Introduction
Airway fibrosis (AF) is a common, chronic and
progressive disease with an unclear etiology (1). AF can be caused by several
pathogenic factors and is characterized by repetitive airway
epithelial cell injury, fibroblast activation and increased
extracellular matrix deposition, resulting in airway destruction
(2). Despite the development of
various detection methods and clinical treatments, the morbidity
and mortality of AF remain high and AF remains a serious threat to
public health (3). Therefore, the
need to determine the underlying mechanism of AF is urgent.
Several studies have suggested that one possible
pathogenesis mechanism of AF involves epithelial-mesenchymal
transition (EMT), which is defined by the loss of epithelial
features and the gain of mesenchymal features (4,5);
therefore, EMT serves an important role in fibrosis pathology
(6). Increasing evidence
indicates that TGF-β1 is a master switch for the induction of
fibrosis in various organs (7).
TGF-β1 can upregulate profibrotic gene expression, such as
N-cadherin, vimentin and α-smooth muscle actin (α-SMA); and
simultaneously downregulate the expression of epithelial
cell-related genes, such as E-cadherin, promoting AF (8).
MicroRNAs (miRNAs or miRs) are a class of small,
noncoding RNAs that inhibit gene expression by binding target gene
3′-untranslated regions (3′-UTRs) (9). Increasing evidence shows that miRNAs
participate in various biological processes, including cell
proliferation, differentiation and apoptosis (10). Additionally, accumulated evidence
indicates that miRNAs are vital to the EMT process in AF (11). For example, Li et al
(12) found that ‘miR-184 targets
TP63 to block idiopathic pulmonary fibrosis by inhibiting
proliferation and epithelial-mesenchymal transition of airway
epithelial cells’. Moreover, the miRNA miR-423-5p is known to be
involved in the regulation of the EMT process in various diseases
(13). Nevertheless, to the best
of the authors' knowledge, the possible role of miR-423-5p in AF
development has not been studied.
The aim of the present study was to detect
miR-423-5p expression in normal airway tissue and AF tissue. In
addition, the role of miR-423-5p in TGF-β1-induced EMT was
determined in the BEAS-2B cell line. Finally, the underlying
mechanism by which miR-423-5p affects TGF-β1-induced EMT was
examined.
Materials and methods
Ethics statement
The First Affiliated Hospital of Chongqing Medical
University Ethics Committee approved the present study
(institutional approval number. 2020-147) and the research was
performed in accordance with the Declaration of Helsinki; all
subjects provided written informed consent.
Tissues, cells and cell culture
A total of 10 AF tissue samples were collected from
patients undergoing fiberoptic bronchoscopy biopsy (7 females and 3
males). The scar tissue samples were collected from patients
undergoing fiberoptic bronchoscopy biopsy via an electrocautery
needle knife (VIO 300S; Erbe Elektromedizin GmbH). A total of 10
control airway tissue samples (from 4 females and 6 males) were
obtained from normal areas of the central airway removed during
nasopharyngeal cancer resection. The 64-slice helical
high-resolution computed tomography (HRCT) system (Siemens AG) was
used to obtain HRCT chest images and two radiologists blinded on
clinical data interpreting all HRCT findings by consensus. Tracheal
stenosis and bronchomalacia were diagnosed based on lesions
detection by a bronchoscope (CV-290; Olympus Corporation). Detailed
information on the control subjects and AF patients who were
diagnosed with AF by pathology examination is provided in Table I. The inclusion criteria for
control subjects were as follows: nasopharyngeal cancer resection
and the absence of other airway diseases. From the included
subjects, normal areas of the central airway were collected. All
tissue samples were collected between June 2020 and December 2020
at The First Affiliated Hospital of Chongqing Medical University.
Human airway epithelial cells (BEAS-2B) were purchased from the
Cell Bank of the Chinese Academy of Sciences. The cells were
maintained in DMEM/F12 medium (Gibco; Thermo Fisher Scientific,
Inc.) containing 10% FBS (PAN-Biotech GmbH), 100 U/ml penicillin
and 100 µg/ml streptomycin at 37°C in a humidified 5%
CO2 atmosphere. BEAS-2B cells were treated with 10 ng/ml
TGF-β1 (PeproTech, Inc.) at 37°C for 24 h to establish the EMT cell
model as previously described (8).
 | Table I.Clinical features of patients and
controls. |
Table I.
Clinical features of patients and
controls.
| Number | Type | Age (years) | Sex | Smoking | Cough | Dyspnoea | Wheezing | Atelectasis (chest
HRCT features) | Tracheal stenosis
(Broncho scopic features) | Bronchomalacia
(Bronchoscopic features) | Diagnose |
|---|
| 1 | AF | 38 |
Female | No | Yes | No | No | Yes | No | Yes | TBTB |
| 2 | AF | 40 |
Male | Yes | Yes | No | No | No | No | Yes | TBTB |
| 3 | AF | 44 |
Female | No | Yes | No | Yes | No | No | Yes | TBTB |
| 4 | AF | 34 |
Female | No | Yes | Yes | No | No | Yes | Yes | TBTB |
| 5 | AF | 47 |
Female | Yes | No | Yes | No | Yes | No | No | TBTB |
| 6 | AF | 34 |
Male | No | Yes | No | Yes | No | Yes | No | TBTB |
| 7 | AF | 37 |
Female | No | No | Yes | No | No | No | No | TBTB |
| 8 | AF | 41 |
Female | No | Yes | No | No | No | No | No | TBTB |
| 9 | AF | 46 |
Female | No | Yes | Yes | No | No | No | No | TBTB |
| 10 | AF | 31 |
Male | Yes | Yes | Yes | No | No | No | Yes | TBTB |
| 11 | Control | 51 |
Male | Yes | No | No | No | No | No | No |
NPC |
| 12 | Control | 63 |
Female | No | No | Yes | No | No | No | No |
NPC |
| 13 | Control | 62 |
Male | Yes | Yes | No | Yes | No | No | No |
NPC |
| 14 | Control | 58 |
Female | No | Yes | Yes | No | Yes | No | No |
NPC |
| 15 | Control | 47 |
Male | Yes | No | No | No | No | No | No |
NPC |
| 16 | Control | 50 |
Female | No | No | No | No | No | No | No |
NPC |
| 17 | Control | 48 |
Female | No | No | Yes | No | No | No | No |
NPC |
| 18 | Control | 49 |
Male | Yes | Yes | No | No | No | No | No |
NPC |
| 19 | Control | 55 |
Male | No | No | Yes | No | No | No | No |
NPC |
| 20 | Control | 52 |
Female | Yes | No | No | No | No | No | No |
NPC |
Cell transfection
BEAS-2B cells were transfected with a miR-423-5p
mimic or inhibitor (Shanghai GenePharma Co, Ltd.) at a final
concentration of 50 nM and FOXP4 short interfering (si)RNA
(Guangzhou RiboBio Co., Ltd.) or negative control (NC) siRNA using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.). The sequences of the miR-423-5p mimic and
inhibitor, the corresponding NCs, FOXP4 siRNA and NC siRNA are
provided in Table II. After 24 h
of transfection, the cells were treated with or without 10 ng/ml
TGF-β1 for 24 h at 37°C and then collected for further
experiments.
 | Table II.Sequences of miR-423-5p mimic,
inhibitor, NC, FOXP4 siRNA and siRNA NC. |
Table II.
Sequences of miR-423-5p mimic,
inhibitor, NC, FOXP4 siRNA and siRNA NC.
| Gene | Sense | Antisense |
|---|
| hsa-miR-423-5p
mimics |
UGAGGGGCAGAGAGCGAGACUUU |
AGUCUCGCUCUCUGCCCCUCAUU |
| hsa-miR-423-5p
inhibitor |
AAAGUCUCGCUCUCUGCCCCUCA |
|
| Mimic NC |
UUCUCCGAACGUGUCACGUTT |
ACGUGACACGUUCGGAGAATT |
| Inhibitor NC |
CAGUACUUUUGUGUAGUACAA |
|
| genOFFTM
st-h-FOXP4_001 |
|
CCTCAGCTATGGAGCACTT |
| genOFFTM
st-h-FOXP4_002 |
|
GCAGACAGCAATGGTGAGA |
| genOFFTM
st-h-FOXP4_003 |
|
TTCGCCTATTTCCGCAGAA |
| NC-siRNA |
|
GGCUCUAGAAAAGCCUAUGCdTdT |
Histological analysis
Tissue sections (5 µm thick) were subjected to
hematoxylin and eosin (H&E) and Masson staining to enable
histological evaluation of airway tissue fibrosis. Paraffin
embedding was used following dehydration with 70 and 90% alcohol
then transparency with xylene and permeation with paraffin.
Finally, Hematoxylin was used for 1 min and eosin for 2 min at room
temperature to stain the slices. Furthermore, Masson staining was
performed for 3 min at room temperature.
Immunohistochemistry
Paraffin-embedded airway tissue sections were
dewaxed in xylene, dehydrated in graded alcohol and incubated in 3%
H2O2 for 0.5 h. The tissue sections were then
blocked with normal goat serum (Wuhan Servicebio Technology Co.,
Ltd.) and incubated with anti-TGF-β1 (1:100; Wuhan Servicebio
Technology Co., Ltd. cat. no. GB13028), anti-E-cadherin (1:200;
Wuhan Servicebio Technology Co., Ltd. cat. no. GB12082),
anti-N-cadherin (1:100; Wuhan Servicebio Technology Co., Ltd. cat.
no. GB13447), anti-α-SMA (1:500; Wuhan Servicebio Technology Co.,
Ltd. cat. no. GB111364), anti-vimentin (1:500; Wuhan Servicebio
Technology Co., Ltd. cat. no. GB111308), anti-FOXP4 (1:200;
ProteinTech, Wuhan, China; cat. no. 16772-1-AP), anti-PI3K
(1:1,000; Wuhan Servicebio Technology Co., Ltd. no.GB113360),
anti-Akt (1:500; Wuhan Servicebio Technology Co., Ltd. cat. no.
GB11629) and anti-mTOR (1:200; Wuhan Servicebio Technology Co.,
Ltd. cat. no. GB111839) antibodies overnight at 4°C. After
incubation with an HRP-labelled secondary antibody (goat-anti
rabbit IgG) for 2 h in room temperature, the sections were observed
under a light microscope (Nikon Eclipse 80i; Nikon
Corporation).
Wound healing assay
BEAS-2B cells were seeded into a six-well plate and
transfected as described above. A 10-µl pipette tip was used to
scrape the cell monolayer, after which the cells were washed three
times with PBS to remove cellular debris and floating cells. Then,
1.000 µl of serum-free medium containing 10 ng/ml TGF-β1 or the
equivalent amount of PBS was added, the migration ability was
measured by capturing the images at the beginning and in the same
position after 24 h.
Transwell migration assay
Transwell inserts (pore size: 8.0 µm; Corning, Inc.)
were used for the migration assay. The ability of the various
groups of cells to migrate was detected according to the
manufacturer's instructions.
Flow cytometry
Cellular apoptosis was performed by flow cytometry
(CytoFLEX; Beckman Coulter, Brea) using an Annexin V-FITC/Propidium
Iodine double staining kit (Tianjin Sungene Biotech Co., Ltd.)
according to manufacturer's instructions. Cells (1×106)
were stained for 15 min at room temperature in the dark. After
washing twice, the cells were resuspended in 200 µl PBS and
analyzed using a FACSCalibur flow cytometer (BD Biosciences). The
apoptotic rate was calculated by counting early and late apoptotic
cells percentage.
ELISA
Culture medium was collected from each group and
centrifuged at 3,000 × g for 10 min at 4°C, after which the
supernatant was collected for further examination. The levels of
FOXP4, E-cadherin, N-cadherin, vimentin and α-SMA were detected
using ELISA kits (Meike Jiangsu Sumeike Biological Technology Co.,
Ltd.) according to the manufacturer's instructions. At least three
parallel wells were analyzed for each sample.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA (1 µg) from airway tissues or BEAS-2B
cells (1×106) was used to synthesize complementary DNA
(cDNA) with the PrimeScript RT Reagent kit (Takara Biotechnology
Co., Ltd.). Subsequently, cDNA was amplified via the TB Green
Premix EX Taq II PCR kit (Takara Biotechnology Co., Ltd.).
Amplification cycle included an initial 30 sec incubation at 95°C
for denaturation, followed by 40 cycles of 5 sec at 95°C for
annealing and 30 sec at 60°C for elongation. The small nuclear RNA
U6 was utilized for the internal normalization of miR-423-5p levels
and GAPDH was utilized as a control for mRNA expression. Relative
quantification was carried out via the 2−ΔΔCq method
(14). The primer sequences used
are listed in Table SI. RNA
extraction, cDNA synthesis and qPCR were performed according to the
manufacturer's protocols and these experiments were replicated
three times.
Western blot assay
Total protein was extracted from the BEAS-2B cells
using RIPA lysis buffer (Beyotime Institute of Biotechnology)
mixing with protease and phosphatase inhibitor. The proteins were
quantified by bicinchoninic acid kit (Beyotime Institute of
Biotechnology). Proteins (25 µg) extracted from BEAS-2B cells were
separated via 10% SDS-PAGE and transferred onto PVDF membranes (EMD
Millipore). The membranes were blocked with 5% nonfat milk at room
temperature for 2 h and then incubated overnight with specific
primary antibodies at 4°C. The next day, the membranes were washed
three times with Tris-buffered saline containing 1% Tween-20 and
then incubated with secondary antibodies (1:8,000; Zhongshan Golden
Bridge Biological Technology Co., Ltd.; cat. no. ZB-2306) for 1 h
at room temperature. The following primary antibodies were
utilized: anti-E-cadherin (1:10,000; Abcam; cat. no. ab40772),
anti-N-cadherin (1:1,000; CST; cat. no. 13116T), anti-α-SMA
(1:2,000; Abcam; cat. no. ab32575), anti-vimentin (1:3,000; Abcam;
cat. no. ab92547), anti-FOXP4 (1:600; Affinity; cat. no. AF4057),
anti-phosphorylated (p)-PI3K (1:800; Affinity; cat. no. AF3241),
anti-PI3K (1:1,000; CST; cat. no. 4257T), anti-p-Akt (1:800; CST;
cat. no. 4060T), anti-Akt (1:1,000; CST; cat. no. 4691T),
anti-p-mTOR (1:1,000; CST; cat. no. 5536T), anti-mTOR (1:1,000;
CST; cat. no. 2983T) and anti-β-actin (1:2,000; Chongqing Biospes
Co., Ltd.; cat. no. BTL1026). The western blots results were
analyzed by Bio-Rad gel imaging system (170–8170, Bio-Rad
Laboratories, Inc.) and western blot kit (Beyotime Institute of
Biotechnology).
Immunofluorescence staining
Precooled PBS was used to rinse BEAS-2B cells three
times for 5 min each and then the cells were fixed with 4%
paraformaldehyde for 20 min at room temperature. Goat serum was
used to block the cells for 1 h at 37°C. The BEAS-2B cells were
incubated with primary antibodies against E-cadherin (1:200; CST),
N-cadherin (1:200; CST), vimentin (1:200; CST), α-SMA (1:200; CST)
and FOXP4 (1:200; ProteinTech Group, Inc.) at 4°C overnight in a
wet box, rinsed with precooled PBS three times, incubated with
donkey anti-rabbit IgG (Alexa Fluor 64) (1:500; Abcam, USA) for 1 h
in the dark at 37°C and then rinsed three times with PBS. The
BEAS-2B cells were then treated with DAPI for 20 min at 4°C in the
dark to stain the cell nuclei.
RNA fluorescence in situ hybridization
(FISH) analysis
The Ribo FISH kit (Guangzhou RiboBio Co., Ltd.) was
used for FISH experiments. In brief, BEAS-2B cells were cultured to
60% confluence prior to the experiments. Prechilled permeate at 4°C
for 5 min and 4% paraformaldehyde at room temperature for 10 min
were used for cell permeation and fixation, respectively. Fixed
cells were incubated with a FISH Probe Mix stock solution. Nuclei
were stained with DAPI for 20 min at 4°C in the dark.
RNA pull-down assay
A potential target gene of miR-423-5p was identified
using TargetScan (version 7.2; http://www.targetscan.org/). The FOXP4 and FOXP4-NC
sequences were labelled with biotin by Shanghai GenePharma Co.,
Ltd. The biotinylated oligonucleotides were transfected into
BEAS-2B cells at 4°C for 24 h and the cells were lysed with RIP
lysis buffer (MilliporeSigma), followed by incubation with
Streptavidin MagneSphere magnetic beads (MilliporeSigma) at room
temperature for 3 h. The magnetic beads were washed and the RNA and
mRNA sequences pulled down by the antibodies were assessed by
RT-qPCR.
Statistical analyses
The data were analyzed via SPSS 23.0 software (IBM
Corp.) and are presented as the mean ± SD (n=3). Comparisons
between two groups were carried out via unpaired Student's t-tests.
Multiple comparisons were carried out via one-way ANOVA followed by
the homogeneity test of variance and then Tukey's post hoc test was
performed. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-423-5p is upregulated in AF
tissues and TGF-β1-treated BEAS-2B cells
The principal histological findings based on the
H&E and Masson staining results were squamous epithelialization
of the bronchial epithelial cells and fibrotic lesions with
thickened submucosal layers. In addition, the fibrosis presence was
verified by Masson staining (Fig.
1A). To investigate whether miR-423-5p participates in EMT
development, miR-423-5p expression in AF tissue and during
TGF-β1-induced EMT was detected. The expression of miR-423-5p,
TGF-β1, N-cadherin, vimentin and α-SMA was significantly increased
in AF tissue compared with control tissue; conversely, E-cadherin
expression was decreased (Figs. 1B,
C, E and F and S1A).
miR-423-5p expression in BEAS-2B cells treated with TGF-β1 was then
detected, which showed that TGF-β1 upregulated miR-423-5p
expression compared with that in the control group (Fig. 1D). Taken together, these findings
indicated that miR-423-5p expression was upregulated in both AF
tissues and TGF-β1-stimulated BEAS-2B cells.
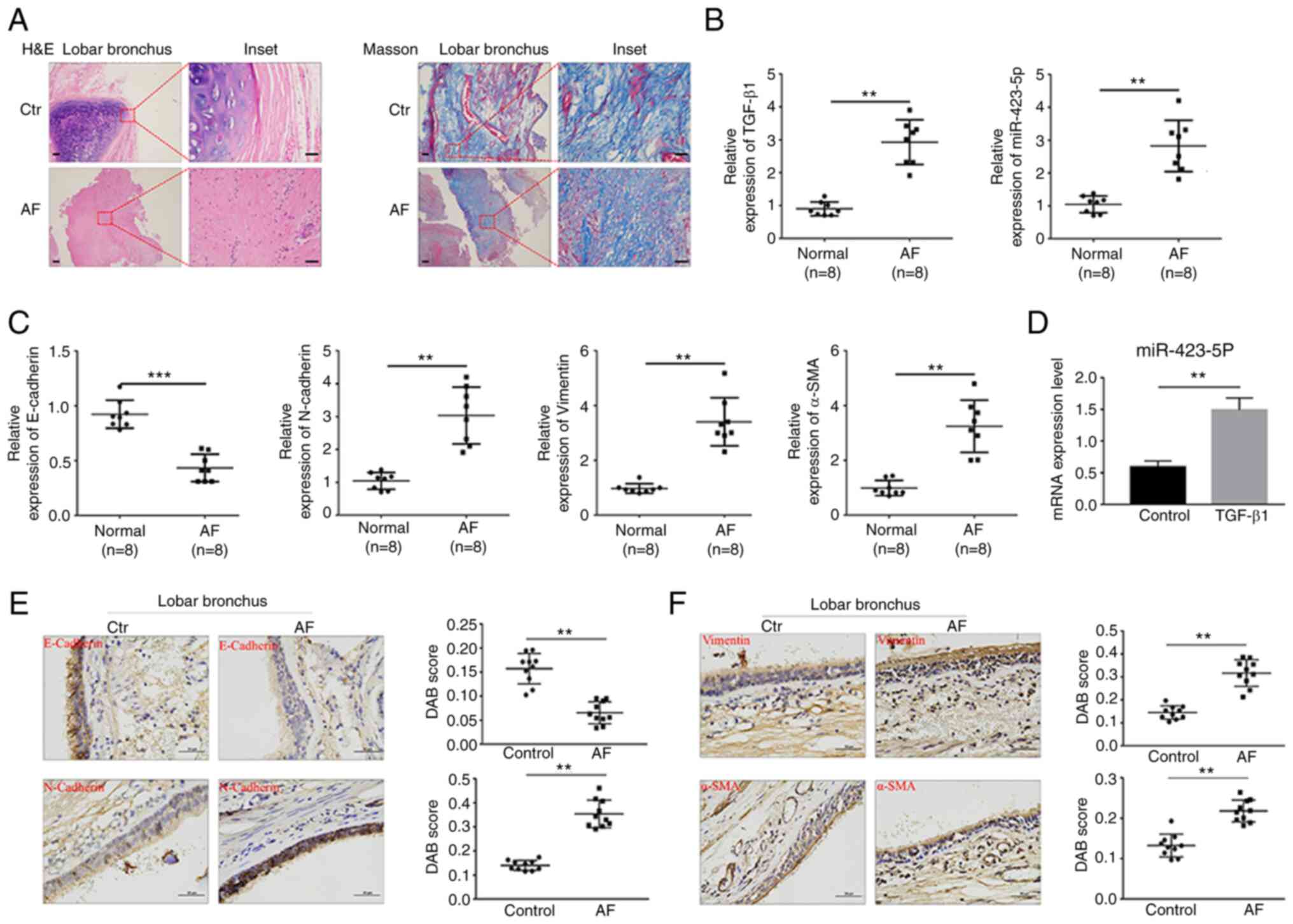 | Figure 1.miR-423-5p expression in human AF
tissue and BEAS-2B cells treated with TGF-β1. (A) H&E and
Masson staining of the control group and AF group. Scale bar=500
and 100 µm (inset) respectively. (B and C) The relative expression
of TGF-β1, miR-423-5p, E-cadherin, N-cadherin, vimentin and α-SMA
in normal airway tissues (n=8) and AF tissues (n=8) was determined
by RT-qPCR. (D) The relative expression of miR-423-5p in BEAS-2B
cells treated with 10 ng/ml TGF-β1 for 24 h was determined by
RT-qPCR. (E and F) The protein expression of E-cadherin,
N-cadherin, vimentin and α-SMA in normal airway tissues (n=10) and
AF tissues (n=10) was detected by immunohistochemistry. Scale
bar=50 µm. **P<0.01 and ***P<0.001. miR, microRNA; AF, airway
fibrosis; H&E, hematoxylin and eosin; α-SMA, α-smooth muscle
actin; RT-qPCR, reverse transcription-quantitative PCR; Ctr,
control. |
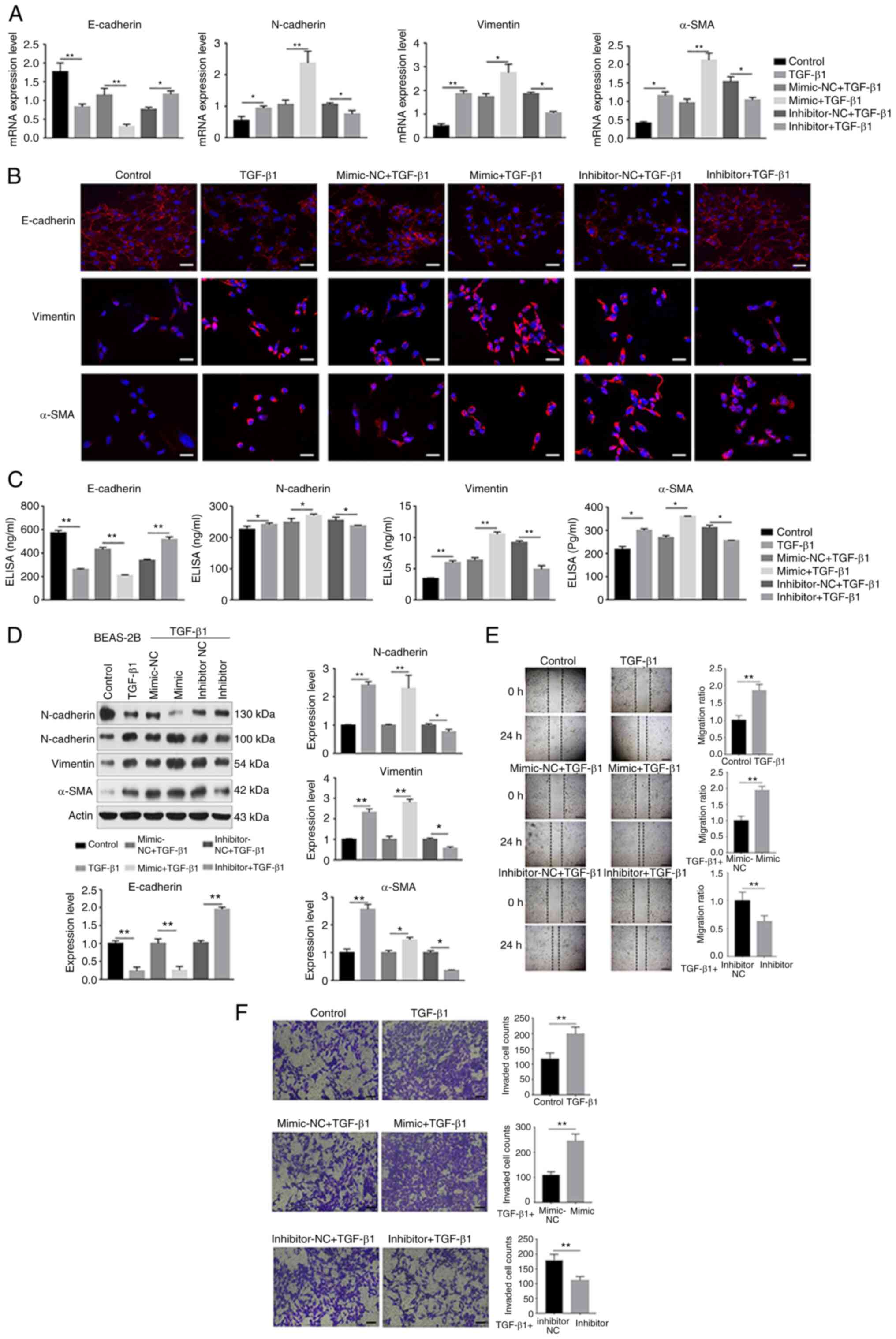
miR-423-5p promotes TGF-β1-induced
EMT
To assess the role of miR-423-5p in TGF-β1-induced
EMT in BEAS-2B cells, a miR-423-5p mimic or inhibitor was
transfected into BEAS-2B cells, which were then treated with
TGF-β1. The miRNA transfection efficiency was verified using
RT-qPCR. After 24 h of transfection, the miR-423-5p level increased
by ~20-fold in the miR-423-5p mimic group compared with the
mimic-NC group. Additionally, the miR-423-5p level decreased by
~6-fold in the miR-423-5p inhibitor group compared with the
inhibitor-NC group (Fig. S1B).
These results demonstrated that the transfection was efficient. To
reveal the functional effect of miR-423-5p on the BEAS-2B cell
model of EMT induced by TGF-β1, the EMT biomarkers E-cadherin,
N-cadherin, vimentin and α-SMA were detected via RT-qPCR,
immunofluorescence staining, ELISA and western blot analysis. In
addition, a wound healing assay and Transwell migration assay were
used to examine the ability of the cells to migrate. Furthermore,
the apoptosis of BEAS-2B cells was determined by flow cytometry.
The results showed that upregulation of miR-423-5p decreased
TGF-β1-stimulated E-cadherin expression but increased the
expression of N-cadherin, vimentin and α-SMA at both the mRNA and
protein levels. In contrast, downregulation of miR-423-5p increased
TGF-β1-stimulated E-cadherin expression but decreased N-cadherin,
vimentin and α-SMA expression at both the mRNA and protein levels
(Fig. 2A-D). Additionally,
miR-423-5p upregulation increased BEAS-2B cell migration and
suppressed BEAS-2B cell apoptosis. In contrast, miR-423-5p
downregulation decreased the migration of BEAS-2B cells and
promoted BEAS-2B cell apoptosis (Figs. 2E and F and S1C). Collectively, these findings
suggested that upregulation of miR-423-5p promoted TGF-β1-induced
EMT, whereas downregulation of miR-423-5p suppressed TGF-β1-induced
EMT.
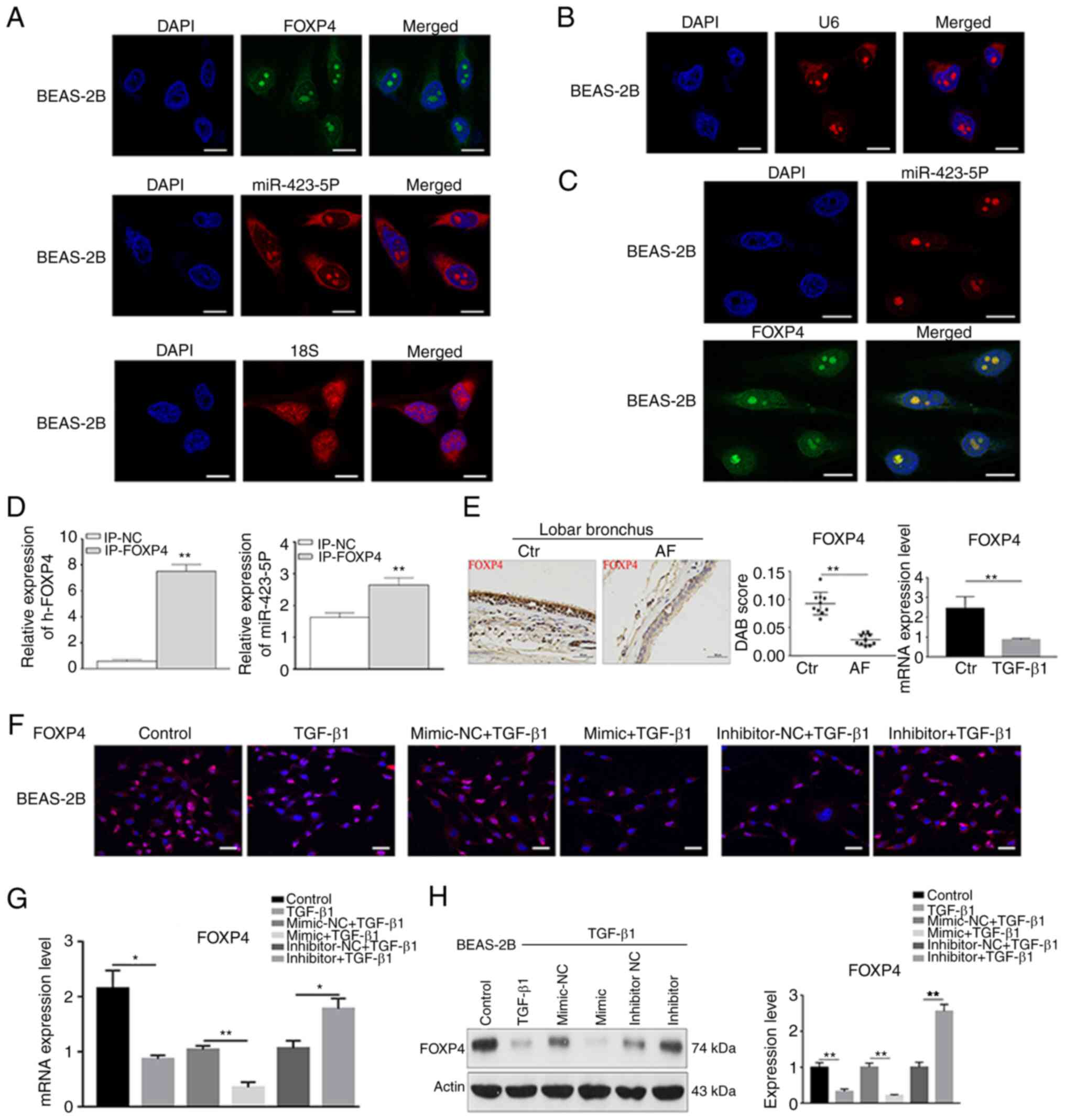
FOXP4 is a target gene of
miR-423-5p
Target genes of miR-423-5p were predicted via the
online database TargetScan 7.2 and the results identified FOXP4
(Fig. S1D). To confirm FOXP4 as
a miR-423-5p target gene, RNA FISH analysis and an RNA pull-down
assay were performed. The results of FISH analysis indicated that
miR-423-5p was located in the cytoplasm or nucleus and that FOXP4
mRNA was located in the nucleus. The results also revealed the
colocalization of miR-423-5p and FOXP4 mRNAs in the nuclei of
BEAS-2B cells, suggesting an interaction between them (Fig. 3A-C). FOXP4 mRNA expression in the
input-FOXP4 group was higher than that in the input-NC groups,
showing that the FOXP4 gene was bound by the probe. Moreover,
miR-423-5p expression in the input-FOXP4 group was higher than that
in the input-NC group, indicating that miR-423-5p and FOXP4 were
bound (Fig. 3D). Furthermore,
FOXP4 expression was detected in AF tissues and TGF-β1-treated
BEAS-2B cells. The results showed that FOXP4 expression was clearly
decreased in the AF tissues and BEAS-2B cells treated with TGF-β1
at both the mRNA and protein levels (Figs. 3E and S1E). In addition, the association
between miR-423-5p and FOXP4 was detected by RT-qPCR,
immunofluorescence staining and western blot analysis. FOXP4
expression was decreased in the miR-423-5p mimic group compared
with the mimic-NC group at both the mRNA and protein levels but
increased in the miR-423-5p inhibitor group compared with the
inhibitor-NC group, indicating a negative association between
miR-423-5p and FOXP4 (Fig. 3F-H).
These findings suggested that FOXP4 is directly regulated by
miR-423-5p.
FOXP4 silencing eliminates miR-423-5p
inhibitor-induced effects on TGF-β1-induced EMT
To confirm that the miR-423-5p inhibitor suppressed
TGF-β1-induced EMT through a FOXP4-dependent pathway, the
miR-423-5p inhibitor was transfected into BEAS-2B cells in which
FOXP4 had been knocked down. First, the knockdown efficiency in
BEAS-2B cells was tested. The FOXP4 expression was significantly
downregulated in the siRNA-FOXP4 group compared with the siRNA-NC
group at both the mRNA and protein levels, which demonstrated
efficient knockdown (Fig.
S1F-H). The results of RT-qPCR, immunofluorescence staining,
ELISA and western blot analysis indicated that FOXP4 and E-cadherin
expression was decreased, but N-cadherin, vimentin and α-SMA
expression was increased in the TGF-β1 + miR-423-5p inhibitor +
siRNA-FOXP4 group compared with the TGF-β1 + miR-423-5p inhibitor +
siRNA-NC group (Fig. 4A-D).
Additionally, the ability of the cells to migrate was increased and
apoptosis was decreased in the TGF-β1 + miR-423-5p inhibitor +
siRNA-FOXP4 group compared with the TGF-β1 + miR-423-5p inhibitor +
siRNA-NC group (Figs. 4E and F
and S1I). These findings
indicated that FOXP4 knockdown partly eliminated miR-423-5p
inhibitor-mediated effects on EMT induced by TGF-β1.
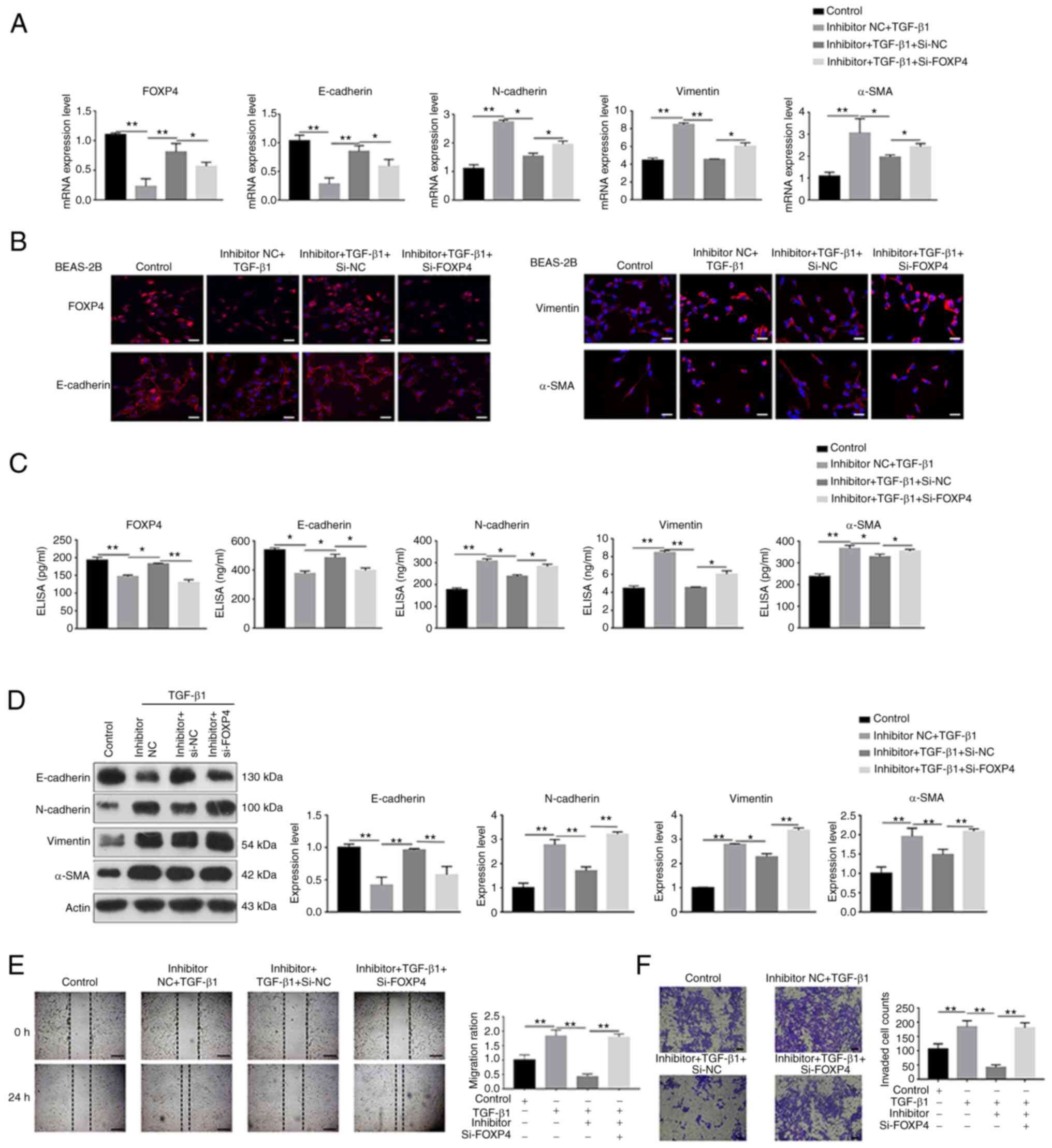 | Figure 4.FOXP4 silencing eliminated the
miR-423-5p inhibitor-mediated effect on TGF-β1-induced EMT. BEAS-2B
cells transfected with siRNA-FOXP4 to silence FOXP4 were
transfected with a miR-423-5p inhibitor for 24 h and then treated
with TGF-β1 for 24 h. The relative expression of FOXP4, E-cadherin,
N-cadherin, vimentin and α-SMA was determined by (A) reverse
transcription-quantitative PCR, (B) immunofluorescence staining
(scale bar=50 µm), (C) ELISA and (D) western blot analysis.
Migration and the apoptosis rate were detected by (E) wound healing
assay (scale bar=200 µm) and (F) Transwell migration assay (scale
bar=100 µm). *P<0.05 and **P<0.01. FOXP4, forkhead box p4;
miR, microRNA; EMT, epithelial-mesenchymal transition; si, short
interfering; α-SMA, α-smooth muscle actin. |
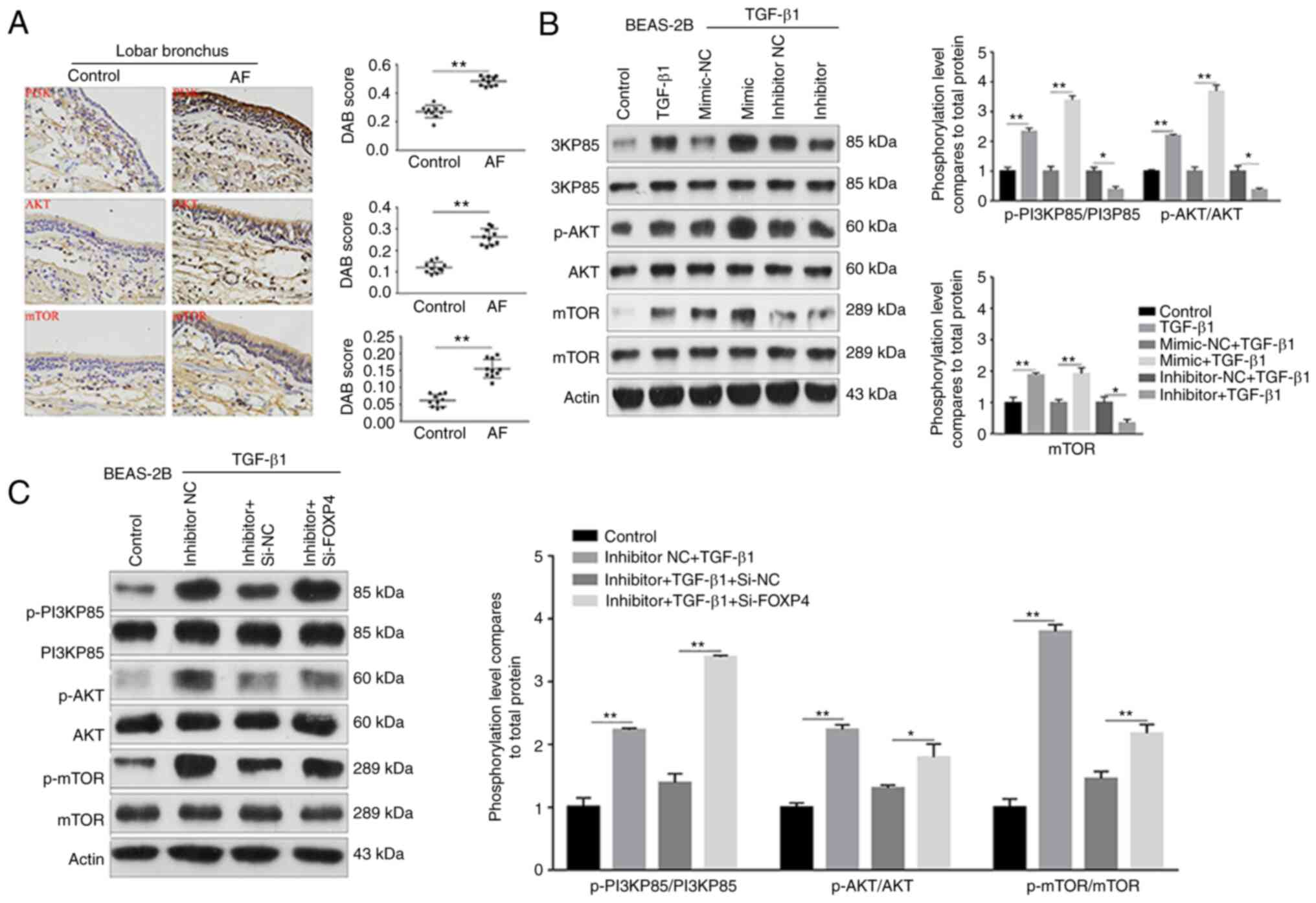
miR-423-5p promotes EMT through the
PI3K/AKT/mTOR signaling pathway
To reveal the underlying mechanism by which the
miR-423-5p mimic promoted TGF-β1-induced EMT, the signaling pathway
downstream of FOXP4 was detected. Existing evidence shows that the
PI3K/AKT/mTOR pathway participates in the lung fibrosis process;
however, the role of the PI3K/AKT/mTOR pathway in the airway is
unclear. Therefore, the expression levels of PI3K/AKT/mTOR
pathway-associated indicators were evaluated. The expression of
PI3K, AKT and mTOR was significantly higher in the AF group than in
the control group (Fig. 5A).
Moreover, p-PI3K, p-AKT and p-mTOR expression was significantly
increased in the TGF-β1 group compared with the control group.
Conversely, in the TGF-β1 + miR-423-5p inhibitor + siRNA-NC group,
the expression levels of p-PI3K, p-AKT and p-mTOR were lower than
those in the TGF-β1 group (Fig.
5B). Additionally, p-PI3K, p-AKT and p-mTOR expression was
clearly upregulated in the TGF-β1 + miR-423-5p inhibitor +
siRNA-FOXP4 group compared with the TGF-β1 + miR-423-5p inhibitor +
siRNA-NC group (Fig. 5C). Taken
together, these findings suggested that miR-423-5p promotes
TGF-β1-induced EMT through the PI3K/AKT/mTOR signaling pathway in
AF.
Discussion
AF is a progressive airway disease characterized by
airway epithelial injury, fibroblast activation and extracellular
matrix deposition (15,16). miRNAs are endogenous, small
noncoding RNAs that modulate genes at the posttranscriptional level
in various physiological and pathological processes (17). Accumulating evidence indicates
that miRNAs trigger the onset and progression of AF (18,19).
In the present study, miR-423-5p was found to be
upregulated in human AF tissue; additionally, the functional role
of miR-423-5p and the underlying mechanism of its effect on AF were
identified. The results collected from BEAS-2B cells demonstrated
that TGF-β1 increased the expression of miR-423-5p and decreased
FOXP4 expression. Additionally, overexpression of miR-423-5p
facilitated TGF-β1-induced EMT in BEAS-2B cells; by contrast,
downregulation of miR-423-5p suppressed TGF-β1-induced EMT in
BEAS-2B cells. In addition, FOXP4 silencing eliminated the
miR-423-5p inhibitor-mediated effect on TGF-β1-induced EMT. These
results verified that FOXP4 is a potential target of miR-423-5p.
EMT serves a critical role in organ fibrosis, including liver and
kidney fibrosis (20,21). Moreover, TGF-β1 is an important
cytokine involved in the initiation of EMT (22). As AF progresses, alveolar
epithelial cell features are gradually replaced by mesenchymal cell
features with the development of EMT (23). BEAS-2B cells are an airway
alveolar epithelial cell type that shows the general
characteristics of alveolar epithelial cells (24), such as a common morphology and
specific biomarkers, and this cell line has been used to
investigate the mechanisms of EMT in other airway diseases
(25,26). Therefore, the BEAS-2B cell line
was utilized to establish an EMT model in the present study.
Generally, downregulated E-cadherin expression and upregulated
N-cadherin, vimentin and α-SMA expression are reliable markers of
EMT in epithelial cells (4). In
addition, increasing evidence reveals that the EMT process can
increase cell migration and suppress apoptosis (27,28). As BEAS-2B cells are an epithelial
cell line, they exhibit high basal expression of E-cadherin, which
is an epithelial cell marker (29). After BEAS-2B cells were stimulated
with 10 ng/ml TGF-β1 for 24 h, E-cadherin expression was
significantly decreased and N-cadherin, vimentin and α-SMA
expression was increased at the mRNA and protein levels. Moreover,
compared with normal control airway tissues, AF tissues exhibited
decreased E-cadherin expression, whereas TGF-β1, N-cadherin,
vimentin and α-SMA expression was increased at the protein level.
These findings indicated that TGF-β1 induced EMT, which is
consistent with a previous study (30). Furthermore, overexpression of
miR-423-5p promoted TGF-β1-induced EMT. Reciprocally,
downregulation of miR-423-5p suppressed TGF-β1-induced EMT.
FOXP, which is a member of the forkhead
transcription factor family, is involved in various cell processes,
including migration, proliferation, ageing and the cell cycle, by
its function in regulating transcription (31). FOXP consists of four members:
FOXP1, FOXP2, FOXP3 and FOXP4. FOXP4 is ubiquitously expressed in
different types of cells and has been studied in various types of
disease (32). For instance, Tao
et al (33) reported that
‘FOXP4-AS1 promotes mantle cell lymphoma progression through the
upregulation of NACC1 expression by inhibiting miR-423-5p’. Xiong
et al (34) found that the
‘circRNA ZNF609 functions as a competitive endogenous RNA to
regulate FOXP4 expression by sponging miR-138-5p in renal
carcinoma’. More notably, several studies have demonstrated that
FOXP4 participates in the development of the EMT process (35,36). In the present study, FOXP4 was
downregulated in AF tissues and BEAS-2B cells with TGF-β1-induced
EMT and FOXP4 was also directly regulated by miR-423-5p.
Furthermore, FOXP4 knockdown eliminated the miR-423-5p
inhibitor-induced effect on TGF-β1-induced EMT. These results
indicated that FOXP4 participated in AF via the EMT process and is
a target gene of miR-423-5p.
Several studies have highlighted the critical role
of the PI3K/AKT/mTOR pathway in the development of fibrosis in
various organs, including the lungs, liver and myocardial tissue
(37–39). When exogenous cytokines trigger
cellular signals, the AKT signaling cascade is activated by
receptors via PI3K phosphorylation, which immediately activates
mTOR, resulting in the transcription of activation-related
molecules (40). The results of
the present study indicated that TGF-β1 increased the levels of
p-PI3K, p-AKT and p-mTOR and that the miR-423-5p inhibitor
decreased the levels of p-PI3K, p-AKT and p-mTOR. In addition,
FOXP4 knockdown reversed the effect of the miR-423-5p inhibitor on
the expression of p-PI3K, p-AKT and p-mTOR, showing that the
miR-423-5p mimic promoted TGF-β1-induced EMT by targeting FOXP4 via
PI3K/AKT/mTOR pathway activation.
However, the following limitations of the present
study should be considered. Due to various factors, an in
vivo experiment was not completed. Therefore, the effects of
miR-423-5p in animal models of AF will be explored in the future.
In addition, the present study did not investigate the effect of
miR-423-5p overexpression on EMT (e.g., markers expression,
migration and invasion) in BEAS-2B cells without TGF-β1 treatment
because the aim was to investigate miR-423-5p function and its
possible underlying mechanism in the EMT process of BEAS-2B cells.
Unfortunately, the chest HRCT and bronchoscope images were lost due
to the Coronavirus pandemic and attempts to find such data failed.
Furthermore, caspase should be further detected to investigated
cell apoptosis and rescue experiments should be done by
overexpressing miR-423-5p or its target genes to make the results
more solid. Additionally, it would be more effective to
comprehensively estimate the cell transcriptome level by RNA-seq.
Moreover, because a total of 10 AF scar tissue samples were
collected from patients undergoing fiberoptic bronchoscopy biopsy
via an electrocautery needle knife (VIO 300S; Erbe Elektromedizin
GmbH) which led to destruction of normal airway tissue structure so
the histopathological images could not be confirmed to originated
from airway tissue in Fig. 1A.
These defects will be remedied in the future.
In conclusion, the present study suggested that
miR-423-5p promoted TGF-β1-induced EMT in AF by depressing FOXP4
expression and this effect may partly be attributed to the
activation of the PI3K/AKT/mTOR pathway. Therefore, miR-423-5p
inhibition may be a target for the prevention and treatment of
AF.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Major Science
and Technology Projects of China (grant no. 2018ZX10302302003).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YC and SG designed the experiments and revised the
manuscript. YW analyzed the data and revised the manuscript. YC and
XL performed the experiments and wrote the manuscript. YL, GH and
XW analyzed the data. YC and SG confirm the authenticity of all the
raw data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The First Affiliated Hospital of Chongqing Medical
University Ethics Committee approved the present study
(institutional approval number. 2020-147) and the research was
performed in accordance with the Declaration of Helsinki.
Patient consent for publication
All subjects provided written informed consent.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AF
|
airway fibrosis
|
|
FOXP4
|
forkhead box p4
|
|
miR
|
microRNA
|
|
α-SMA
|
α-smooth muscle actin
|
|
H&E
|
hematoxylin and eosin
|
|
ELISA
|
enzyme-linked immunosorbent assay
|
|
FISH
|
fluorescence in situ
hybridization.
|
References
|
1
|
Rao W, Wang S, Duleba M, Niroula S, Goller
K, Xie J, Mahalingam R, Neupane R, Liew AA, Vincent M, et al:
Regenerative metaplastic clones in COPD lung drive inflammation and
fibrosis. Cell. 181:848–864.e18. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Van Dyken SJ, Liang HE, Naikawadi RP,
Woodruff PG, Wolters PJ, Erle DJ and Locksley RM: Spontaneous
chitin accumulation in airways and age-related fibrotic lung
disease. Cell. 169:497–509.e13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Swatek AM, Lynch TJ, Crooke AK, Anderson
PJ, Tyler SR, Brooks L, Ivanovic M, Klesney-Tait JA, Eberlein M,
Pena T, et al: Depletion of airway submucosal glands and
TP63+KRT5+ basal cells in obliterative
bronchiolitis. Am J Respir Crit Care Med. 197:1045–1057. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tian B, Hosoki K, Liu Z, Yang J, Zhao Y,
Sun H, Zhou J, Rytting E, Kaphalia L, Calhoun WJ, et al: Mucosal
bromodomain-containing protein 4 mediates aeroallergen-induced
inflammation and remodeling. J Allergy Clin Immunol.
143:1380–1394.e9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Di Campli MP, Azouz A, Assabban A,
Scaillet J, Splittgerber M, Van Keymeulen A, Libert F, Remmelink M,
Le Moine A, Lemaitre P and Goriely S: The mononuclear phagocyte
system contributes to fibrosis in post-transplant obliterans
bronchiolitis. Eur Respir J. 57:20003442021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang J, Tian B, Sun H, Garofalo RP and
Brasier AR: Epigenetic silencing of IRF1 dysregulates type III
interferon responses to respiratory virus infection in epithelial
to mesenchymal transition. Nat Microbiol. 2:170862017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Su J, Morgani SM, David CJ, Wang Q, Er EE,
Huang YH, Basnet H, Zou Y, Shu W, Soni RK, et al: TGF-β
orchestrates fibrogenic and developmental EMTs via the RAS effector
RREB1. Nature. 577:566–571. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Piera-Velazquez S and Jimenez SA:
Endothelial to mesenchymal transition: Role in physiology and in
the pathogenesis of human diseases. Physiol Rev. 99:1281–1324.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jones D: Setbacks shadow microRNA
therapies in the clinic. Nat Biotechnol. 36:909–910. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang ZC, Qu ZH, Yi MJ, Shan YC, Ran N, Xu
L and Liu XJ: MiR-448-5p inhibits TGF-β1-induced
epithelial-mesenchymal transition and pulmonary fibrosis by
targeting Six1 in asthma. J Cell Physiol. 234:8804–8814. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li J, Pan C, Tang C, Tan W, Zhang W and
Guan J: MiR-184 targets TP63 to block idiopathic pulmonary fibrosis
by inhibiting proliferation and epithelial-mesenchymal transition
of airway epithelial cells. Lab Invest. 101:142–154. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hou Y, Zhang Y, Lin S, Yu Y, Yang L, Li L
and Wang W: Protective mechanism of apigenin in diabetic
nephropathy is related to its regulation of miR-423-5P-USF2 axis.
Am J Transl Res. 13:2006–2020. 2021.PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−delta delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Michalik M, Wójcik-Pszczoła K, Paw M, Wnuk
D, Koczurkiewicz P, Sanak M, Pękala E and Madeja Z:
Fibroblast-to-myofibroblast transition in bronchial asthma. Cell
Mol Life Sci. 75:3943–3961. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zaiss DMW: Amphiregulin as a driver of
tissue fibrosis. Am J Transplant. 20:631–632. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao Y, Shen X, Tang T and Wu CI: Weak
regulation of many targets is cumulatively powerful-an evolutionary
perspective on microRNA functionality. Mol Biol Evol. 34:3041–3046.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guiot J, Cambier M, Boeckx A, Henket M,
Nivelles O, Gester F, Louis E, Malaise M, Dequiedt F, Louis R, et
al: Macrophage-derived exosomes attenuate fibrosis in airway
epithelial cells through delivery of antifibrotic miR-142-3p.
Thorax. 75:870–881. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pommier A, Varilh J, Bleuse S, Delétang K,
Bonini J, Bergougnoux A, Brochiero E, Koenig M, Claustres M and
Taulan-Cadars M: miRNA repertoires of cystic fibrosis ex vivo
models highlight miR-181a and miR-101 that regulate WISP1
expression. J Pathol. 253:186–197. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Bi X, Xiong J, Han W, Xiao T, Xu X,
Yang K, Liu C, Jiang W, He T, et al: MicroRNA-34a Promotes renal
fibrosis by downregulation of klotho in tubular epithelial cells.
Mol Ther. 27:1051–1065. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song L, Chen TY, Zhao XJ, Xu Q, Jiao RQ,
Li JM and Kong LD: Pterostilbene prevents hepatocyte
epithelial-mesenchymal transition in fructose-induced liver
fibrosis through suppressing miR-34a/Sirt1/p53 and TGF-β1/Smads
signalling. Br J Pharmacol. 176:1619–1634. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yeh HW, Hsu EC, Lee SS, Lang YD, Lin YC,
Chang CY, Lee SY, Gu DL, Shih JH, Ho CM, et al: PSPC1 mediates
TGF-β1 autocrine signalling and Smad2/3 target switching to promote
EMT, stemness and metastasis. Nat Cell Biol. 20:479–491. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun Y, Shi Z, Liu B, Li X, Li G, Yang F
and Tang H: YKL-40 mediates airway remodeling in asthma via
activating FAK and MAPK signaling pathway. Cell Cycle.
19:1378–1390. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang J, Tian X, Zhang J, Tan L, Ouyang N,
Jia B, Chen C, Ge C and Li J: Postchronic single-walled carbon
nanotube exposure causes irreversible malignant transformation of
human bronchial epithelial cells through DNA methylation changes.
ACS Nano. 15:7094–7104. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Benedikter BJ, Bouwman FG, Heinzmann ACA,
Vajen T, Mariman EC, Wouters EFM, Savelkoul PHM, Koenen RR, Rohde
GGU, van Oerle R, et al: Proteomic analysis reveals procoagulant
properties of cigarette smoke-induced extracellular vesicles. J
Extracell Vesicles. 8:15851632019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sundar IK, Li D and Rahman I: Small
RNA-sequence analysis of plasma-derived extracellular vesicle
miRNAs in smokers and patients with chronic obstructive pulmonary
disease as circulating biomarkers. J Extracell Vesicles.
8:16848162019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao Y, Shang S, Guo S, Li X, Zhou H, Liu
H, Sun Y, Wang J, Wang P, Zhi H, et al: Lnc2Cancer 3.0: An updated
resource for experimentally supported lncRNA/circRNA cancer
associations and web tools based on RNA-seq and scRNA-seq data.
Nucleic Acids Res. 49(D1):D1251–D1258. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Serresi M, Kertalli S, Li L, Schmitt MJ,
Dramaretska Y, Wierikx J, Hulsman D and Gargiulo G: Functional
antagonism of chromatin modulators regulates epithelial-mesenchymal
transition. Sci Adv. 7:eabd79742021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bernstein DI, Lummus ZL, Kesavalu B, Yao
J, Kottyan L, Miller D, Cartier A, Cruz MJ, Lemiere C, Muñoz X, et
al: Genetic variants with gene regulatory effects are associated
with diisocyanate-induced asthma. J Allergy Clin Immunol.
142:959–969. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang X, Liu L, Deng X, Li D, Cai H, Ma Y,
Jia C, Wu B, Fan Y and Lv Z: MicroRNA 483-3p targets Pard3 to
potentiate TGF-β1-induced cell migration, invasion and
epithelial-mesenchymal transition in anaplastic thyroid cancer
cells. Oncogene. 38:699–715. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim JH, Hwang J, Jung JH, Lee HJ, Lee DY
and Kim SH: Molecular networks of FOXP family: Dual biologic
functions, interplay with other molecules and clinical implications
in cancer progression. Mol Cancer. 18:1802019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Snijders Blok L, Vino A, den Hoed J,
Underhill HR, Monteil D, Li H, Reynoso Santos FJ, Chung WK, Amaral
MD, Schnur RE, et al: Heterozygous variants that disturb the
transcriptional repressor activity of FOXP4 cause a developmental
disorder with speech/language delays and multiple congenital
abnormalities. Genet Med. 23:534–542. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tao HF, Shen JX, Hou ZW, Chen SY, Su YZ
and Fang JL: lncRNA FOXP4-AS1 predicts poor prognosis and
accelerates the progression of mantle cell lymphoma through the
miR-423-5p/NACC1 pathway. Oncol Rep. 45:469–480. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xiong Y, Zhang J and Song C: CircRNA
ZNF609 functions as a competitive endogenous RNA to regulate FOXP4
expression by sponging miR-138-5p in renal carcinoma. J Cell
Physiol. 234:10646–10654. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rajarajan D, Selvarajan S, Charan Raja MR,
Kar Mahapatra S and Kasiappan R: Genome-wide analysis reveals
miR-3184-5p and miR-181c-3p as a critical regulator for
adipocytes-associated breast cancer. J Cell Physiol.
234:17959–17974. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
E C, Yang J, Li H and Li C: LncRNA
LOC105372579 promotes proliferation and epithelial-mesenchymal
transition in hepatocellular carcinoma via activating
miR-4316/FOXP4 signaling. Cancer Manag Res. 11:2871–2879. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Han B, Chu C, Su X, Zhang N, Zhou L, Zhang
M, Yang S, Shi L, Zhao B, Niu Y and Zhang R:
N6-methyladenosine-dependent primary microRNA-126
processing activated PI3K-AKT-mTOR pathway drove the development of
pulmonary fibrosis induced by nanoscale carbon black particles in
rats. Nanotoxicology. 14:1–20. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kong D, Zhang Z, Chen L, Huang W, Zhang F,
Wang L, Wang Y, Cao P and Zheng S: Curcumin blunts
epithelial-mesenchymal transition of hepatocytes to alleviate
hepatic fibrosis through regulating oxidative stress and autophagy.
Redox Biol. 36:1016002020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liang M, Lv J, Jiang Z, He H, Chen C,
Xiong Y, Zhu X, Xue Y, Yu Y, Yang S, et al: Promotion of
myofibroblast differentiation and tissue fibrosis by the
leukotriene B4 -leukotriene B4 receptor axis
in systemic sclerosis. Arthritis Rheumatol. 72:1013–1025. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Janku F, Yap TA and Meric-Bernstam F:
Targeting the PI3K pathway in cancer: Are we making headway. Nat
Rev Clin Oncol. 15:273–291. 2018. View Article : Google Scholar : PubMed/NCBI
|