This SNP, SNP2, was only
present in one patient in the present study and should therefore be
interpreted with caution. NCBI dbSNP accession number rs904759782
(chr 5:143404514; SNP3) was previously reported to have a highest
population MAF <0.01 but was not further investigated and
therefore its clinical significance is unknown. SNP3 was fairly
common in the present study with over 40% of the patients being
carriers of this minor allele. The minor alleles of SNPs 1–3 all
result in depletions of CpG sites. Bioinformatics data on
transcription factor (TF) binding sites at these four specific
positions were obtained, revealing five different TFs: EGR1,
SMARCA4, TFAP2C, STAT1, and RBL2. These are listed in Table III along with their
mechanisms.
 | Table II.Summary of genomic loci significant
in PLS. |
Table II.
Summary of genomic loci significant
in PLS.
| ID | Position
(chr5) | % cons. | NCBI dbSNP
accession no. | Allele freq. in
ref. seq. | m.a. effect | m.a. CDs (≥1) | TF binding
sites |
|---|
| SNP1 | 143402837
(353a) | 0 | rs10482614 | G/A (−) strand
(88,5/11,5 (C 0,04)) | CpG del | 2 of 14 (2/24) | SMARCA4, TFAP2,
STAT1 |
| SNP2 | 143404507
(2023a) | 99 | rs1039242888 | G/C
unknownb | CpG del | 1 of 14 (1/24) | SMARCA4, TFAP2C,
STAT1, RBL2, EGR1 |
| SNP3 | 143404514
(2030a) | 95 | rs904759782 | G/C
unknownb | CpG del | 6 of 14
(10/24) | SMARCA4, TFAP2C,
STAT1, RBL2 |
| SNP4 | 143404564
(2080a) | 99 | rs3806855 | A/C
(88,5/11,5) | Unknown | 7 of 14 (8/24) | SMARCA4, TFAP2C,
STAT1, RBL2 |
 | Table III.Summary of TFs. |
Table III.
Summary of TFs.
| TF | Definition | Function |
|---|
| EGR1 | Early growth
response protein 1 | Transcriptional
regulator of genes involved in differentiation and mitogenesis |
| RBL2 | RB transcriptional
corepressor like 2 | Tumor suppressor
gene |
| SMARCA4 | SWI/SNF related,
matrix associated, actin dependent regulator of chromatin,
subfamily a, member 4 | Regulation of
chromatin structure |
| STAT1 | Signal transducer
and activator of transcription 1 | Transcription
activator (upon cytokine and growth factor signaling) |
| TFAP2C | TF AP-2γ | Sequence specific
DNA-binding TF implicated in gene regulation development and
differentiation |
Major vs. minor SNP alleles
The PLS analysis was repeated containing each allele
(major or minor) for each SNP, one at a time and the findings from
the pooled PLS analysis, as presented above were confirmed. Both
homozygotes and heterozygotes for the minor alleles were included
in the minor allele group. The influence of major vs. minor allele
carriership on postoperative complications was compared and of the
four SNPs which emerged significant in our PLS (above), only SNP4
was significantly different between the major and minor alleles
(p=0.016) when analysing one factor, i.e. major vs minor allele
carriership for this one SNP, at a time (Fig. 3).
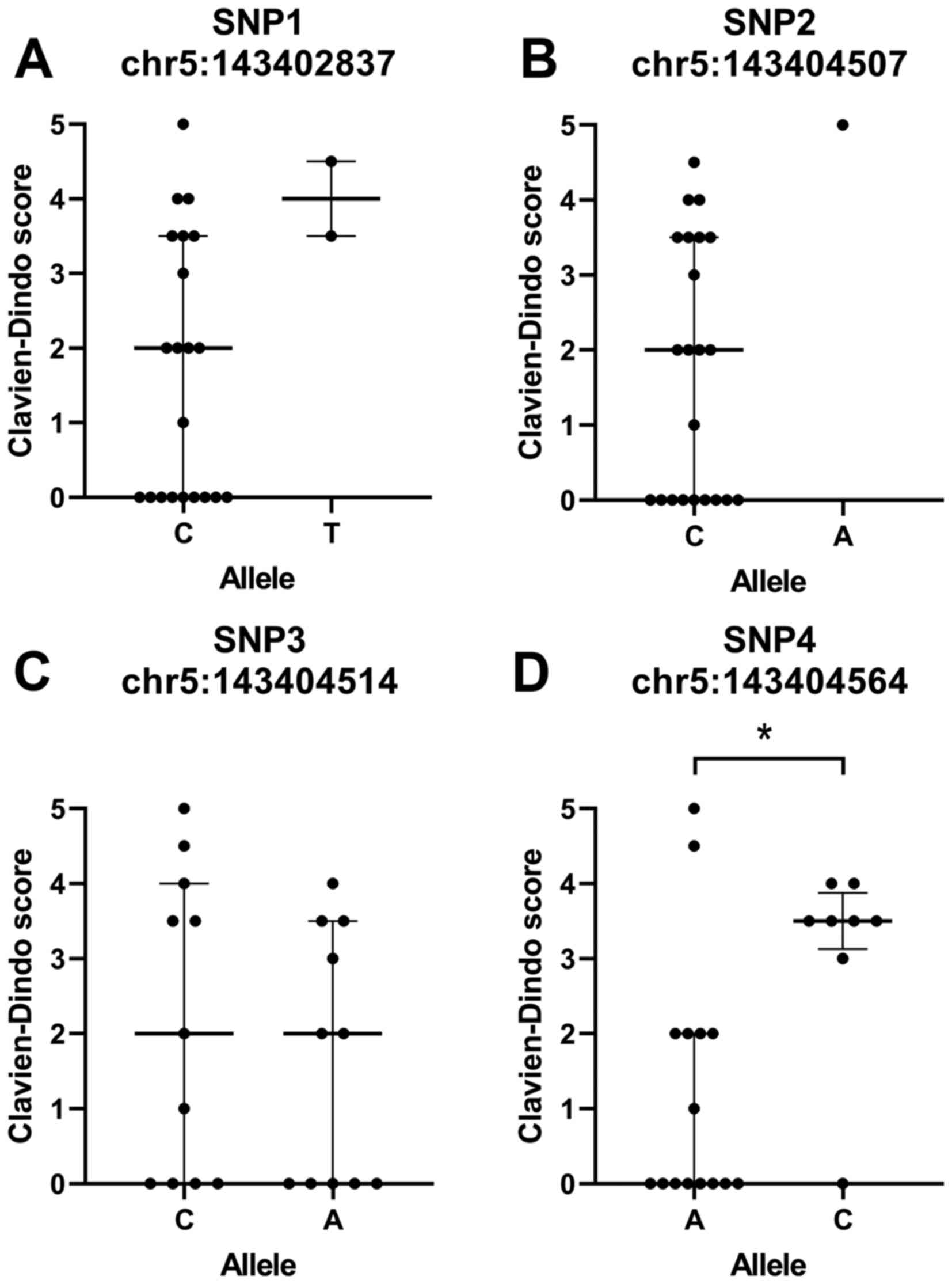 | Figure 3.Major vs. minor allele carriership
and postoperative complications. Postoperative complications,
classified according to the Clavien-Dindo system, in carriers of
the major vs. minor alleles of SNP1-4. Patients with no
Clavien-Dindo score are labelled as zero. SNP1,2 and 4, n=24; SNP3,
n=23. (A) chr5:143402837, SNP1; (B) chr:143404507, SNP2; (C)
chr5:143404514, SNP3; and (D) chr5:143404564, SNP4. Data are
presented as the mean ± inter-quartile range. SNP4 is significantly
different between the major and minor alleles (P=0.016). Major
alleles are shown on the left, minor alleles in the right. Chr,
chromosome; A, adenine; C, cytosine; T, Thymine; *P<0.05. |
Discussion
In the present study, it was demonstrated that GR
gene NR3C1 exon 1 genotype was associated with postoperative
complications (using the Clavien-Dindo classification) in patients
undergoing major abdominal surgery. However, the results also
demonstrated that DNA methylation in the CpG island of the
NR3C1 exon 1 had no impact on clinical outcome. Moreover,
this region proved to be highly demethylated in all patients
included in the study. This unexpected finding contradicted the
hypothesis and contrasts with the high variance in methylation of
this region found in a population of healthy individuals (13). The observed enrichment in
demethylation of the CpG island in the NR3C1 gene in the
studied cohort merits further investigation, for example, by
comparing healthy volunteers.
The sequenced region (chr5:143,402,505–143,405,805)
contained 13 previously described SNPs (with a frequency of >1%
in the population). Of these common SNPs, four were noted as being
associated with postoperative complications in the present study.
SNP1 (NCBI dbSNP accession number rs10482614), located in-between
exon 1H and 1C, has previously been investigated (16). SNP1 can eliminate a CpG site;
however, it was not found to be correlated with the methylation of
the 1H promotor, which is connected to haemodynamic stress
responses (17). Furthermore, SNP1
is associated with psychiatric disorders and affects the relative
expression levels of different mRNA isoforms (25). In the present study, SNP1 was also
correlated with SNP4 (NCBI dbSNP accession number rs3806855), in an
allelic associative manner, as previously reported (16). There is therefore evidence that
these SNPs have a biological function and could possibly harbour
clinically relevant functions. Furthermore, in addition to SNP1 and
SNP4, two more SNPs emerged in the present study. SNP3 was
significantly associated with postoperative complications. This SNP
has previously been recorded but not studied (rs1039242888). In the
present study SNP2 was identified in a single patient with a major
complication (biliary anastomosis leakage).
SNP2-4 are closely situated within 60 bp from one
another, making this region particularly interesting when
investigating the regulation of the NR3C1 gene. The
frequencies of SNP3 and SNP4 were remarkably high in the small
patient cohort of the present study, >40% and 33%, respectively,
compared with the previously reported highest population MAF of
carriers of the minor allele (GRCh38/hg38). Similar to the marked
enrichment of demethylation of the CpG site, the high frequency of
SNP3 and 4 in our patient cohort merits further investigation.
The high degree of conservation in the region of
SNP2-4, along with the multiple binding sites of TFs, has proven
this to be an important area of regulation of GR expression. Other
studies have reported SNPs and demonstrated their different GR
transactivation potentials (30–33).
Associations between GR polymorphisms and disease activity in
chronic inflammation have previously been reported (34). Both risk and severity of acute graft
vs. host disease following transplantation of haematopoietic stem
cells, are shown to be affected by the presence of certain SNPs in
either recipients or donors (35).
GR SNP results from numerous studies have been compiled in a review
concluding that there is substantial evidence of GR SNP effects on
clinical phenotypes (36).
The altered frequency of SNPs reported in the
present study may have impacted the binding affinity and avidity of
TFs and thus affected subsequent GR mRNA transcription. It has
previously been demonstrated that methylation in this region can
counteract transcriptional effects of certain common SNPs, possibly
compensating for genotype alterations (17). Furthermore, there was a trend
towards an association with postoperative complications for
carriers of the SNP1 minor allele (P=0.087), which in the present
study was correlated with SNP4. This finding is similar to previous
reports (16,32). Furthermore, for SNP2, which is
located close to SNP4, there was a similar trend for patients with
the minor allele (P=0.083).
Overall, the results suggested that multiple levels
of GR expression regulation may be associated with the severity of
postoperative complications, including the genetic variants in exon
1, and that this regulatory region within the NR3C1 gene may
be of relevance in a clinical setting. Furthermore, genetic
variability of the NR3C1 gene is not infrequent in the
general population and it is therefore unlikely that it has a
noticeable impact on healthy individuals (www.ncbi.nlm.nih.gov/SNP) (37). However, in the present study an
association has been demonstrated with an adverse clinical outcome
after surgically induced inflammatory stress.
A limitation of the present study was the lack of
comparisons to healthy volunteers. There were also shortcomings in
statistical power, but these findings can be useful for power
calculations in future studies. However, the present study was an
exploratory study and to the best of our knowledge is the first of
its kind, combining gene sequencing, clincal data on surgical
outcomes and advanced statistics. Furthermore, similar
translational studies are needed to elucidate patterns of GR
regulation in patients with normal as well as complicated outcomes
in the context of inflammatory responses.
In conclusion, the association between genetic
variability in GR gene NR3C1 exon 1 and postoperative
complications in patients undergoing major abdominal surgery
indicates that the GC response may be of importance for
inflammatory responses that affect clinical outcomes. The observed
enrichment of demethylation of the CpG island in exon 1 of the
NR3C1 gene in patients planned for major surgery warrants
further clinical studies.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Language editing was generously provided by Dr Ahl
Hulme, Division of Emergency and Trauma Surgery, Department of
Traumatology, Karolinska University Hospital Solna and Örebro
University School of Medicine, who otherwise had no part in this
project.
Funding
This paper was supported by the Swedish Cancer Foundation (grant
number CAN 2014/537, Regional Research Council in the
Uppsala-Örebro region (grant number 313841), the Swedish Research
Council (funding for clinical research in medicine, grant numbers
2004-05821 and 2004-06898, the Cancer Research Foundation in
Norrland (grant number: AMP 16-813), and The Emil Andersson
Foundation for Medical Research Foundation.
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request. According to Swedish law, clinical individual data can be
shared only if Ethical Approval is obtained as human subjects are
involved. The sequencing data generated in the present study may be
found in the European Genome-phenome Archive (EGA), under accession
number EGAS00001005737 (ega-archive.org).
Authors' contributions
TG, LS, ACW and OW conceived the study. TG, LS, MW,
EAB and LKS were responsible for data curation. MW, EAB and TG
confirm the authenticity of all raw data, MW and TG performed data
analysis. OW, ACW, LS and EAB acquired funding for the project. TG,
EAB and MW performed the investigation. TG, MW, EAB, OW, ACW and
LKS designed the methodology. LS, ACW and OW provided resources.
LS, ACW and OW were responsible for project administration. MW was
responsible for the software used in the project. LS, ACW, OW and
LKS supervised the project. EAB and MW validated the results. MW
and EAB visualized the study. EAB and TG prepared the original
manuscript draft. TG, MW, ACW and LS wrote, reviewed and edited the
manuscript. All authors read and agreed to the final
manuscript.
Ethics approval and consent to
participate
Ethical approval for this study was granted by the
Stockholm Regional Ethical Review Board (Stockholm, Sweden;
approval no. 2009/366-31). The patients recruited for the present
study were enrolled after informed consent was obtained, orally and
in writing, in accordance with the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GR
|
glucocorticoid receptor
|
|
GC
|
glucocorticoid
|
|
NR3C1
|
nuclear receptor subfamily 3 group C
member 1
|
|
SNP
|
single nucleotide polymorphisms
|
|
CpG
|
cytosine-phosphate-guanine
|
|
PLS
|
partial least squares
|
|
chr
|
chromosome
|
|
BiS
|
bisulfite
|
|
TF
|
transcription factor
|
|
GRCh38/hg38
|
Human Genome Reference Consortium,
human (build) 38 (human genome 38)
|
References
|
1
|
Rettig TC, Verwijmeren L, Dijkstra IM,
Boerma D, van de Garde EM and Noordzij PG: Postoperative
interleukin-6 level and early detection of complications after
elective major abdominal surgery. Ann Surg. 263:1207–1212. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Desborough JP: The stress response to
trauma and surgery. Br J Anaesth. 85:109–117. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Barnes PJ: Anti-inflammatory actions of
glucocorticoids: Molecular mechanisms. Clin Sci (Lond). 94:557–572.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Annane D, Bellissant E, Bollaert PE,
Briegel J, Confalonieri M, De Gaudio R, Keh D, Kupfer Y, Oppert M
and Meduri GU: Corticosteroids in the treatment of severe sepsis
and septic shock in adults: A systematic review. JAMA.
301:2362–2375. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marik PE, Pastores SM, Annane D, Meduri
GU, Sprung CL, Arlt W, Keh D, Briegel J, Beishuizen A, Dimopoulou
I, et al: Recommendations for the diagnosis and management of
corticosteroid insufficiency in critically ill adult patients:
Consensus statements from an international task force by the
American college of critical care medicine. Crit Care Med.
36:1937–1949. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cain DW and Cidlowski JA: Immune
regulation by glucocorticoids. Nat Rev Immunol. 17:233–247. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hearing SD, Norman M, Smyth C, Foy C and
Dayan CM: Wide variation in lymphocyte steroid sensitivity among
healthy human volunteers. J Clin Endocrinol Metab. 84:4149–4154.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Graberg T, Strommer L, Hedman E, Uzunel M,
Ehrenborg E and Wikstrom AC: An ex vivo RT-qPCR-based assay for
human peripheral leukocyte responsiveness to glucocorticoids in
surgically induced inflammation. J Inflamm Res. 8:149–160. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Oakley RH and Cidlowski JA: The biology of
the glucocorticoid receptor: New signaling mechanisms in health and
disease. J Allergy Clin Immunol. 132:1033–1044. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Carvalho LA, Bergink V, Sumaski L,
Wijkhuijs J, Hoogendijk WJ, Birkenhager TK and Drexhage HA:
Inflammatory activation is associated with a reduced glucocorticoid
receptor alpha/beta expression ratio in monocytes of inpatients
with melancholic major depressive disorder. Transl Psychiatry.
4:e3442014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sanchez-Vega B, Krett N, Rosen ST and
Gandhi V: Glucocorticoid receptor transcriptional isoforms and
resistance in multiple myeloma cells. Mol Cancer Ther. 5:3062–3070.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Turner JD and Muller CP: Structure of the
glucocorticoid receptor (NR3C1) gene 5′untranslated region:
Identification, and tissue distribution of multiple new human exon
1. J Mol Endocrinol. 35:283–292. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Turner JD, Pelascini LP, Macedo JA and
Muller CP: Highly individual methylation patterns of alternative
glucocorticoid receptor promoters suggest individualized epigenetic
regulatory mechanisms. Nucleic Acids Res. 36:7207–7218. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Palma-Gudiel H, Cordova-Palomera A, Leza
JC and Fananas L: Glucocorticoid receptor gene (NR3C1) methylation
processes as mediators of early adversity in stress-related
disorders causality: A critical review. Neurosci Biobehav Rev.
55:520–535. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Leenen FA, Muller CP and Turner JD: DNA
methylation: Conducting the orchestra from exposure to phenotype?
Clin Epigenetics. 8:922016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cao-Lei L, Leija SC, Kumsta R, Wüst S,
Meyer J, Turner JD and Muller CP: Transcriptional control of the
human glucocorticoid receptor: Identification and analysis of
alternative promoter regions. Hum Genet. 129:533–543. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li-Tempel T, Larra MF, Sandt E, Mériaux
SB, Schote AB, Schächinger H, Muller CP and Turner JD: The
cardiovascular and hypothalamus-pituitary-adrenal axis response to
stress is controlled by glucocorticoid receptor sequence variants
and promoter methylation. Clin Epigenetics. 8:122016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Suderman M, McGowan PO, Sasaki A, Huang
TC, Hallett MT, Meaney MJ, Turecki G and Szyf M: Conserved
epigenetic sensitivity to early life experience in the rat and
human hippocampus. Proc Natl Acad Sci USA. 109 Suppl 2(Suppl
2):S17266–S17272. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Oberlander TF, Weinberg J, Papsdorf M,
Grunau R, Misri S and Devlin AM: Prenatal exposure to maternal
depression, neonatal methylation of human glucocorticoid receptor
gene (NR3C1) and infant cortisol stress responses. Epigenetics.
3:97–106. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Weaver IC, Cervoni N, Champagne FA,
D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M and Meaney MJ:
Epigenetic programming by maternal behavior. Nat Neurosci.
7:847–854. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dindo D, Demartines N and Clavien PA:
Classification of surgical complications: A new proposal with
evaluation in a cohort of 6336 patients and results of a survey.
Ann Surg. 240:205–213. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
R Core Team (2017), . R: A language and
environemnt for statistical computing. R Foundation for Statistical
Computing Vienna, Austria: https://www.r-project.orgJanuary 172022
|
|
23
|
Bodenhofer U, Bonatesta E, Horejs-Kainrath
C and Hochreiter S: msa: An R package for multiple sequence
alignment. Bioinformatics. 31:3997–3999. 2015.PubMed/NCBI
|
|
24
|
Lê S, Josse J and Husson F: FactoMineR: An
R package for multivariate analysis. Journal of Statistical
Software. 25:1–18. 2008. View Article : Google Scholar
|
|
25
|
Mayor C, Brudno M, Schwartz JR, Poliakov
A, Rubin EM, Frazer KA, Pachter LS and Dubchak I: VISTA:
Visualizing global DNA sequence alignments of arbitrary length.
Bioinformatics. 16:1046–1047. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
1000 Genomes Project Consortium, . Auton
A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini
JL, McCarthy S, McVean GA and Abecasis GR: A global reference for
human genetic variation. Nature. 526:68–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
NHLBI GO Exome Sequencing Project (ESP), .
Exome Variant Server. NHLBI; Seattle, WA: http://evs.gs.washington.edu/EVS/June 18–2018
|
|
28
|
Karczewski KJ, Francioli LC, Tiao G,
Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A,
Birnbaum DP, et al: The mutational constraint spectrum from
variation in 141,456 humans. Nature. 581:434–443. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Howe KL, Achuthan P, Allen J, Allen J,
Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J,
et al: Ensembl 2021. Nucleic Acids Res. 49(D1):D884–D891. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sinclair D, Fullerton JM, Webster MJ and
Shannon Weickert C: Glucocorticoid receptor 1B and 1C mRNA
transcript alterations in schizophrenia and bipolar disorder, and
their possible regulation by GR gene variants. PLoS One.
7:e317202012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lasker MV, Leventhal SM, Lim D, Green TL,
Tung K, Cho K and Greenhalgh DG: Hyperactive human glucocorticoid
receptor isoforms and their implications for the stress response.
Shock. 43:228–232. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Leventhal SM, Lim D, Green TL, Cantrell
AE, Cho K and Greenhalgh DG: Uncovering a multitude of human
glucocorticoid receptor variants: An expansive survey of a single
gene. BMC Genet. 20:162019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Baker AC, Chew VW, Green TL, Tung K, Lim
D, Cho K and Greenhalgh DG: Single nucleotide polymorphisms and
type of steroid impact the functional response of the human
glucocorticoid receptor. J Surg Res. 180:27–34. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Quax RAM, Koper JW, Huisman AM, Weel A,
Hazes JM, Lamberts SW and Felders RA: Polymorphisms in the
glucocorticoid receptor gene and in the glucocorticoid-induced
transcript 1 gene are associated with disease activity and response
to glucocorticoid bridging therapy in rheumatoid arthritis.
Rheumatol Int. 35:1325–1333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Norden J, Pearce KF, Irving JAE, Collin
MP, Wang XN, Wolff D, Kolb HJ, Socie G, Kuzmina Z, Greinix H, et
al: The influence of glucocorticoid receptor single nucleotide
polymorphisms on outcome after haematopoietic stem cell
transplantation. Int J Immunogenet. 2018.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Koper JW, van Rossum EFC and van den Akker
EL: Glucocorticoid receptor polymorphisms and haplotypes and their
expression in health and disease. Steroids. 92:62–73. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sherry ST, Ward M and Sirotkin K:
dbSNP-database for single nucleotide polymorphisms and other
classes of minor genetic variation. Genome Res. 9:677–679. 1999.
View Article : Google Scholar : PubMed/NCBI
|