Introduction
Cholangiocarcinoma (CCA) is rare cancer, but its
incidence is rising worldwide. Because CCA is frequently
asymptomatic in the early stages, it is commonly diagnosed at an
advanced stage, which has a significant impact on the therapeutic
selection (1,2). Despite significant advances in CCA
diagnosis and treatment over the last decade, there has been no
significant improvement in patient prognosis with a 5-year survival
of 7–20% (3–10). Surgery is one of the most primary
therapeutic options for CCA. However, the vast majority of patients
(~70%) are not indicated for resection when diagnosed (11). Moreover, even after surgery, the
tumour recurrence rate is quite high (12).
As a result, chemotherapy remains the treatment of
choice for the vast majority of CCA patients. 5-Fluorouracil (5-Fu)
is one of the drugs that can be used to treat CCA. Because of its
low cost, it is a type of chemotherapy drug that is widely used in
the treatment of digestive system neoplasms. Nonetheless, 5-Fu has
some drawbacks, including a short biological half-life (10–20 min),
volatility in absorption, distribution, metabolism and
pharmacokinetics. Approximately 90% of the injected dose will
eventually be converted into inactive materials due to the
metabolism (13), limiting its
efficacy. As a result, maintaining the therapeutic serum
concentrations necessitates long-term administration of a high-dose
of the drug, which results in severe toxic and side effects
(14). Approximately 10–20% of
patients treated with standard 5-Fu exhibit a serious toxic
reaction, such as myelosuppression, gastrointestinal side effects,
neurologic and cutaneous adverse reaction, and others (15). Therefore, other techniques to
improve the effectiveness of 5-Fu treatment are urgently needed to
extend its effective time, improve its drug application efficiency,
and reduce its relevant side effects.
Exosomes, a type of extracellular vesicles (EV), are
now regarded as drug carriers with great potential and have been
extensively studied, owing to their ability to transport
therapeutic molecules such as proteins, nucleic acids, and drugs.
Exosomes can be found in a variety of body fluids and tissues,
including blood, saliva, urine, breast milk, cerebrospinal fluid,
adipose tissue (16–18), and even plants and milk (19). Exosomes play a role in
physiological and pathological processes, as well as serving as a
critical link in cell-cell communication and material interchange.
Exosomes also have distinct advantages, such as natural sources,
excellent biocompatibility, organ propensity, and low
immunogenicity. They can deliver bioactive substances and
therapeutic molecules in a variety of ways and sites, as well as
participate in cell modulation with target cells, which includes
tissue repair, tumour diagnosis and treatment, and immunomodulation
(20–24). Taking into account the properties
of exosomes, this study used exosomes as a chemotherapy drug
carrier, and 5-Fu was loaded into the exosomes (5-Fu-Exos) via
sonication and incubation methods. The experimental results showed
that 5-Fu-Exos are more effective than free 5-Fu in inhibiting CCA
cell viability, implying that exosomes could be used as a new
targeted chemotherapy drug carrier for CCA treatment.
Materials and methods
Cell culture
Human bone marrow mesenchymal stem cells (HBMSCs)
and the human CCA cell line QBC939 were obtained from Shanghai Cell
Bank, Chinese Academy of Sciences. Both cell lines were grown in
DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal
bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc.), 100
U/ml penicillin, and 100 U/ml streptomycin. HBMSCs were found to be
uniformly distributed at the bottom of the culture flask and were
incubated in a constant temperature incubator at 37°C with 5%
CO2. When the cells reached 70–80% confluency, they were
digested with trypsin digestive juice (Gibco; Thermo Fisher
Scientific, Inc.) and passaged at a 1:4 ratio under an inverted
microscope. For this experiment, cells from generations 3–5 were
chosen.
Isolation of the exosomes
Ultracentrifugation was used to separate exosomes
from the conditioned medium. The HBMSC medium was replaced with
high-glucose DMEM containing 10% exosome-depleted FBS and cultured
for 48 h. The cultured fluid was collected and centrifuged at 800 ×
g for 5 min. The supernatant was collected and filtered using a
0.22-µm filter. The supernatant was centrifuged at 300 × g for 10
min to collect the supernatant, 2,000 × g for 10 min to precipitate
dead cells, and 10,000 × g for 30 min to remove cell debris at 4°C.
Following that, the exosomes were pelleted by ultracentrifugation
at 100,000 × g for 70 min and washed with PBS to remove the
contaminated cells. The collected solution was then centrifuged at
100,000 × g for 70 min before being resuspended in PBS, with the
supernatant removed, to obtain final exosomes, which were then
stored in sub-packages at −80°C for the next experimental
stage.
Exosome characterization
The morphology of the exosomes was studied using a
transmission electron microscope (TEM). An amount of 10 µl exosomes
was collected by ultracentrifugation, dropped onto a copper grid,
and dried with filter paper after 1 min. The grid was then stained
with 2% phosphotungstic acid and dried with filter paper after 1
min. Afterward, it was dried with a filament lamp before being
observed and photographed under a TEM. Western blot analysis was
used to identify exosome-specific proteins. To determine the
concentration of proteins derived from the HBMSCs, the BCA Protein
Assay Kit (Beyotime Institute of Biotechnology) was used. After
dissociation by sodium dodecyl sulfate-poly-acrylamide gel
electrophoresis (SDS-PAGE), proteins in the gel were transferred to
a PVDF membrane, sealed with dry skim milk for 1 h at room
temperature, and incubated overnight at 4°C with tumor
susceptibility 101 (TSG101) (dilution 1:500, cat. no. ab125011;
Abcam), and CD9 (dilution 1:500, cat. no. ab236630; Abcam)
monoclonal antibodies. After washing the membrane (10 min × 3
times) with Tris-buffered saline with Tween-20 (TBST), goat
anti-rabbit secondary antibody (dilution 1:5,000, cat. no.
ab150077; Abcam) was applied for 2 h before rewashing with TBST (10
min × 3 times). Finally, the protein was coloured using a
chemiluminescent ECL kit (Beyotime Institute of Biotechnology).
Nanoparticle tracking analysis (NTA) of exosome size distribution
curves was performed using Zetaview (PMX 110, Particle Metrix) in
accordance with the manufacturer's instructions. PBS was used to
properly dilute exosomes (1 mg/ml) (1:1,000). Zetaview Software
8.02.31 was used to record and analyse particle sizes and numbers
from all 11 positions.
Drug entrapment
Exosomes were mixed with 5-Fu in PBS at a protein
concentration of 1 mg/ml. The exosome solution (5 ml/1 mg/ml) and
the 5-Fu solution (5 ml/1 mg/ml) were mixed uniformly. Two loading
methods were used: direct incubation and sonication. The mixture
was incubated in an incubation shaker at 37°C for 1 h for the
direct incubation method. The ultrasonic probe (Ultrasonic Cell
Breaker 705, Thermo Fisher Scientific, Inc.) was immersed in the
mixture for the sonication method. The entire procedure was carried
out in an ice bath in six cycles (frequency 20 kHz, amplitude 20%,
30 sec on and 30 sec off for each cycle, 2-min interval between
each cycle). The solution was sonicated and then incubated at 37°C
for 30 min to recover the exosomal membrane. The two types of
solutions were centrifuged and washed with PBS to collect exosomes
and 5-Fu-loaded exosomes (5-Fu-Exos).
Determination of 5-Fu
concentration
A high-performance liquid chromatography (HPLC)
system (High-Performance Liquid Chromatography Analyser, Agilent
Technologies, Inc.) was used to determine the amount of 5-Fu loaded
into exosomes. Bond Elut LRC C-18 solid-phase disposable extraction
column (Varian) was loaded with 5-Fu solution and exosome
dispersion containing 100 µg protein. The extract was evaporated
after extraction, and the residue was mixed with 200 µl methanol.
HPLC was used to determine the 5-Fu content under the following
chromatographic conditions: chromatographic column (Zorbax RX-C18;
Agilent Technologies, Inc.); sample volume, 20 µl; move phase, 40%
water-60% methanol; detecting wavelength, 265 nm; column
temperature, 30°C. PBS standard solutions with 5-Fu concentrations
of 0.5, 1.0, 2.5, 5.0, 10.0, and 20.0 µg/ml were prepared, and the
5-Fu standard solutions were determined by HPLC to obtain varying
concentrations and their corresponding peak areas, resulting in a
standard curve. The peak area in the standard curve was used to
calculate the concentration of 5-Fu in the sample.
The uptake of exosomes by CCA QBC939
cells
PKH26 (cat. no. PKH26GL-1KT red, Sigma-Adrich; Merck
KGaA) was used to label exosomes that were collected. After
co-culturing the stained exosomes and the QBC929 cell line for 24
h, the cell nucleus was stained with DAPI dye (cat. no C1002,
Beyotime Institute of Biotechnology) for 10 min. In the subsequent
observation, a laser scanning confocal microscope was used.
Cytotoxicity of 5-Fu
The CCK-8 assay was used to determine the
cytotoxicity of 5-Fu-Exos and free 5-Fu on the CCA QBC939 cell
line. After being resuspended in PBS and medium, the QBC939 cells
were counted and were seeded at a density of 5×104/ml in
96-well plates with 100 µl (5×103 cells) per well, three
compound wells in each group. For 24 h, the cells were grown in
complete DMEM containing 10% (v/v) FBS at 37°C. After three washes
with PBS, the DMEM with 10% (v/v) exosome-depleted FBS was added.
5-Fu-Exos were filtered through a 0.22-µm aseptic filter and
diluted into nutrient solutions with varying 5-Fu concentrations
(0, 0.1, 0.5, 1.0, 2.0, 5.0 and 10.0 µg/ml) using DMEM. One hundred
microliters of the above-mentioned 5-Fu-Exos at various
concentrations were added to the seeded QBC939 cells. The drug
control group received DMEM containing varying concentrations of
5-Fu (0, 0.1, 0.5, 1.0, 2.0, 5.0 and 10.0 µg/ml). The culture plate
was incubated at 37°C for 24 h. Incubation was carried out for 1 h
after adding 20 µl CCK-8 solvent. Following colour change,
absorbance at 450 nm was detected to measure the cell viability
using an enzyme labelling instrument (BIO-TekELx800 automatic
enzyme labelling instrument, Bio-Rad Laboratories, Inc.). Exosome
toxicity against QBC939 cells was determined using the same method
as described above. QBC939 cells were co-incubated with pure DMEM,
pure HBMSC-Exos, and 5-Fu-Exos.
Statistical analysis
All data were statistically analysed using GraphPad
Prism 7.0 (GraphPad Software, Inc.). The data are presented as a
mean value ± standard deviation (SD). For inter-group and
multiple-group comparisons, the t-test and one-way analysis of
variance (ANOVA) followed by Bonferroni's post hoc test was used
with a significance level of P<0.05.
Results
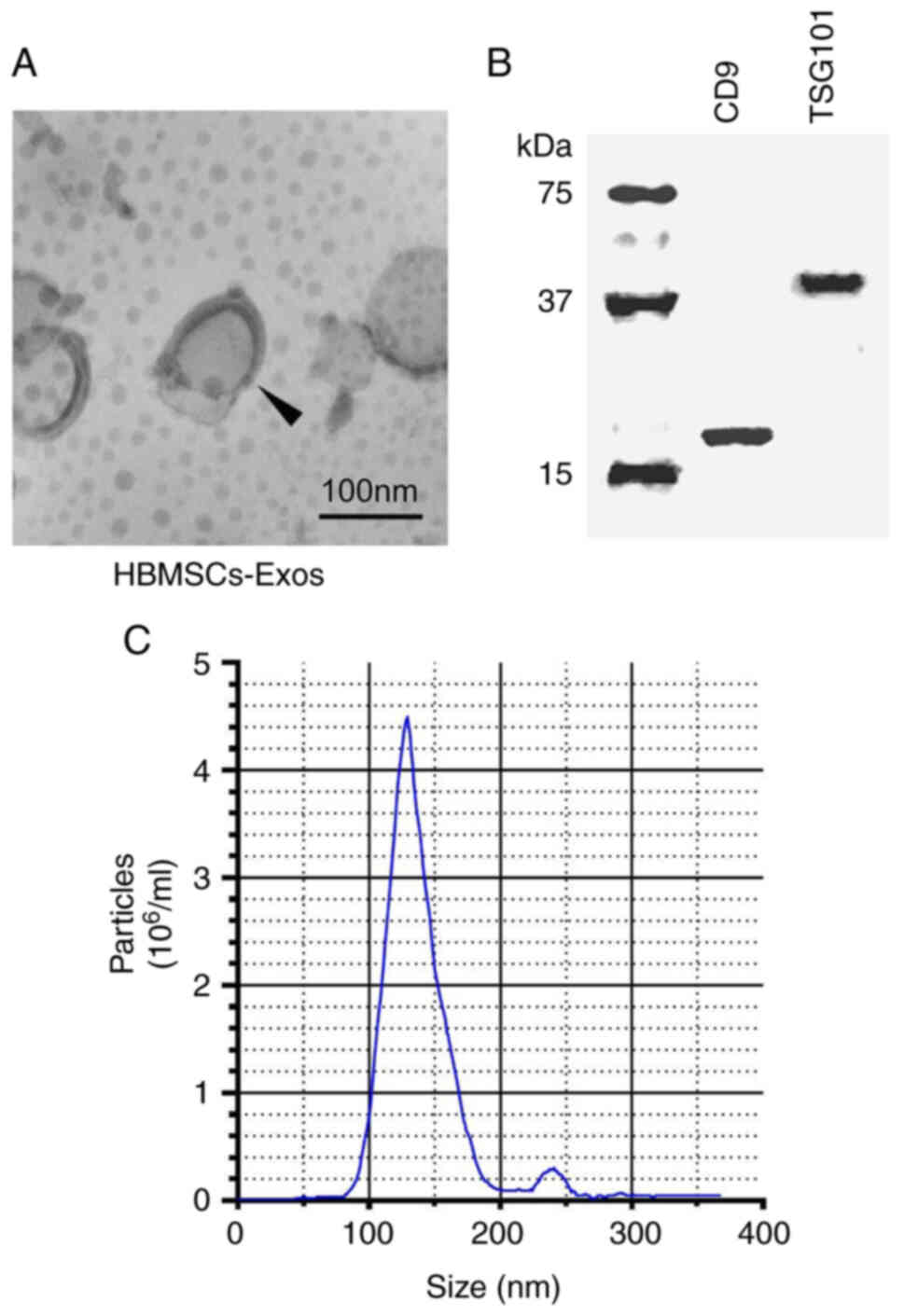
Identification of the exosomes
Exosomes were isolated from the HBMSCs using
ultracentrifugation, and the morphology and size of the exosomes
were examined using a transmission electron microscope. Round and
small vesicles with a double-layer structure were detected, with a
size of approximately 100 nm (Fig.
1A), which was consistent with previous reports (25). Then, exosome-specific proteins
were identified using western blot analysis. The findings revealed
that CD9 and TSG101 were highly expressed (Fig. 1B). The aforementioned findings
indicated that exosomes were successfully purified and separated.
Exosomes from the HBMSCs had a narrow size distribution, with an
average diameter of 127.8±2.3 nm, according to NTA measurements
(Fig. 1C).
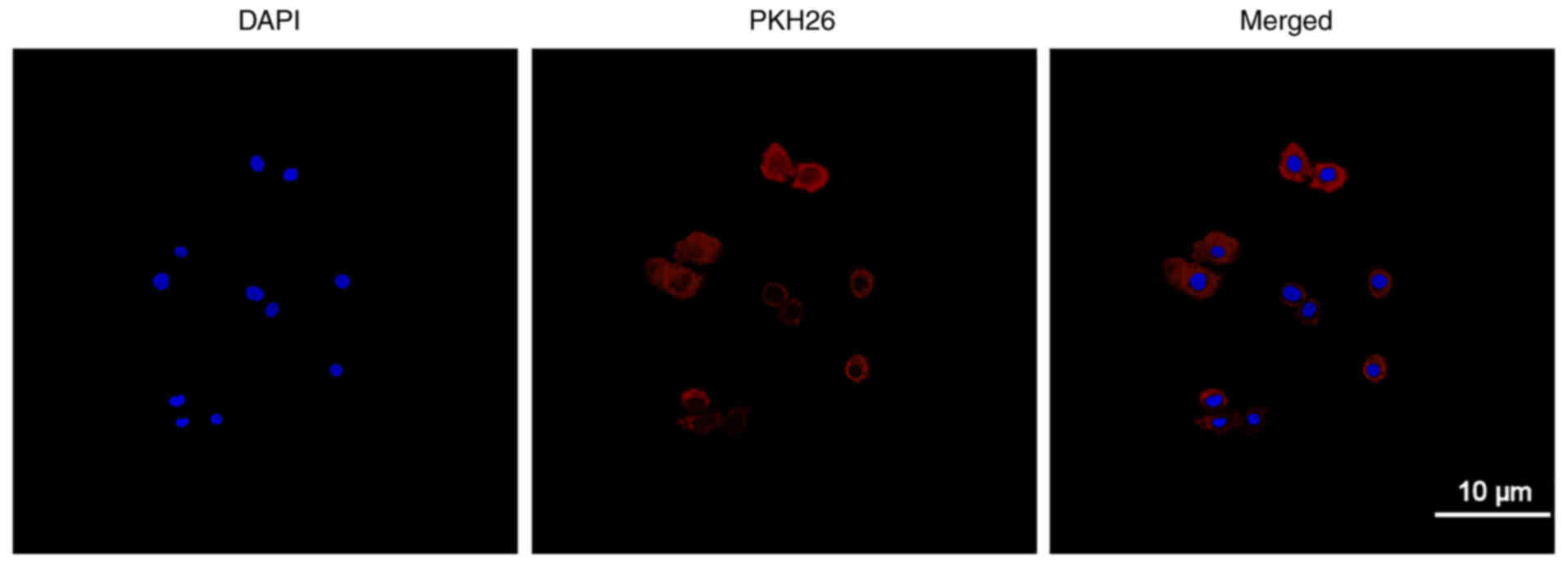
Uptake of exosomes
Exosomes were labelled with PKH26 and incubated with
QBC939 cells before being observed using laser confocal scanning
microscopy to determine the efficiency of the QBC939 cellular
uptake of exosomes. Fig. 2 shows
the cell nucleus stained with DAPI (blue) and exosomes labelled
with PKH26 (red). Red fluorescence can be seen in the cytoplasm
after confluence, revealing the QBC939 cellular uptake of the
exosomes.
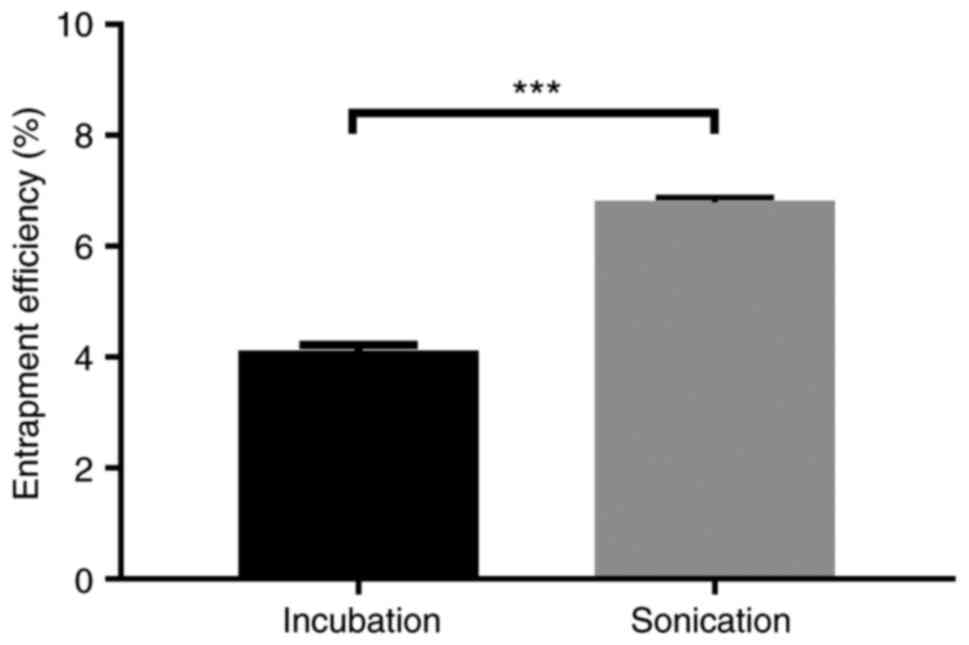
Efficiency of 5-Fu loaded in the
exosomes
The protein content in exosomes was determined using
a BCA kit, and the amount of 5-Fu encapsulated in the exosome was
determined using HPLC to determine the loading efficiency of
different loading methods (sonication and incubation). The
following equation was used to calculate the drug loading rate
(DL%): DL (%)=(Weight of drug-loaded/Weight of exosomes) ×100%.
Fig. 3 depicts the result. In
comparison to the incubation DL of 4.10%, the DL of sonication
reached 6.79%.
Effect of 5-Fu-Exos on the cell
viability of QBC939 cells
The CCK-8 test was used to assess the effect of the
drug-loaded exosomes on tumour proliferation. Fig. 4 depicts the cytotoxic properties
of 5-Fu and 5-Fu-Exos. After 24 h of incubation with QBC939 cells,
the 5-Fu-Exos group significantly inhibited tumour cell viability
more than the 5-Fu group (Fig.
4A) and had a lower half-maximal inhibitory concentration
(IC50) (Fig. 4B). The
cytotoxicity of the exosome control groups (DMEM) revealed that
naive exosomes (Exos) had no cytotoxic effect on QBC939 cells
(Fig. 4C).
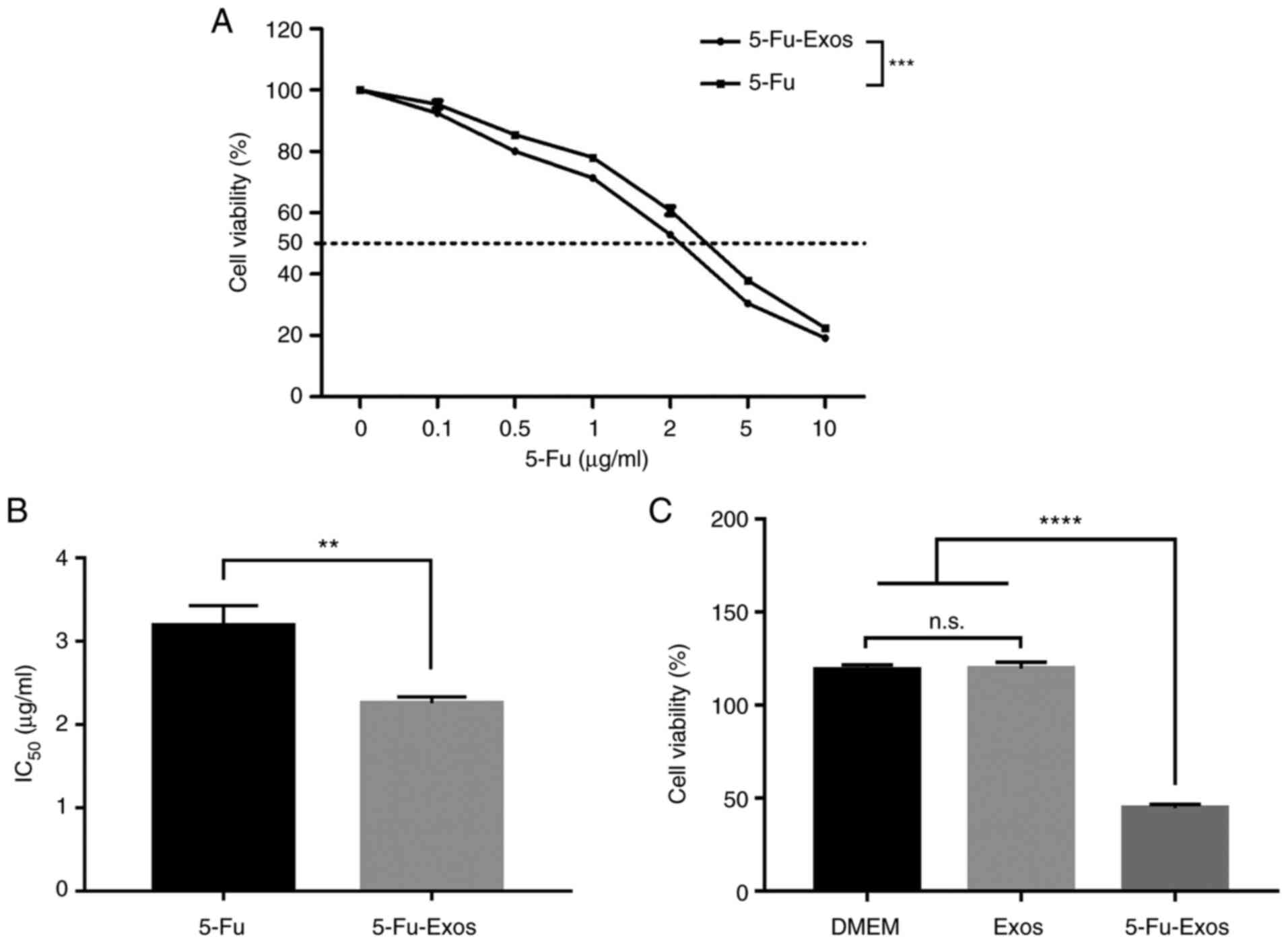 | Figure 4.Effect of HBMSC-derived Exo-5-Fu, free
5-Fu, DMEM and naive exosomes on the viability of CCA QBC939 cells.
(A) QBC939 cells were treated with free 5-Fu, and Exo-5-Fu. The
cells were further incubated for 24 h, and the viability assay was
performed following CCK-8 assay protocol. (B) The half-maximal
inhibitory concentration (IC50) of free 5-Fu and
Exo-5-Fu, where the value was calculated according to A. (C) The
naive exosomes from HBMSCs had no cytotoxic effect against QBC939
cells. Data were analyzed by one-way ANOVA with Bonferroni's post
hoc test. Significance level; **P<0.01, ***P<0.001,
****P<0.0001, n.s., not significant. CCA, cholangiocarcinoma;
HBMSCs, human bone marrow mesenchymal stem cells; Exos, exoxomes;
5-Fu, 5-fluorouracil. |
Discussion
5-Fluorouracil (5-Fu) is currently one of the most
commonly used chemotherapy drugs for cholangiocarcinoma (CCA)
treatment, but its efficacy and clinical application are limited
due to its dose-dependent nature. As a classic chemotherapy drug,
5-Fu promotes cell apoptosis by releasing metabolites after
entering cells to disrupt RNA synthesis and the action of
thymidylate synthase (26).
Therefore, the apoptosis experiment was not repeated in this study.
As for whether 5-Fu-Exos have any other different modulation
actions, we will investigate them in future research.
Exosomes have been widely studied in targeted
delivery systems in recent years due to their high
biocompatibility, low cytotoxicity, and low immunogenicity.
Exosomes can be loaded with a variety of therapeutic molecules to
improve drug efficacy and inhibit tumour cell proliferation.
According to Wei et al, loading doxorubicin (DOX) into
exosomes derived from mesenchymal stem cells to inhibit
osteosarcoma cell viability in vitro exhibited a better
antitumor effect than free DOX (27). Exosomes are also important in the
treatment of drug-resistant tumours. Liang et al discovered
that electroporation and transfection of tumour-derived exosomes
loaded with miR-21 inhibitor and 5-Fu could effectively reverse
drug resistance and improve therapeutic outcomes in 5-Fu-resistant
colon carcinoma cells (28). In
the present study, exosomes derived from human bone marrow
mesenchymal stem cells (HBMSCs) and loaded with 5-Fu were used to
treat CCA. The exosome medication group significantly increased
cytotoxicity in CCA cells and decreased drug concentration when
compared to free 5-Fu.
Exosomes are gaining popularity as a carrier for
drug and gene therapy. However, the problem of mass production of
exosome loading systems remains difficult, with drug loading being
an important component. Sonication is a popular loading method
because it is simple and efficient. Exosomes have been found to be
capable of carrying small-molecule drugs, proteins and small
nucleic acids (22). Kim et
al loaded paclitaxel into macrophage-derived engineered
exosomes with a loading efficiency of 33% (29). The sonication and standard
incubation methods were used to load 5-Fu in this experiment, and
it was discovered that the sonication method had a higher loading
efficiency. The sonication method can also be used to load small
nucleic acids. According to research, the sonication approach has
higher loading efficiency and less siRNA aggregation when loading
siRNAs by electroporation and sonication (30). Furthermore, Thakur et al
demonstrated that sonication and microfluidic technology are more
efficient than traditional incubation and electroporation methods
in loading DOX into exosomes. In addition, it was discovered that
the sigmoid-type microfluidic structure outperformed the linear
microfluidic structure when it came to drug loading (31). As a result, further research into
optimising loading methods to improve exosome loading efficiency is
warranted.
In conclusion, the present study successfully loaded
the anti-cholangiocarcinoma drug 5-Fu into exosomes derived from
HBMSCs using sonication and incubation methods. In terms of loading
efficiency, sonication outperformed incubation, and 5-Fu-Exos had
higher antineoplastic activity than free 5-Fu. Therefore, the goal
of this study was to develop an exosome-based drug delivery system
for the targeted delivery of 5-Fu for CCA treatment. It is expected
that further research on mouse models of CCA will be conducted in
the future to confirm the safety and efficacy of exosome-loading
drugs in the treatment of CCA.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MC and JZ designed the research, performed the
experiments, analyzed the data and wrote the manuscript. YL and NM
collected the data. MC and JZ confirm the authenticity of all the
raw data. All authors read and approved the manuscript and agree to
be accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work (including the
provided data) are appropriately investigated and resolved. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics Review
Committee at Taizhou People's Hospital, Taizhou, China.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Andersen JB, Spee B, Blechacz BR, Avital
I, Komuta M, Barbour A, Conner EA, Gillen MC and Roskams T: Genomic
and genetic characterization of cholangiocarcinoma identifies
therapeutic targets for tyrosine kinase inhibitors.
Gastroenterology. 142:1021–1031.e15. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Banales JM, Cardinale V, Carpino G,
Marzioni M, Andersen JB, Invernizzi P, Lind GE, Folseraas T, Forbes
SJ, Fouassier L, et al: Expert consensus document:
Cholangiocarcinoma: Current knowledge and future perspectives
consensus statement from the European network for the study of
cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol.
13:261–280. 2016. View Article : Google Scholar
|
|
3
|
Strijker M, Belkouz A, van der Geest LG,
van Gulik TM, van Hooft JE, de Meijer VE, Haj Mohammad N, de Reuver
PR, Verheij J, de Vos-Geelen J, et al: Treatment and survival of
resected and unresected distal cholangiocarcinoma: A nationwide
study. Acta Oncol. 58:1048–1055. 2019. View Article : Google Scholar
|
|
4
|
Kamsa-Ard S, Luvira V, Suwanrungruang K,
Kamsa-Ard S, Luvira V, Santong C, Srisuk T, Pugkhem A,
Bhudhisawasdi V and Pairojkul C: Cholangiocarcinoma trends,
incidence, and relative survival in Khon Kaen, Thailand from 1989
through 2013: A population-based cancer registry study. J
Epidemiol. 29:197–204. 2019. View Article : Google Scholar
|
|
5
|
Lindnér P, Rizell M and Hafström L: The
impact of changed strategies for patients with cholangiocarcinoma
in this millenium. HPB Surg. 2015:7360492015. View Article : Google Scholar
|
|
6
|
Alabraba E, Joshi H, Bird N, Griffin R,
Sturgess R, Stern N, Sieberhagen C, Cross T, Camenzuli A, Davis R,
et al: Increased multimodality treatment options has improved
survival for hepatocellular carcinoma but poor survival for biliary
tract cancers remains unchanged. Eur J Surg Oncol. 45:1660–1667.
2019. View Article : Google Scholar
|
|
7
|
Groot Koerkamp B, Wiggers JK, Allen PJ,
Besselink MG, Blumgart LH, Busch OR, Coelen RJ, D'Angelica MI,
DeMatteo RP, Gouma DJ, et al: Recurrence rate and pattern of
perihilar cholangiocarcinoma after curative intent resection. J Am
Coll Surg. 221:1041–1049. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Komaya K, Ebata T, Yokoyama Y, Igami T,
Sugawara G, Mizuno T, Yamaguchi J and Nagino M: Recurrence after
curative-intent resection of perihilar cholangiocarcinoma: Analysis
of a large cohort with a close postoperative follow-up approach.
Surgery. 163:732–738. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cambridge WA, Fairfield C, Powell JJ,
Harrison EM, Søreide K, Wigmore SJ and Guest RV: Meta-analysis and
meta-regression of survival after liver transplantation for
unresectable perihilar cholangiocarcinoma. Ann Surg. 273:240–250.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Spolverato G, Kim Y, Alexandrescu S,
Marques HP, Lamelas J, Aldrighetti L, Clark Gamblin T, Maithel SK,
Pulitano C, Bauer TW, et al: Management and outcomes of patients
with recurrent intrahepatic cholangiocarcinoma following previous
curative-intent surgical resection. Ann Surg Oncol. 23:235–243.
2016. View Article : Google Scholar
|
|
11
|
Forner A, Vidili G, Rengo M, Bujanda L,
Ponz-Sarvisé M and Lamarca A: Clinical presentation, diagnosis and
staging of cholangiocarcinoma. Liver Int. 39 (Suppl 1):S98–S107.
2019. View Article : Google Scholar
|
|
12
|
Banales JM, Marin JJG, Lamarca A,
Rodrigues PM, Khan SA, Roberts LR, Cardinale V, Carpino G, Andersen
JB, Braconi C, et al: Cholangiocarcinoma 2020: The next horizon in
mechanisms and management. Nat Rev Gastroenterol Hepatol.
17:557–588. 2020. View Article : Google Scholar
|
|
13
|
Lollo G, Matha K, Bocchiardo M, Bejaud J,
Marigo I, Virgone-Carlotta A, Dehoux T, Rivière C, Rieu JP,
Briançon S, et al: Drug delivery to tumours using a novel 5-FU
derivative encapsulated into lipid nanocapsules. J Drug Target.
27:634–645. 2019. View Article : Google Scholar
|
|
14
|
Lee JJ, Beumer JH and Chu E: Therapeutic
drug monitoring of 5-fluorouracil. Cancer Chemother Pharmacol.
78:447–464. 2016. View Article : Google Scholar
|
|
15
|
Yang CG, Ciccolini J, Blesius A, Dahan L,
Bagarry-Liegey D, Brunet C, Varoquaux A, Frances N, Marouani H,
Giovanni A, et al: DPD-based adaptive dosing of 5-FU in patients
with head and neck cancer: Impact on treatment efficacy and
toxicity. Cancer Chemother Pharmacol. 67:49–56. 2011. View Article : Google Scholar
|
|
16
|
Bell BM, Kirk ID, Hiltbrunner S,
Gabrielsson S and Bultema JJ: Designer exosomes as next-generation
cancer immunotherapy. Nanomedicine. 12:163–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Watson DC, Bayik D, Srivatsan A,
Bergamaschi C, Valentin A, Niu G, Bear J, Monninger M, Sun M,
Morales-Kastresana A, et al: Efficient production and enhanced
tumor delivery of engineered extracellular vesicles. Biomaterials.
105:195–205. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vojtech L, Woo S, Hughes S, Levy C,
Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R,
Tewari M and Hladik F: Exosomes in human semen carry a distinctive
repertoire of small non-coding RNAs with potential regulatory
functions. Nucleic Acids Res. 42:7290–7304. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhong J, Xia B, Shan S, Zheng A, Zhang S,
Chen J and Liang XJ: High-quality milk exosomes as oral drug
delivery system. Biomaterials. 277:1211262021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dai J, Su Y, Zhong S, Cong L, Liu B, Yang
J, Tao Y, He Z, Chen C and Jiang Y: Exosomes: Key players in cancer
and potential therapeutic strategy. Signal Transduct Target Ther.
5:1452020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liang Y, Duan L, Lu J and Xia J:
Engineering exosomes for targeted drug delivery. Theranostics.
11:3183–3195. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Chen D and Ho EA: Challenges in
the development and establishment of exosome-based drug delivery
systems. J Control Release. 329:894–906. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gatti S, Bruno S, Deregibus MC, Sordi A,
Cantaluppi V, Tetta C and Camussi G: Microvesicles derived from
human adult mesenchymal stem cells protect against
ischaemia-reperfusion-induced acute and chronic kidney injury.
Nephrol Dial Transplant. 26:1474–1483. 2011. View Article : Google Scholar
|
|
24
|
Farooqi AA, Desai NN, Qureshi MZ,
Librelotto DRN, Gasparri ML, Bishayee A, Nabavi SM, Curti V and
Daglia M: Exosome biogenesis, bioactivities and functions as new
delivery systems of natural compounds. Biotechnol Adv. 36:328–334.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jeppesen DK, Fenix AM, Franklin JL,
Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q,
Evans R, et al: Reassessment of exosome composition. Cell.
177:428–445.e18. 2019. View Article : Google Scholar
|
|
26
|
Longley DB, Harkin DP and Johnston PG:
5-fluorouracil: Mechanisms of action and clinical strategies. Nat
Rev Cancer. 3:330–338. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wei H, Chen J, Wang S, Fu F, Zhu X, Wu C,
Liu Z, Zhong G and Lin J: A nanodrug consisting of doxorubicin and
exosome derived from mesenchymal stem cells for osteosarcoma
treatment in vitro. Int J Nanomedicine. 14:8603–8610. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liang G, Zhu Y, Ali DJ, Tian T, Xu H, Si
K, Sun B, Chen B and Xiao Z: Engineered exosomes for targeted
co-delivery of miR-21 inhibitor and chemotherapeutics to reverse
drug resistance in colon cancer. J Nanobiotechnology. 18:102020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim MS, Haney MJ, Zhao Y, Yuan D, Deygen
I, Klyachko NL, Kabanov AV and Batrakova EV: Engineering
macrophage-derived exosomes for targeted paclitaxel delivery to
pulmonary metastases: In vitro and in vivo evaluations.
Nanomedicine. 14:195–204. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lamichhane TN, Jeyaram A, Patel DB,
Parajuli B, Livingston NK, Arumugasaamy N, Schardt JS and Jay SM:
Oncogene knockdown via active loading of small RNAs into
extracellular vesicles by sonication. Cell Mol Bioeng. 9:315–324.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Thakur A, Sidu RK, Zou H, Alam MK, Yang M
and Lee Y: Inhibition of glioma cells' proliferation by
doxorubicin-loaded exosomes via microfluidics. Int J Nanomedicine.
15:8331–8343. 2020. View Article : Google Scholar : PubMed/NCBI
|