Introduction
Neuropathic pain (NP), which is characterized by
spontaneous pain, allodynia and hyperalgesia, is one of the most
intractable diseases (1,2). NP may be caused by the dysfunction or
primary injury of the somatosensory nervous system (3,4). NP
adversely affects the quality of life of the patients and may lead
to depression (5). Furthermore, the
lack of satisfactory treatment and limited understanding of the
underlying molecular mechanisms make NP one of the most serious
health problems. Thus, it is crucial to explore the mechanisms
underlying the occurrence and progression of NP.
Long non-coding RNAs (lncRNAs) are a class of
non-coding RNAs that are >200 nucleotides in length and are
involved in the regulation of several cellular processes (6,7), such
as cell proliferation, apoptosis and differentiation, via
regulating target gene expression (8). The lncRNA small nucleolar RNA host
gene 5 (SNHG5) is a recently identified lncRNA that has yet to be
extensively investigated, but has been found to be correlated with
tumor range, metastasis, pathological stage and prognosis in
various solid tumors (9). Moreover,
SNHG5 has been indicated to play a key role in NP by sponging
microRNA (miRNA/miR)-154-5p (10).
However, the mechanism underlying the function of SNHG5 in the
progression of NP is yet to be elucidated.
miRNAs/miRs are a group of single-stranded,
non-coding RNAs that are 18–22 nucleotides in length (11) and play important roles in a variety
of physiological and pathological processes, including cell
proliferation and differentiation, apoptosis, metabolism and
tumorigenesis (12) via negatively
regulating the expression of their target genes (13). Numerous studies have indicated the
critical role of miRs in the progression of NP (14).
The aim of the present study was to determine
whether the expression of SNHG5 was dysregulated in chronic
constriction injury (CCI) model mice. Functional studies were
performed to evaluate the effect of SNHG5 on CCI-induced NP.
Mechanistically, the interaction of SNHG5 with miR-142-5p and the
downstream gene calcium/calmodulin-dependent protein kinase II α
(CAMK2A) was investigated.
Materials and methods
Animals
A total of 48 female BALB/c mice (weight, 18–20 g;
age, 5–6 weeks) were purchased from Beijing Vital River Laboratory
Animal Technology Co., Ltd., and housed at 23°C with a 12-h
light/dark cycle under 40–50% humidity. The animals had access to
food and water ad libitum. All animal experiments were
approved by the Hubei University of Arts and Science (approval no.
HUAS-A20191128; Xiangyang, China). This study was performed in
accordance with the guidelines (15) for the Care and Use of Laboratory
Animals. To evaluate the CCI model, 12 mice were randomly divided
into the following two groups (n=6 per group): Sham and CCI model
groups. To investigate SNHG5 function, 18 mice were randomly
divided into the following three groups (n=6/group): Sham,
CCI+adenovirus (Ad)-short hairpin (sh)RNA-negative control (NC) and
CCI+Ad-sh-SNHG5 groups. To investigate the function of CAMK2A, 18
mice were randomly divided into the following three groups (n=6 per
group): Sham, CCI+Ad-sh-NC and CCI+Ad-sh-CAMK2A groups.
CCI model establishment
After anesthesia with intraperitoneal injection of
1% sodium pentobarbital (50 mg/kg), an incision was made in the
skin on the lateral surface of the thigh of the mice. The left
common sciatic nerve was exposed at the mid-thigh level and was
tightly ligated with a 5.0 silk suture at four sites along the
nerve with a 1.0-1.5-mm distance between them. The muscle and skin
were then sutured. The same procedures were performed in the mice
of the sham group, but the sciatic nerve was left untied. At the
end of the study, the mice were euthanized with an intraperitoneal
injection of 3% sodium pentobarbital (160 mg/kg). When mice stopped
breathing and the righting reflex disappeared, their death was
verified and the dorsal root ganglia (DRG) were removed for further
investigation.
Construction of adenovirus and
administration
The 3rd generation of adenoviruses used to knock
down SNHG5, CAMK2A and the negative control were prepared by
RayBiotech, Inc. The shRNA and the whole sequence for SNHG5 along
with their negative controls were synthetized. Recombinant
adenoviruses (1×108 pfu, MOI=200:1) were injected using
a microinjection syringe that was connected to the intrathecal
catheter. Injections were performed 4 days before CCI surgery, and
on days 0 and 7 after CCI surgery. The virus infection was
performed 4 days before CCI operation because expression of SNHG5
started to reduce at day 4 after virus injection whereas the
present study was investigating the effects of CCI treatment when
SNHG5 expression was silenced.
Mechanical allodynia test
Mechanical allodynia was detected by measuring the
paw withdrawal threshold (PWT) in response to a series of von Frey
hair stimulations (Stoelting Co.). A series of von Frey filaments,
starting with the filament 0.5 g, were applied to the dorsal
surface of the hind paw with a sufficient force. PWT was defined as
the pressure (g) at which the mouse withdrew its paw. The PWT was
automatically recorded when the paw was withdrawn. Each trial was
repeated six times at ~3-min intervals.
Thermal hyperalgesia test
Thermal hyperalgesia was detected by measuring paw
withdrawal thermal latency (PWTL). Briefly, the plantar surface of
the hind paw was irradiated using an infrared light beam generated
by a modified Hargreaves device (Ugo Basile SRL). Subsequently, the
PWTL (sec) was automatically recorded as soon as the mouse withdrew
its paw.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR) analysis
Total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's instructions, and reverse-transcribed into cDNA
using an EasyScript® First-Strand cDNA Synthesis
SuperMix according to the manufacture's protocols (TransGen Biotech
Co., Ltd.). qPCR was performed using SYBR Premix Ex Taq (Takara
Biotechnology Co., Ltd.) on a Light Cycler® 480 System
(Roche Diagnostics). qPCR was performed under the following
thermocycling conditions: 95°C for 30 sec, 40 cycles of 95°C for 5
sec and 60°C for 34 sec. mRNA GAPDH and U6 were used as the
internal controls for mRNA or miRNA, respectively. The relative
abundance of mRNA was calculated according to the 2−ΔΔCq
method (16). The qPCR primer
sequences used were as follows: SNHG5 forward,
5′-TACTGGCTGCGCACTTCG-3′ and reverse, 5′-TACCCTGCACAAACCCGAAA-3′;
CAMK2A forward, 5′-TGACCTCAACTACATGGTCTACA-3′ and reverse,
5′-CTTCCCATTCTCGGCCTTG-3′; GAPDH forward,
5′-TATCCGCATCACTCAGTACCTG-3′ and reverse,
5′-GAAGTGGACGATCTGCCATTT-3′; miR-142-5p RT primer,
GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACTCATCAC, forward,
5′-TGCGGGTATTTCATCTTTCGT-3′ and reverse, 5′-CCAGTGCAGGGTCCGAGGT-3′;
and U6 forward, 5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse,
5′-ACGCTTCACGAATTTGCGTGTC-3′.
Western blot analysis
The L4-L6 spinal cord segments of each mouse were
removed and treated with lysis buffer (Beyotime Institute of
Biotechnology) containing protease inhibitor and phosphatase
inhibitor cocktail (Sigma-Aldrich; Merck KGaA). BCA assay was used
to detect the amount of protein. Proteins extracted from the spinal
cord (40 µg) were separated via 10% SDS-PAGE followed by transfer
onto PVDF membranes (EMD Millipore) and incubation with 5% non-fat
dry milk in TBS containing 0.3% Tween-20 buffer for 2 h at room
temperature. Subsequently, the blots were incubated with primary
antibodies against CAMK2A (1:500, ab22609, Abcam) and GAPDH
(1:1,000, ab245357, Abcam) at 4°C for 12 h, followed by incubation
with horseradish peroxidase-conjugated secondary antibody (1:1,000;
cat. no. 6927-100, Amyjet) for another 2 h at room temperature.
Finally, the blots were visualized using an enhanced
chemiluminescence kit (Beyotime Institute of Biotechnology). The
bands were semi-quantified using ImageJ software V1.8.0 (National
Institutes of Health).
ELISA
All spinal cord samples were homogenized in RIPA
lysis buffer (Beyotime Institute of Biotechnology) and, after
centrifugation at 16,009.2 × g g at 4°C for 15 min, the
supernatants were collected. Total protein concentration was
evaluated by a bicinchoninic acid assay protein assay kit (Thermo
Fisher Scientific, Inc.). ELISA for tumor necrosis factor (TNF)-α
(cat. no. 900-TM54), interleukin (IL)-6 (900-M50), IL-10 (900-T53)
and IL-1β (900-K47) was performed following the manufacturer's
protocol (PeproTech EC Ltd.).
Bioinformatics analysis
Bioinformatics website StarBase
(starbase.sysu.edu.cn/) was used to predict the potential targets
for SNHG5 and the target gene of miR-142-5p.
Cell transfection
293 cells in the logic growth phase were
transfected with miR-142-5p mimics (5′CATAAAGTAGTTTGCACTACT3′),
mimic control (5′GTCAGTGGTCAAGTCAGTCAT3′), miR-142-5p inhibitor
(5′AGTAGTGCAAACTACTTTATG3′), inhibitor nc
(5′CACGATTGAGATGACCAGCAT′; all Genewiz, Suzhou, China), as well as
pcDNA3.1 vector, pcDNA3.1/SNHG5, sh-nc (Top strand:
CACCGTCCCAGGATTGTCAGCTGACCGAAGTCAGCTGACAATCCTGGGAC, Bottom strand:
AAAAGTCCCAGGATTGTCAGCTGACTTCGGTCAGCTGACAATCCTGGGAC) and sh-SNHG5
(Top strand: CACCGGGTTTGTGCAGGGTACAATGCGAACATTGTACCCTGCACAAACCC,
Bottom strand: AAAAGGGTTTGTGCAGGGTACAATGTTCGCATTGTACCCTGCACAAACCC)
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacture's instructions at
room temperature. Forty-eight hours after transfection, the cells
can be used for the further experiments.
Dual-luciferase reporter assay
The wild-type (Wt) and mutant (Mut) SNHG5 or CAMK2A
were obtained by chemical synthesis and subcloned into the pGL3
vector (Promega Corporation). Then, 293 cells obtained from
American Type Culture Collection were co-transfected with the
plasmid carrying Wt or Mut SNHG5/CAMK2A along with miR-142-5p
mimics (5′CATAAAGTAGTTTGCACTACT3′) or mimic control
(5′GTCAGTGGTCAAGTCAGTCAT3′) (Genewiz) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Subsequently, cells were transfected with 0.1 µg
PRL-TK (TK-driven Renilla luciferase expression vector) as
an internal control. Luciferase activities were measured 48 h after
transfection with a dual-luciferase reporter assay kit (Promega
Corporation).
RNA pull down assay
RNA pull down kit (cat. no. P0202, Geenseed) was
used to perform RNA pull down assay. According to the
manufacturer's instructions, after transfection for 48 h, the 293
cells were treated with cell lysate for 10 min. The cleavage was
incubated with beads precoated with Streptavidin Magnetic Beads at
4°C for 3 h. Then, the bound RNA was purified using Trizol regent
(Boyetime) according to the protocols. The expression of LINC00917
or NLRP1 levels were measured by RT-qPCR as mentioned above.
Statistical analysis
All experiments were performed independently in
triplicate. Data were analyzed using SPSS 17.0 (SPSS Inc.) and are
presented as the mean ± standard deviation. Shapiro-Wilk test was
used to detect normal distribution. Differences between two groups
were determined using a unpaired Student's t-test or one-way ANOVA
followed by Tukey's post hoc test. Pearson's correlation analysis
was performed to analyze the correlation between miR-142-5p and
SNHG5, miR-142-5p and CAMK2A and between SNHG5 and CAMK2A.
P<0.05 was considered to indicate a statistically significant
difference.
Results
SNHG5 is significantly upregulated in
CCI mice
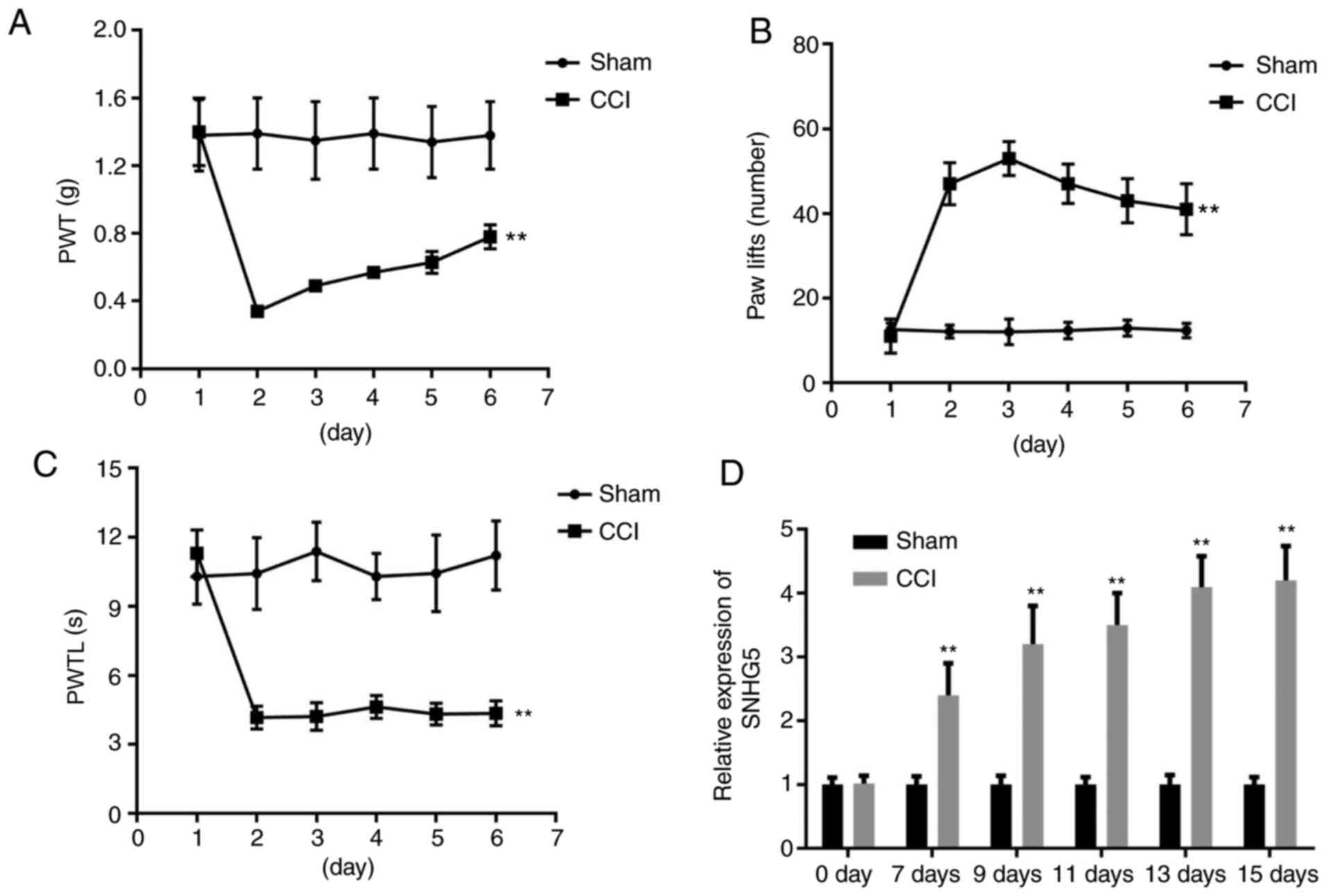
As shown in Fig.
1A-C, CCI decreased the PWT and the PWTL, whereas an increased
number of paw lifts was observed after CCI surgery compared with
the sham group, confirming that the CCI model was successfully
established. Subsequently, the expression of SNHG5 was evaluated
using RT-qPCR analysis. The results revealed that the expression of
SNHG5 was significantly elevated in the DRG of the mice 7 days
after CCI surgery, and reached the highest level at 15 days
post-CCI surgery (Fig. 1D).
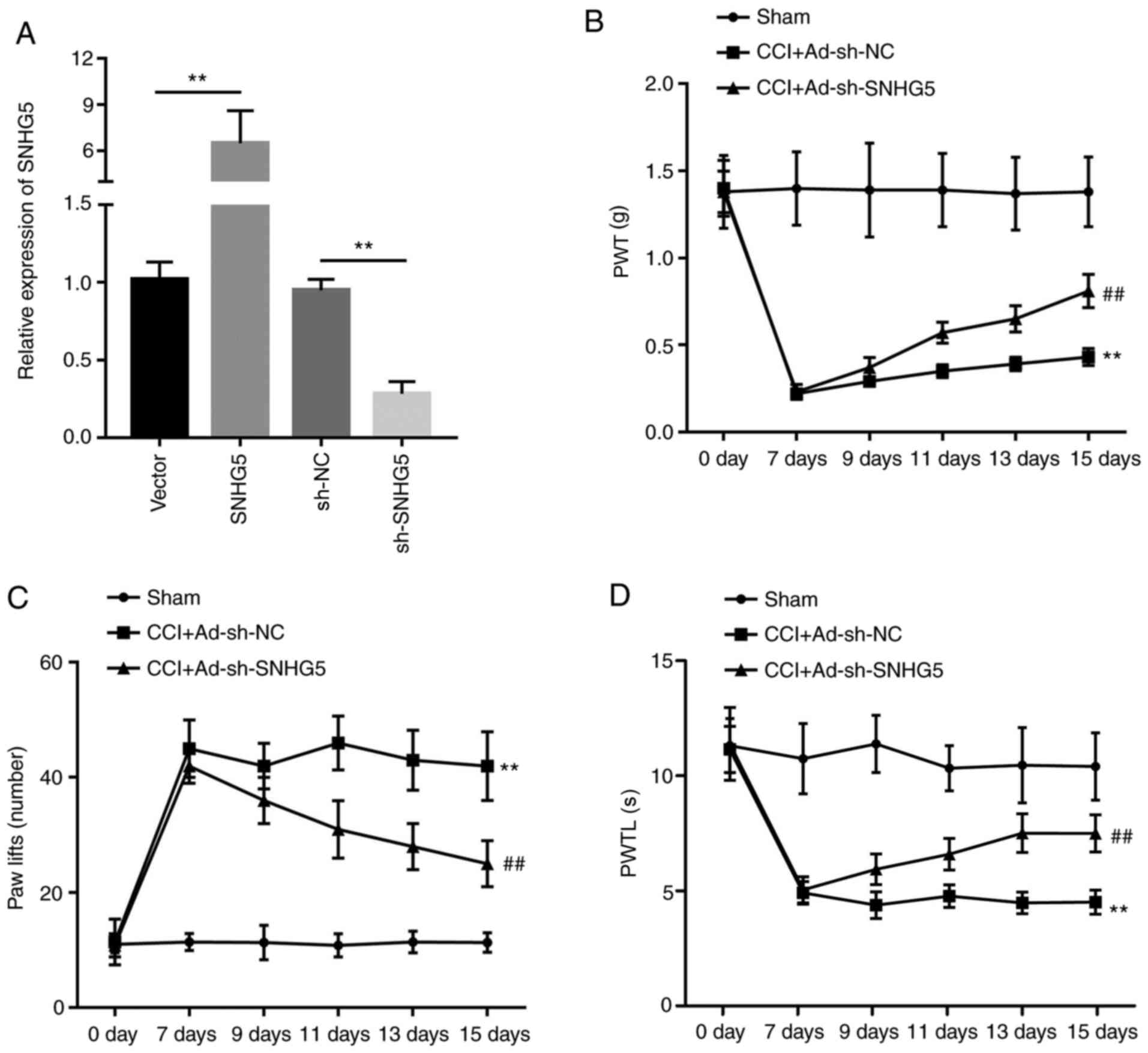
SNHG5 knockdown inhibits NP
As SNHG5 was found to be upregulated in mice with
CCI-induced NP, whether SNHG5 plays a role in the regulation of NP
was next investigated. Adenovirus vector was constructed to
overexpress or silence the expression of SNHG5. RT-qPCR results
indicated that the expression of SNHG5 was significantly
upregulated in the SNHG5 overexpression group compared with the
empty vector group, whereas SNHG5 expression was significantly
inhibited in the sh-SNHG5 group compared with the sh-NC group
(Fig. 2A). The mechanical allodynia
test and thermal hyperalgesia test indicated that in the
CCI+Ad-sh-SNHG5 group, PWT and the PWTL were increased, while the
number of paw lifts was reduced compared with the CCI+Ad-sh-NC
group (Fig. 2B-D). These results
confirmed the critical role of SNHG5 in the development of NP.
SNHG5 knockdown inhibits the release
and mRNA expression of IL-1β, IL-6 and TNF-α, but increases that of
IL-10 induced by CCI
The levels of IL-1β, IL-6, IL-10 and TNF-α in DRG
tissues and their mRNA expression were evaluated using ELISA and
RT-qPCR analysis, respectively. The results revealed that the
release and mRNA expression levels of IL-1β, IL-6 and TNF-α were
significantly increased in the CCI+Ad-sh-NC group, while IL-10 was
reduced in spinal cord tissue, compared with the sham group.
Furthermore, in the CCI+Ad-sh-SNHG5 group, these increases in
IL-1β, IL-6 and TNF-α expression levels were reversed, while the
release and mRNA expression of IL-10 was significantly increased,
compared with the CCI+Ad-sh-NC group (Fig. 3A and B).
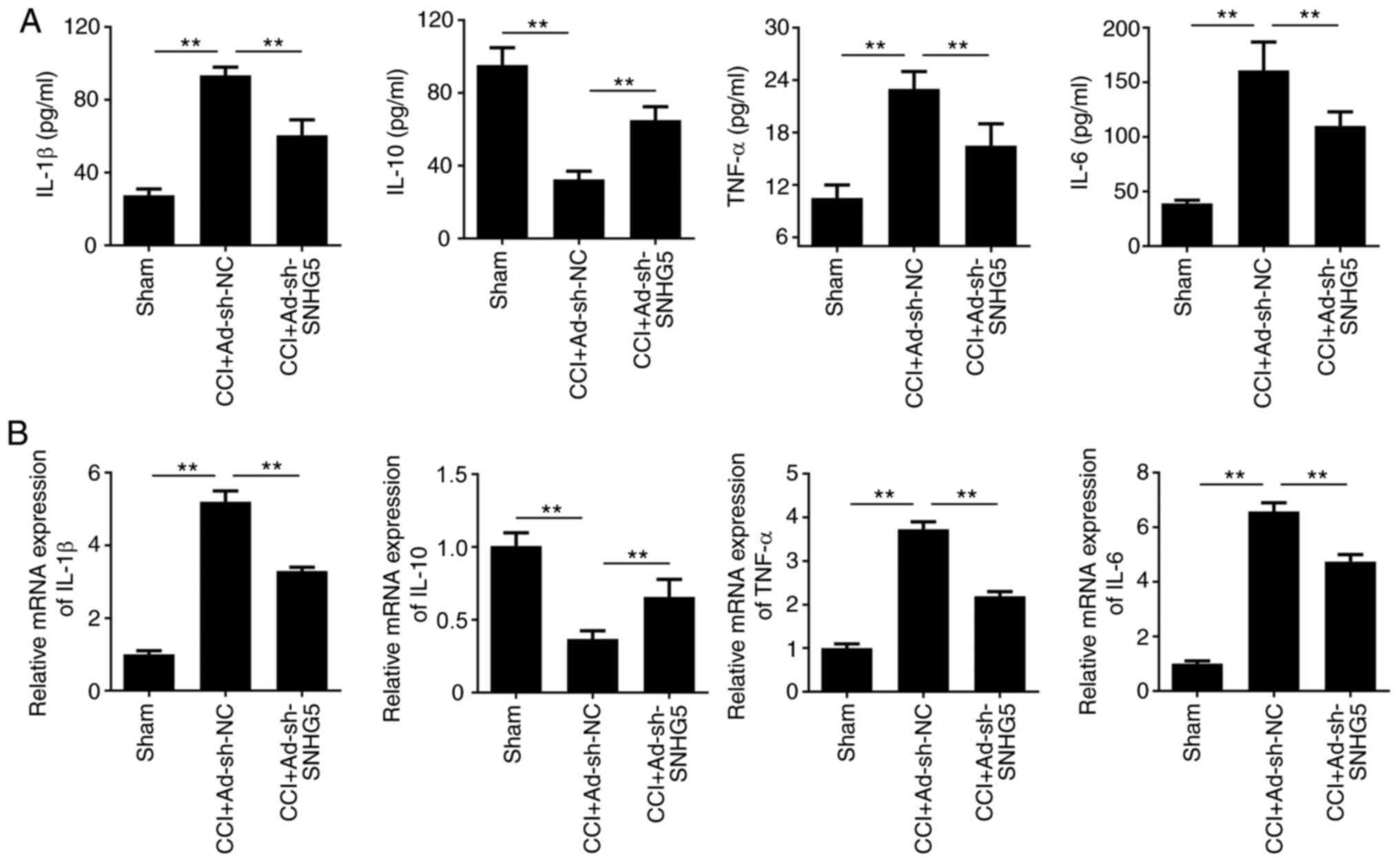 | Figure 3.SNHG5 knockdown inhibits the release
and mRNA expression of IL-1β, IL-6, IL-10 and TNF-α. (A) ELISA was
used to evaluate the release of the inflammatory factors IL-1β,
IL-6, IL-10 and TNF-α in the spinal cord (n=6). (B) Reverse
transcription-quantitative PCR analysis was performed to evaluate
the mRNA expression of IL-1β, IL-6, IL-10 and TNF-α in the spinal
cord (n=6). **P<0.01. SNHG5, small nucleolar RNA host gene 5;
IL-, interleukin; TNF, tumor necrosis factor; sh-, short hairpin
RNA; NC, negative control; Ad-, adenovirus; CCI, chronic
constriction injury. |
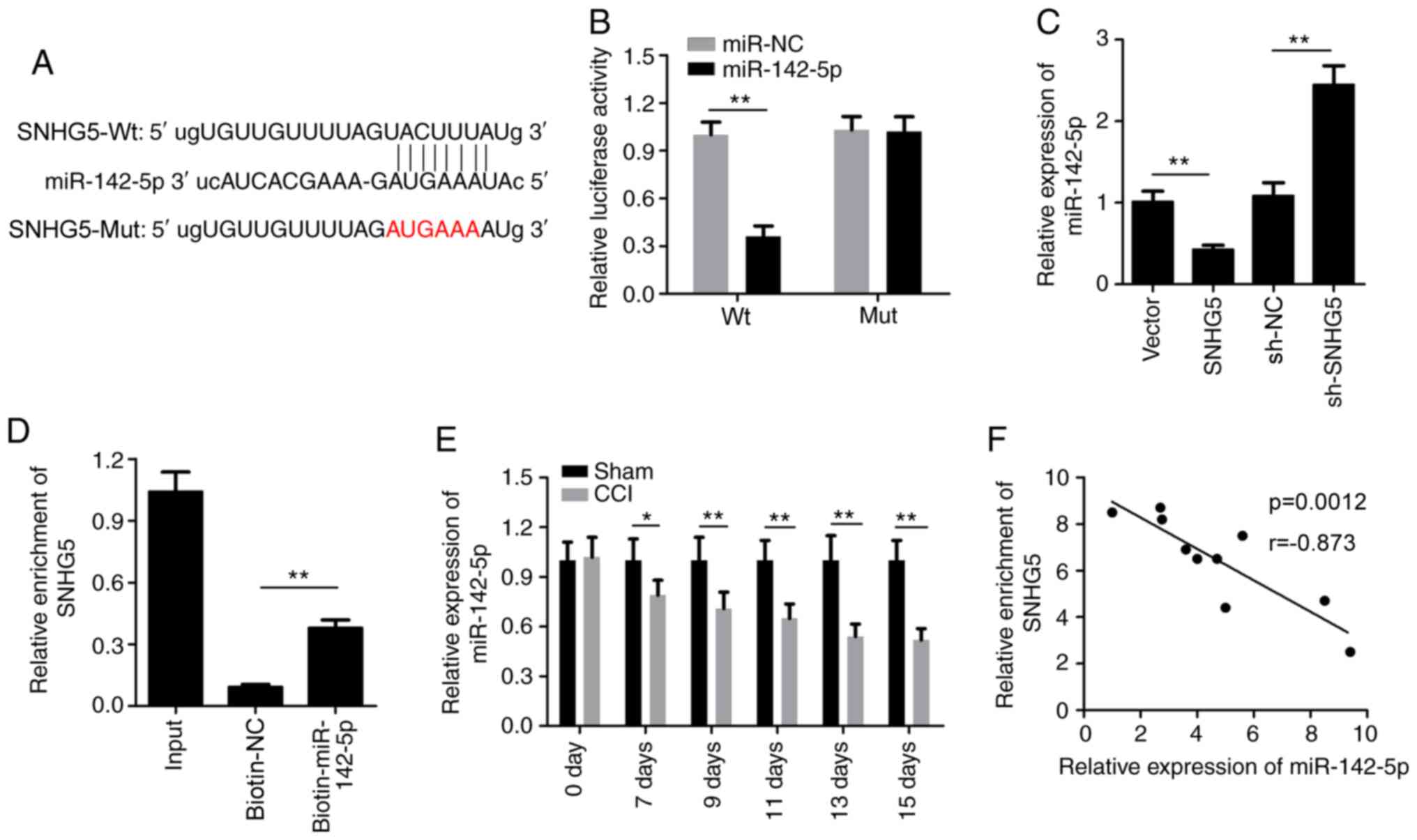
SHNG5 sponges miR-142-5p
The bioinformatics website StarBase (http://starbase.sysu.edu.cn/) was used to predict the
potential targets for SNHG5. As shown in Fig. 4A, binding sites for miR-142-5p were
found within SNHG5. Subsequently, the dual-luciferase reporter
assay confirmed that co-transfection of HEK-293 cells with Wt SMHG5
and miR-142-5p mimics resulted in significantly decreased
luciferase activity compared with the miR-NC group (Fig. 4B). In addition, the relative
expression of miR-142-5p was significantly decreased in the SNHG5
overexpression group and increased in the sh-SNHG5 group (Fig. 4C). Then, the RNA pull-down assay
verified that biotinylated-miR-142-5p, but not biotinylated-NC,
could enrich SNHG5 (Fig. 4D). The
results of the RT-qPCR analysis indicated that miR-142-5p
expression was significantly downregulated after CCI (Fig. 4E). The Pearson's correlation
analysis demonstrated that there was a negative correlation between
miR-142-5p and SNHG5 (Fig. 4F).
miR-142-5p targets CAMK2A
It is well known that miRs bind to their target
genes to inhibit their expression. Thus, the present study
attempted to identify the target gene of miR-142-5p in order to
elucidate its underlying mechanism of action. The bioinformatics
website StarBase was used to predict the potential targets of
miR-142-5p. As shown in Fig. 5A,
binding sites for miR-142-5p were found within CAMK2A.
Subsequently, the dual-luciferase reporter assay confirmed that
co-transfection of HEK-293 cells with Wt SAMK2A and the miR-142-5p
mimics resulted in significantly decreased luciferase activity
compared with the miR-NC group (Fig.
5B). miR-142-5p mimic significantly increased the expression of
miR-142-5p, while miR-142-5p inhibitor reduced the expression of
miR-142-5p (Fig. 5C). In addition,
the relative expression of CAMK2A was significantly decreased in
the miR-142-5p mimic group, and it increased in the miR-142-5p
inhibitor group (Fig. 5D and E).
Then, the RNA pull-down assay verified that
biotinylated-miR-142-5p, but not biotinylated-NC, could enrich
CAMK2A (Fig. 5F). The results of
the RT-qPCR analysis indicated that CAMK2A was significantly
upregulated after CCI (Fig. 5G).
The Pearson's correlation analysis demonstrated that there was a
strong negative correlation between miR-142-5p and CAMK2A (Fig. 5H) and a strong positive correlation
between CAMK2A and SNHG5 (Fig.
5I).
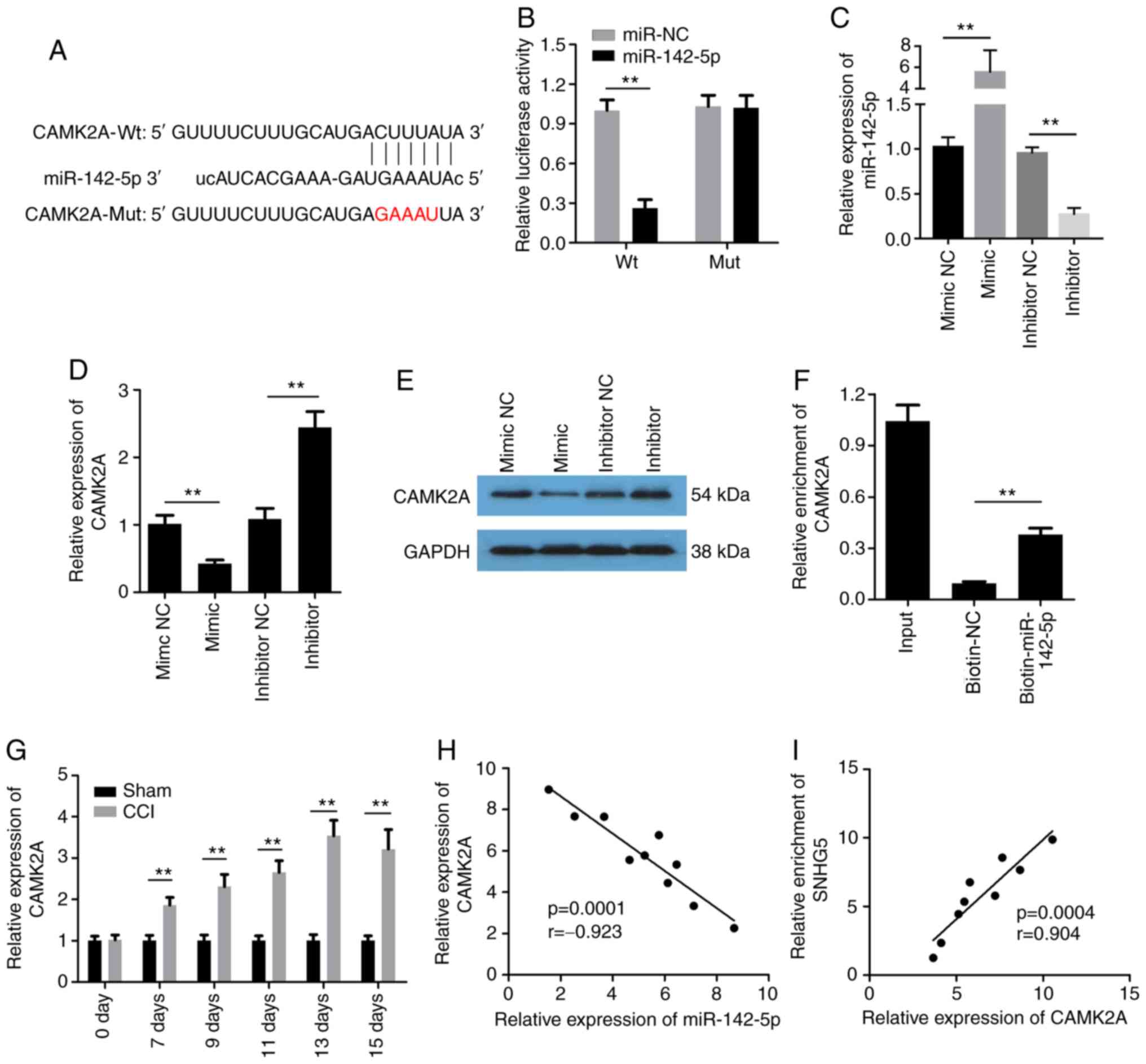 | Figure 5.miR-142-5p directly targets CAMK2A.
(A) Bioinformatics analysis predicted the binding cite between
miR-142-5p and CAMK2A. (B) Dual-luciferase assay was used to detect
the interaction between miR-142-5p and CAMK2A (n=6). (C-E) RT-qPCR
analysis and western blotting were performed to evaluate the
expression of miR-142 or CAMK2A (n=6). (F) RNA pull-down assay was
used to detect the direct interaction between miR-142-5p and CAMK2A
(n=3). (G) RT-qPCR analysis was performed to detect the expression
of CAMK2A (n=6). (H and I) Pearson's correlation analysis was
conducted to assess the correlation between miR-142-5p and CAMK2A,
as well as between SNHG5 and CAMK2A. **P<0.01. CAMK2A,
calcium/calmodulin-dependent protein kinase II α; miR, microRNA;
SNHG5, small nucleolar RNA host gene 5; RT-qPCR, reverse
transcription-quantitative PCR; Wt, wild-type; Mut, mutant; NC,
negative control; sh-, short hairpin RNA; CCI, chronic constriction
injury. |
CAMK2A knockdown inhibits NP and the
inflammatory response induced by CCI
To further elucidate the role of CAMK2A, adenovirus
for silencing CAMK2A was established. RT-qPCR and western blot
analysis indicated that transfection with sh-CAMK2A significantly
reduced the expression of CAMK2A (Fig.
6A and B). The mechanical allodynia and thermal hyperalgesia
tests indicated that PWT and PWTL were increased in the
CCI+Ad-sh-CAMK2A group, while the number of paw lifts was reduced,
compared with the CCI+Ad-sh-NC group (Fig. 6C). Furthermore, the levels of IL-1β,
IL-6, IL-10 and TNF-α in DRG tissues and their mRNA expression were
evaluated using ELISA and RT-qPCR analysis, respectively. The
results revealed that in the CCI+Ad-sh-NC group, the release and
mRNA expression levels of IL-1β, IL-6 and TNF-α were significantly
increased, while IL-10 was reduced in spinal cord tissue, compared
with in the sham group. Furthermore, in the CCI+Ad-sh-CAMK2A group,
this increase in IL-1β, IL-6 and TNF-α was reversed, while the
inhibition of the release and mRNA expression of IL-10 was markedly
reversed, compared with the CCI+Ad-sh-NC group (Fig. 6D and E).
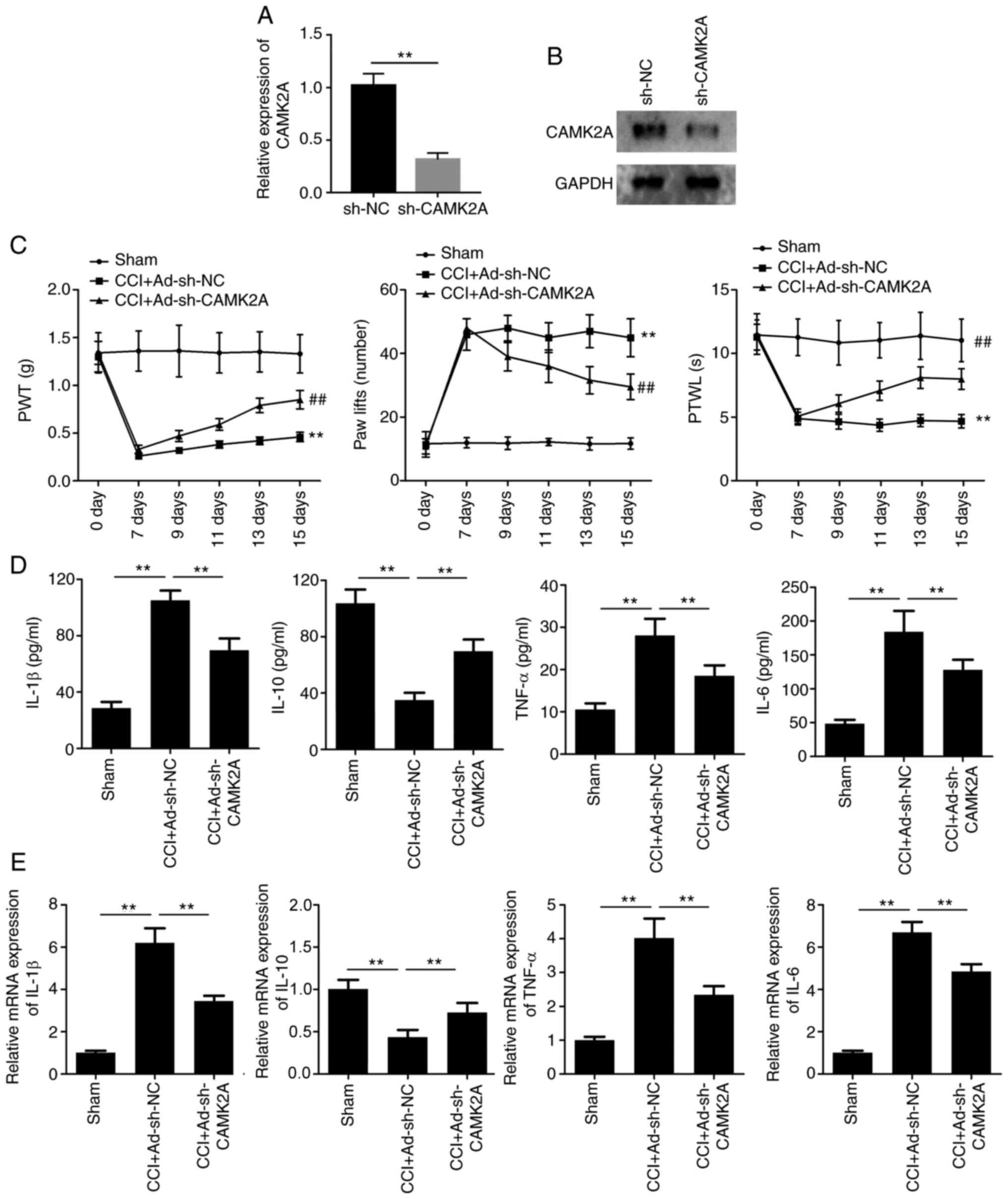 | Figure 6.CAMK2A knockdown inhibits neuropathic
pain and inflammatory response induced by CCI. (A) RT-qPCR and (B)
western blotting was performed to detect the expression of CAMK2A.
**P<0.01 vs. sh-NC. (C) PWT, number of paw lifts and PWTL were
detected to evaluate mechanical allodynia, thermal allodynia and
thermal hyperalgesia (n=6). (D) ELISA was used to evaluate the
levels of inflammatory factors, including IL-1β, IL-6, IL-10 and
TNF-α, in the spinal cord (n=6). (E) RT-qPCR analysis was performed
to evaluate the mRNA expression of IL-1β, IL-6, IL-10 and TNF-α in
the spinal cord (n=6). **P<0.01 vs. sham; ##P<0.01
vs. CCI+Ad-sh-NC. CCI, chronic constriction injury; CAMK2A,
calcium/calmodulin dependent protein kinase II α; RT-qPCR, reverse
transcription-quantitative PCR; NC, negative control; sh-, short
hairpin RNA; PWT, paw withdrawal threshold; PWTL, paw withdrawal
threshold latency; IL-, interleukin; TNF, tumor necrosis factor;
Ad-, adenovirus. |
Discussion
The aim of the present study, was to explore the
anti-nociceptive effect of the lncRNA SNHG5 on CCI-induced
inflammatory response and NP. The results revealed that SNHG5
knockdown markedly inhibited mechanical and thermal hyperalgesia.
Further investigation was undertaken to determine the effects of
SNHG5 on the release of inflammatory factors, such as IL-1β, IL-6,
TNF-α and IL-10. The results of the present study were consistent
with previously reported findings (9). The majority of the studies of SNHG5
are in the field of human cancer, including gastric, breast, liver
and esophageal cancer (17–21). In addition, SNHG5 has been reported
to regulate cell apoptosis and the inflammatory response in chronic
obstructive pulmonary disease via sponging miR-132 (22).
Mechanistically, the results of the current study
indicated that SNHG5 can sponge miR-142-5p to upregulate the
expression CAMK2A. SNHG5 has been demonstrated to be capable of
sponging various miRs, such as miR-32, miR-212-3p, miR-26-5p,
miR-154-5p, miR-23a and miR-26a (17,18,23–26).
To the best of our knowledge, the present study was the first to
reveal that SNHG5 interacts with miR-142-5p.
miR-142-5p has been investigated in the context of
NP. It was reported that miR-142-3p could inhibit NP development
via inhibiting HMGB1 expression (27). Adeno-associated virus vector
overexpressing miR-142-5p was able to inhibit NP in rats through
binding to the 3′-untranslated region of the mRNA and inhibiting
its expression (28). In the
present study, it was also observed that miR-142-5p was
downregulated in CCI rats, which was consistent with these previous
findings. Although, the direct effect of miR-142-5p on CCI was not
evaluated, which is a limitation of the present study. As it is
known that miRs bind to their target mRNAs to inhibit their
expression, we attempted to identify the target gene of miR-142-5p.
Finally, CAMK2A was predicted and verified to be the target of
miR-142-5p.
CAMK2A is a serine/threonine protein kinase, which
can be found in sensory axons and is involved in neuroplasticity
(29–32). It was previously demonstrated that
CAMK2A silencing can inhibit complete Freund's adjuvant-induced NP
(33). Ryanodine receptors, which
is capable of producing a focal high concentration of calcium and
is found in close proximity to CAMK2A, activates CAMK2A to produce
hyperalgesic priming (34).
Furthermore, CAMK2A knockdown was also found to suppress NP induced
by spinal nerve ligation (35). In
addition, it was reported that CAMK2A plays a key role during the
transition from acute to chronic pain (35). These findings indicated that CAMK2A
may be intricately involved in the regulation of NP. Of note, a
selective inhibitor of CAMK2A, KN93, has been developed, and it can
reduce both mechanical and thermal hypersensitivity (36). The present study further confirmed
that CAMK2A was upregulated in CCI-induced NP and that silencing of
CAMK2A could alleviate the pain induced by CCI.
The downstream genes regulated by CAMK2A were not
investigated in the present study. CAMK2A was previously reported
to regulate the function of the cytoplasmic polyadenylation
element-binding protein by phosphorylating the regulatory site
threonine 171 (37). Activated
CAMK2A can phosphorylate the N-methyl-D-aspartate receptor and
promote its activation (38). The
downstream gene(s) modulated by CAMK2A will be investigated in
future studies.
In the present study, we did not investigate the
direct effect of miR-142-5p to support the conclusions, and more
studies are required to fully elucidate the interaction between
miR-142-5p and CAMK2A. Rescue experiments should be performed to
further confirm the interaction between SNHG5 and miR-142-5p, as
well as between miR-142-5p and CAMK2A. These limitations will be
studied in our further research.
Coronavirus disease 2019 has affected tens of
millions of individuals globally. At present, the mRNA vaccine
BNT162b2 has been used for the prevention of this epidemic
(39). In the future, more
therapeutic methods based on RNA will likely be utilized. With
further investigation of the effects and underlying mechanisms of
the SNHG5/miR-142-5p/CAMK2A signaling pathway, these molecules
could be used as a therapeutic or diagnostic target for NP.
However, further studies are required before any issues with
stability, efficacy and side effects are solved.
In conclusion, the present study demonstrated that
the effects of the lncRNA SNHG5 on CCI-induced NP are mediated
through regulation of the miR-142-5p/CAMK2A signaling pathway.
These findings may improve our understanding of the mechanism
underlying the function of SNHG5 in NP.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
HD and ZY designed this study. SJ, ST and ML
performed all the experiments. ML was responsible for data
analysis. SJ and ST drafted the manuscript. SJ, ST, HD and ZY
revised the paper. SJ, ST and ZY confirm the authenticity of all
the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Hubei University of
Arts and Science (Xiangyang, China) and were performed in
accordance with the Guidelines for the Care and Use of Laboratory
Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CCI
|
chronic constriction injury
|
|
lncRNA
|
long non-coding RNA
|
|
IL
|
interleukin
|
|
miRs
|
microRNAs
|
|
NP
|
neuropathic pain
|
|
PWT
|
paw withdrawal threshold
|
|
PWTL
|
paw withdrawal threshold latency
|
|
SD
|
standard deviation
|
|
SNHG5
|
small nucleolar RNA host gene 5
|
|
TNF
|
tumor necrosis factor
|
References
|
1
|
Costigan M, Scholz J and Woolf CJ:
Neuropathic pain: A maladaptive response of the nervous system to
damage. Annu Rev Neurosci. 32:1–32. 2009. View Article : Google Scholar
|
|
2
|
Calvo M, Davies AJ, Hébert HL, Weir GA,
Chesler EJ, Finnerup NB, Levitt RC, Smith BH, Neely GG, Costigan M
and Bennett DL: The genetics of neuropathic pain from model
organisms to clinical application. Neuron. 104:637–653. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Campbell JN and Meyer RA: Mechanisms of
neuropathic pain. Neuron. 52:77–92. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tramullas M, Francés R, de la Fuente R,
Velategui S, Carcelén M, García R, Llorca J and Hurlé MA:
MicroRNA-30c-5p modulates neuropathic pain in rodents. Sci Transl
Med. 10:eaao62992018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Descalzi G, Mitsi V, Purushothaman I,
Gaspari S, Avrampou K, Loh YHE, Shen L and Zachariou V: Neuropathic
pain promotes adaptive changes in gene expression in brain networks
involved in stress and depression. Sci Signal. 471:eaaj15492017.
View Article : Google Scholar
|
|
6
|
Yao RW, Wang Y and Chen LL: Cellular
functions of long noncoding RNAs. Net Cell Biol. 21:542–551. 2019.
View Article : Google Scholar
|
|
7
|
Boon RA, Jaé N, Holdt L and Dimmeler S:
Long noncoding RNAs: From clinical genetics to therapeutic targets?
J Am Coll Cardiol. 67:1214–1226. 2016. View Article : Google Scholar
|
|
8
|
Long Y, Wang X, Youmans DT and Cech TR:
How do lncRNAs regulate transcription? Sci Adv. 3:eaao21102017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Han W, Shi J, Cao J, Dong B and Guan W:
Latest advances of long non-coding RNA SNHG5 in human cancers. Onco
Targets Ther. 13:6393–6403. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen M, Yang Y, Zhang W, Li X, Wu J, Zou X
and Zeng X: Long noncoding RNA SNHG5 knockdown alleviates
neuropathic pain by targeting the miR-154-5p/CXCL13 axis. Neurochem
Res. 45:1566–1575. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shang Q, Yang Z, Jia R and Ge S: The novel
roles of circRNAs in human cancer. Mol Cancer. 18:62019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang T, Wan X, Alvarez AA, James CD, Song
X, Yang Y, Sastry N, Nakano I, Sulman EP, Hu B and Cheng SY: MIR93
(microRNA-93) regulates tumorigenicity and therapy response of
glioblastoma by targeting autophagy. Autophagy. 15:1100–1111. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bhaskaran V, Yao Y, Bei F and Peruzzi P:
Engineering, delivery, and biological validation of artificial
microRNA clusters for gene therapy applications. Nat Protoc.
14:3538–3553. 2019. View Article : Google Scholar
|
|
14
|
Yang Y, Ding L, Hu Q, Xia J, Sun J, Wang
X, Xiong H, Gurbani D, Li L, Liu Y and Liu A: MicroRNA-218
functions as a tumor suppressor in lung cancer by targeting
IL-6/STAT3 and negatively correlates with poor prognosis. Mol
Cancer. 16:1412017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun D, Yue B, Sun R, Wang T, Pang W, Kong
Q, Zhu D, Li N and Qin C: Laboratory animal-Guideline for ethical
review of animal welfare (GBT35892-2018), China. 2018. https://www.chinesestandard.net/PDF.aspx/GBT35892-2018February
6–2018
|
|
16
|
Zhao L, Han T, Li Y, Sun J, Zhang S, Liu
Y, Shan B, Zheng D and Shi J: The lncRNA SNHG5/miR-32 axis
regulates gastric cancer cell proliferation and migration by
targeting KLF4. Faseb J. 31:893–903. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chi JR, Yu ZH, Liu BW, Zhang D, Ge J, Yu Y
and Cao XC: SNHG5 promotes breast cancer proliferation by sponging
the miR-154-5p/PCNA axis. Mol Ther Nucleic Acids. 17:138–149. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Y, Guo D, Zhao Y, Ren M, Lu G, Wang Y,
Zhang J, Mi C, He S and Lu X: Long non-coding RNA SNHG5 promotes
human hepatocellular carcinoma progression by regulating
miR-26a-5p/GSK3β signal pathway. Cell Death Dis. 9:8882018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang QY, Peng L, Chen Y, Liao LD, Chen JX,
Li M, Li YY, Qian FC, Zhang YX, Wang F, et al: Characterization of
super-enhancer-associated functional lncRNAs acting as ceRNAs in
ESCC. Mol Oncol. 14:2203–2230. 2020. View Article : Google Scholar
|
|
21
|
Zhao L, Guo H, Zhou B, Feng J, Li Y, Han
T, Liu L, Li L, Zhang S, Liu Y, et al: Long non-coding RNA SNHG5
suppresses gastric cancer progression by trapping MTA2 in the
cytosol. Oncogene. 35:5770–5780. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shen Q, Zheng J, Wang X, Hu W and Jiang Y
and Jiang Y: lncRNA SNHG5 regulates cell apoptosis and inflammation
by miR-132/PTEN axis in COPD. Biomed Pharmacother. 126:1100162020.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ju C, Zhou R, Sun J, Zhang F, Tang X, Chen
KK, Zhao J, Lan X, Lin S, Zhang Z and Lv XB: lncRNA SNHG5 promotes
the progression of osteosarcoma by sponging the miR-212-3p/SGK3
axis. Cancer Cell Int. 18:1412018. View Article : Google Scholar
|
|
24
|
Gao J, Zeng K, Liu Y, Gao L and Liu L:
lncRNA SNHG5 promotes growth and invasion in melanoma by regulating
the miR-26a-5p/TRPC3 pathway. Onco Targets Ther. 12:169–179. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin H, Shen L, Lin Q, Dong C, Maswela B,
Illahi GS and Wu X: SNHG5 enhances Paclitaxel sensitivity of
ovarian cancer cells through sponging miR-23a. Biomed Pharmacother.
123:1097112020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Z, Wang Z, Liu J and Yang H: Long
non-coding RNA SNHG5 sponges miR-26a to promote the tumorigenesis
of osteosarcoma by targeting ROCK1. Biomed Pharmacother.
107:598–605. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Mou J, Cao L, Zhen S, Huang H and
Bao H: MicroRNA-142-3p relieves neuropathic pain by targeting high
mobility group box 1. Int J Mol Med. 41:501–510. 2018.PubMed/NCBI
|
|
28
|
Xu H, Yue C and Chen L:
Post-transcriptional regulation of soluble guanylate cyclase that
governs neuropathic pain in Alzheimer's disease. J Alzheimers Dis.
71:1331–1338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Giese KP, Fedorov NB, Filipkowski RK and
Silva AJ: Autophosphorylation at Thr286 of the alpha
calcium-calmodulin kinase II in LTP and learning. Science.
279:870–873. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gleason MR, Higashijima S, Dallman J, Liu
K, Mandel G and Fetcho JR: Translocation of CaM kinase II to
synaptic sites in vivo. Nat Neurosci. 6:217–218. 2003. View Article : Google Scholar
|
|
31
|
Geddis MS and Rehder V: The
phosphorylation state of neuronal processes determines growth cone
formation after neuronal injury. J Neurosci Res. 74:210–220. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
VanBerkum MF and Goodman CS: Targeted
disruption of Ca(2+)-calmodulin signaling in Drosophila growth
cones leads to stalls in axon extension and errors in axon
guidance. Neuron. 14:43–56. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Luo F, Yang C, Chen Y, Shukla P, Tang L,
Wang LX and Wang ZJ: Reversal of chronic inflammatory pain by acute
inhibition of Ca2+/calmodulin-dependent protein kinase II. J
Pharmacol Exp Ther. 325:267–275. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen Y, Luo F, Yang C, Kirkmire CM and
Wang ZJ: Acute inhibition of Ca2+/calmodulin-dependent protein
kinase II reverses experimental neuropathic pain in mice. J
Pharmacol Exp Ther. 330:650–659. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ferrari LF, Bogen O and Levine JD: Role of
nociceptor αCaMKII in transition from acute to chronic pain
(hyperalgesic priming) in male and female rats. J Neurosci.
33:11002–11011. 2013. View Article : Google Scholar
|
|
36
|
He Y, Chen Y, Tian X, Yang C, Lu J, Xiao
C, DeSimone J, Wilkie DJ, Molokie RE and Wang ZJ: CaMKIIα underlies
spontaneous and evoked pain behaviors in Berkeley sickle cell
transgenic mice. Pain. 157:2798–2806. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Atkins CM, Nozaki N, Shigeri Y and
Soderling TR: Cytoplasmic polyadenylation element binding
protein-dependent protein synthesis is regulated by
calcium/calmodulin-dependent protein kinase II. J Neurosci.
24:5193–5201. 2004. View Article : Google Scholar
|
|
38
|
Kitamura Y, Miyazaki A, Yamanaka Y and
Nomura Y: Stimulatory effects of protein kinase C and calmodulin
kinase II on N-methyl-D-aspartate receptor/channels in the
postsynaptic density of rat brain. J Neurochem. 61:100–109. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tartof SY, Slezak JM, Fischer H, Hong V,
Ackerson BK, Ranasinghe ON, Frankland TB, Ogun OA, Zamparo JM, Gray
S, et al: Effectiveness of mRNA BNT162b2 COVID-19 vaccine up to 6
months in a large integrated health system in the USA: A
retrospective cohort study. Lancet. 398:1407–1416. 2021. View Article : Google Scholar
|