Introduction
Colorectal cancer is one of the most common
malignancies in Western countries and is the second most common
cause of cancer mortality. Early detection, prediction of
recurrence during the pre-symptomatic phase of the disease and
response to chemotherapy are all factors that improve survival.
Molecular techniques and new methods of detecting metastasis or
recurrent disease in these pre-clinical or pre-symptomatic phases
of the disease may contribute to this strategy. However, the
majority of these methods have limited sensitivity or specificity,
or entail invasive procedures (1).
In recent years, circulating nucleic acids in the plasma of cancer
patients have been studied as a source of tumor information
obtained by non-invasive methods.
The presence of extracellular nucleic acids in human
plasma or serum has been well established (2). In a number of studies, higher
concentrations of circulating nucleic acids were detected in the
plasma or serum of cancer patients as compared to that of healthy
subjects, as well as higher levels in patients with metastases than
in those with localized disease (3,4).
Alterations in DNA found in plasma from patients with diverse types
of cancer were similar to alterations found in primary tumors,
suggesting that plasma and serum nucleic acids originate in tumor
cells (2). More recently, the
extraction of RNA from the plasma of cancer patients and its
subsequent analysis by reverse transcription-polymerase chain
reaction (RT-PCR) was reported (2).
The potential use of plasma RNA for the analysis of cancer is
highly attractive for a number of reasons: it only requires a
minimally invasive method (extraction of a small amount of blood);
it can be obtained repeatedly and at any time during tumor
progression, allowing for an analysis of treatment response; and
its simplicity makes it suitable for use in the asymptomatic
population at risk. In previous studies, a correlation was noted
between circulating tumor cells and circulating tumor mRNA in colon
cancer (5), and it was found that
mRNA is more sensitive than DNA in the plasma of breast cancer
patients (6). Extracellular RNA
exists with sufficient integrity in RT-PCR amplification, making it
a useful tool in determining nucleic acid in the plasma of cancer
patients and assessing its value as a prognostic factor. Thus,
tumor-associated mRNA in plasma from cancer patients was also
detected in various studies and associated with more aggressive
tumors and with poor outcome (5,7–11).
In a 2007 study, cDNA microarray hybridization was
employed to perform genomic profiling of plasma RNA from colorectal
cancer (CRC) patients and from healthy donors (12). Expression analysis identified 40
mRNA differentially up-regulated genes in the cancer group. Using
real-time PCR on a short external set of CRC samples, higher levels
of the three genes (KIAA0101, UBE2D3 and
EPAS1) were confirmed in CRC patients as compared to healthy
donors. Thus, these mRNA may be diagnostic indicators.
KIAA0101, also known as p15PAF, was identified as a
commonly overexpressed gene in a variety of solid tumors by two
independent groups using large-scale meta-analysis of cancer DNA
microarray data (13,14). EPAS1 encodes hypoxia
inducible factor 2-α, an important angiogenic factor whose high
expression in CRC was shown to play a significant role in tumor
progression and to possess prognostic value (15). UBE2D3, another of our
selected markers, encodes an ubiquitin-conjugating enzyme involved
in the regulated degradation of major cellular factors, such as
tumor suppressor p53 and NF-κB regulator (16,17).
The present study aimed to analyze the mRNA levels
of the three genes (KIAA0101, UBE2D3 and
EPAS1) by real-time PCR in the plasma of a large series of
patients with CRC to identify their prognostic value, and
investigate potential associations with parameters indicative of
poor prognosis, disease-free survival (DFS) and overall survival
(OS).
Materials and methods
Plasma samples and clinicopathological
parameters
Informed consent was obtained from all participants
after an explanation of the nature of the study, as approved by the
research ethics board of our hospital. All of the patients were
considered sporadic cases on the basis that no clinical background
of family adenomatous polyposis was reported. Any patient who met
the clinical criteria for hereditary non-polyposis colon cancer
(Amsterdam criteria) was excluded from the present study. Between
February 2003 and January 2009, blood samples were obtained by
venepuncture from 154 patients with CRC prior to intervention on
the day of surgery. Plasma samples were obtained by centrifugation
of peripheral blood at 500 g for 25 min and divided into aliquots,
which were snap-frozen at −80°C until processing.
The following variables were obtained from the
medical record of 154 patients: age, gender, tumor site, tumor
differentiation, lymph node metastases, pathological stage,
histological grade, vascular invasion and evidence of polyps
(defined by the presence of polyps removed during surgery). The
pathological stage was assessed using the tumor-node-metastases
(TNM) classification.
Patient follow-up
Clinical follow-up after surgery and diagnosis was
based on periodic visits (every 3 months during the first year,
every 6 months during the second year and then annually until
relapse in our Medical Oncology Department, complemented by other
periodic controls in Health Centers attached to our hospital), as
well as clinical, biochemical and imaging techniques (chest X-ray,
endoscopy, bone scan and other areas as clinically indicated). In
addition, an ultrasonic study was performed when liver function was
impaired. OS was defined as the period from time of diagnosis until
patients succumbed to the disease. DFS was defined as the interval
between diagnosis and first recurrence.
Nucleic acid isolation and real-time
PCR
Plasma mRNA was obtained from 1 ml of the samples by
Dynabeads mRNA direct kit (Invitrogen Dynal AS, Oslo, Norway).
Plasma was incubated with 100 μl of Dynabeads oligo (dt) for 10 min
at room temperature. mRNA was eluted in 10 mM HCL. For the
synthesis of cDNA, RNA was retro-transcribed using the Gold RNA PCR
core kit (PE Applied Biosystems, Foster City, CA, USA) according to
the manufacturer’s instructions. Random hexamers were used as
primers for cDNA synthesis.
Real-time PCR was performed in a Light Cycler
apparatus (Roche Diagnostics, Mannheim, Germany) using the
LightCycler-FastStart DNA MasterPLUS SYBR-Green I kit
(Roche Diagnostics) according to the manufacturer’s instructions.
The primers and annealing temperatures are shown in Table I. The relative concentrations of the
target and the reference genes were calculated by interpolation,
using a standard curve of each gene plotted from the same serial
dilution of cDNA from tumor tissue. The target mRNA levels
(KIAA0101, UBE2D3 and EPAS1) were normalized
by the housekeeping gene, succinate dehydrogenase complex subunit A
(SDHA), in each sample. At the end of PCR cycles, melting
curve analysis and electrophoresis of the products on
non-denaturing 8% polyacrylamide gels together with a molecular
weight marker were performed to confirm the generation of the
specific PCR product expected. Representative bands were sequenced
in an ABI Prism™ 377 DNA sequencer apparatus (PE Applied
Biosystems).
 | Table ISequences and annealing temperatures
(aT) for each primer used. |
Table I
Sequences and annealing temperatures
(aT) for each primer used.
| mRNA | Sequence | aT (°C) |
|---|
| EPAS1 |
5′-ACGCCACCCAGTACCAGGA3′-F
5′-AATGAGGGCCCGAGCAGC3′-R | 65 |
|
KIAA0101 |
5′-AGGTTGTCCCCTAAAGATTCTG3′-F
5′-CAGGTTGCAAAGGACATGC3′-R | 59 |
| UBE2D3 |
5′-AACCCAGATGACCCCCTAGTG3′-F
5′-CCATTCCCGAGATATTCTGTTG3′-R | 63 |
| SDHA |
5′-TGGGAACAAGAGGGCATCTG3′-F
5′-CCACCACTGCATCAAATTCATG3′-R | 59 |
Statistical analysis
The target mRNA data in the plasma were not normally
distributed (Kolmogorov-Smirnov test). Plasma samples showing the
presence of mRNA for each target gene were divided into tertiles.
The clinicopathological parameters were contrasted with the
tertiles using the Pearson Chi-square test. For the survival
analysis, any patients at pathological stage IV were not included
in the DFS evaluation. The relationship between the cumulative
probability of OS and DFS, as well as analyzed predictors, was
calculated by the Kaplan-Meier method, while significant
differences between curves were evaluated using the Mantel’s
log-rank test. In all statistical tests, two-tailed p-values
<0.05 were considered to be statistically significant.
Statistical analysis was performed using SPSS, version 14.0.
Results
Relationship between KIAA0101, EPAS1 and
UBE2D3 mRNA levels in plasma and clinicopathological
parameters
The mRNA levels of EPAS1, KIAA0101 and
UBE2D3 genes were analyzed by Q-RT-PCR in a large series of
154 plasmas from CRC patients. This large series was independent of
the series analyzed in our previous study (12). The presence of mRNA in plasma was
identified in 64% of patients for EPAS1, in 90% for
UBE2D3 and in 44% for KIAA0101.
To evaluate the prognostic value of these markers,
their possible association with clinicopathological parameters of
the tumors indicative of poor prognosis was analyzed. A total of
43% of the patients were females and 57% were males, with 95% of
the patients at >50 years of age. Vascular or lymphatic invasion
was found in 28% of the patients. A total of 32% of the specimens
were classified as well-differentiated, 57% as moderately
differentiated and 11% as poorly differentiated. The majority of
tumors were at intermediate stages: 11% of cases were classified as
TNM I, 35% were TNM II, 39% were TNM III and 15% were TNM IV. For
this purpose, the plasma mRNA levels of each gene were divided into
tertiles (low, medium and high levels). Statistically significant
associations were not found between mRNA levels of KIAA0101
or UBE2D3 and clinicopathological parameters. In the case of
EPAS1, high levels of mRNA in the plasma correlated
statistically with age (≥50 years) and relapse (p=0.012 and
p=0.045, respectively; Pearson’s Chi-square test). Moreover, we
found a trend towards statistical association between high
expression levels of EPAS1 and patient mortality (p=0.07).
No statistically significant correlations were noted between
EPAS1 levels and the remaining pathological parameters
analyzed. The results are summarized in Table II.
 | Table IIAssociations between
clinicopathological characteristics and mRNAs levels in the plasma
from colorectal cancer patients (Chi-square test). |
Table II
Associations between
clinicopathological characteristics and mRNAs levels in the plasma
from colorectal cancer patients (Chi-square test).
|
KIAA0101 | EPAS1 | UBE2D3 |
|---|
|
|
|
|
|---|
| Low (%) | Medium (%) | High (%) | p-value | Low (%) | Medium (%) | High (%) | p-value | Low (%) | Medium (%) | High (%) | p-value |
|---|
| Age |
| <50 years | 100.0 | 0.0 | 0.0 | 0.353 | 100.0 | 0.0 | 0.0 | 0.012 | 42.9 | 42.9 | 14.2 | 0.501 |
| ≥50 years | 31.7 | 33.3 | 35.0 | | 29.0 | 34.5 | 36.7 | | 31.2 | 32.8 | 36.0 | |
| Gender |
| Male | 35.0 | 27.5 | 37.5 | 0.503 | 33.3 | 31.6 | 35.1 | 0.963 | 32.9 | 31.6 | 35.5 | 0.959 |
| Female | 29.2 | 41.7 | 29.2 | | 31.6 | 34.2 | 34.2 | | 32.2 | 33.9 | 33.9 | |
| Localization |
| Rectum | 12.5 | 37.5 | 50.0 | 0.418 | 25.0 | 43.8 | 31.3 | 0.127 | 25.0 | 45.0 | 30.0 | 0.081 |
| Left | 35.5 | 38.7 | 25.8 | | 31.1 | 35.5 | 33.3 | | 26.2 | 42.6 | 31.2 | |
| Right | 36.8 | 21.1 | 42.1 | | 42.9 | 10.7 | 46.4 | | 36.6 | 17.1 | 46.3 | |
| VI |
| Yes | 29.4 | 24.0 | 47.1 | 0.357 | 24.1 | 41.4 | 34.5 | 0.353 | 24.3 | 35.1 | 40.6 | 0.349 |
| No | 34.7 | 36.7 | 29.0 | | 37.7 | 29.0 | 33.3 | | 37.0 | 32.0 | 31.0 | |
| Tumor size |
| <3 cm | 42.8 | 14.3 | 42.8 | 0.603 | 50.0 | 16.6 | 33.3 | 0.699 | 23.1 | 30.8 | 46.1 | 0.677 |
| ≥3 cm | 37.8 | 32.4 | 29.7 | | 34.8 | 30.4 | 34.8 | | 31.8 | 34.1 | 34.1 | |
| LNM |
| Yes | 26.0 | 26.0 | 48.0 | 0.140 | 32.7 | 30.6 | 37.0 | 0.886 | 29.6 | 33.8 | 36.6 | 0.755 |
| No | 37.8 | 37.8 | 24.3 | | 32.6 | 35.0 | 32.6 | | 35.4 | 32.3 | 32.3 | |
| TD |
| Well | 33.3 | 33.3 | 33.3 | 0.684 | 32.6 | 42.0 | 25.8 | 0.449 | 30.2 | 44.2 | 25.6 | 0.463 |
| Moderate | 40.0 | 30.0 | 30.0 | | 32.0 | 27.7 | 40.4 | | 36.2 | 27.6 | 36.2 | |
| Poor | 12.5 | 50.0 | 37.5 | | 45.5 | 36.4 | 18.2 | | 31.0 | 31.0 | 38.0 | |
| Stage |
| I | 41.7 | 33.3 | 25.5 | 0.459 | 36.4 | 55.0 | 9.1 | 0.498 | 46.7 | 33.3 | 20.0 | 0.629 |
| II | 37.5 | 37.5 | 25.0 | 0.291a | 32.4 | 29.4 | 38.2 | 0.959a | 31.1 | 33.3 | 35.6 | 0.893a |
| III | 25.0 | 20.0 | 55.0 | | 35.9 | 30.8 | 33.3 | | 26.9 | 38.5 | 34.6 | |
| IV | 28.6 | 43.0 | 28.6 | | 20.0 | 30.0 | 50.0 | | 36.8 | 21.1 | 42.1 | |
| Relapse |
| Yes | 23.5 | 41.2 | 35.3 | 0.556 | 16.1 | 42.0 | 42.0 | 0.045 | 27.3 | 38.6 | 34.1 | 0.553 |
| No | 37.8 | 31.1 | 31.1 | | 42.0 | 29.0 | 29.0 | | 36.4 | 31.8 | 31.8 | |
| Death |
| Yes | 15.8 | 36.8 | 47.4 | 0.146 | 16.7 | 40.0 | 43.3 | 0.070 | 25.0 | 37.5 | 37.5 | 0.450 |
| No | 40.0 | 31.1 | 28.9 | | 40.6 | 29.7 | 29.7 | | 36.2 | 31.9 | 31.9 | |
Patients with medium or high levels of EPAS1
mRNA in the plasma were found to have a distribution of data that
was similar to patients with low EPAS1 levels (Table II). Thus, patients exhibiting
medium and high EPAS1 levels were grouped together, and only
two categories were analyzed (low and high). Statistical
significance was evident between high levels of EPAS1 with
the clinicopathological parameters of age and relapse (p=0.003 and
p=0.013, respectively; Pearson’s Chi-square test). Furthermore, a
statistical association was found between high levels of
EPAS1 mRNA and patient mortality (p=0.021).
Association of mRNA levels with survival
of CRC patients
Patients were followed up for an average of 33.5
months (range 1–90). During this period, of the 154 CRC patients
examined in this study, 33.3% had relapsed at the time of the last
follow-up and 30% had succumbed to the disease. At 48 months
follow-up, DFS was 52.9% (95% CI 42.2–63.6) and OS was 64.9% (95%
CI 55.1–74.7).
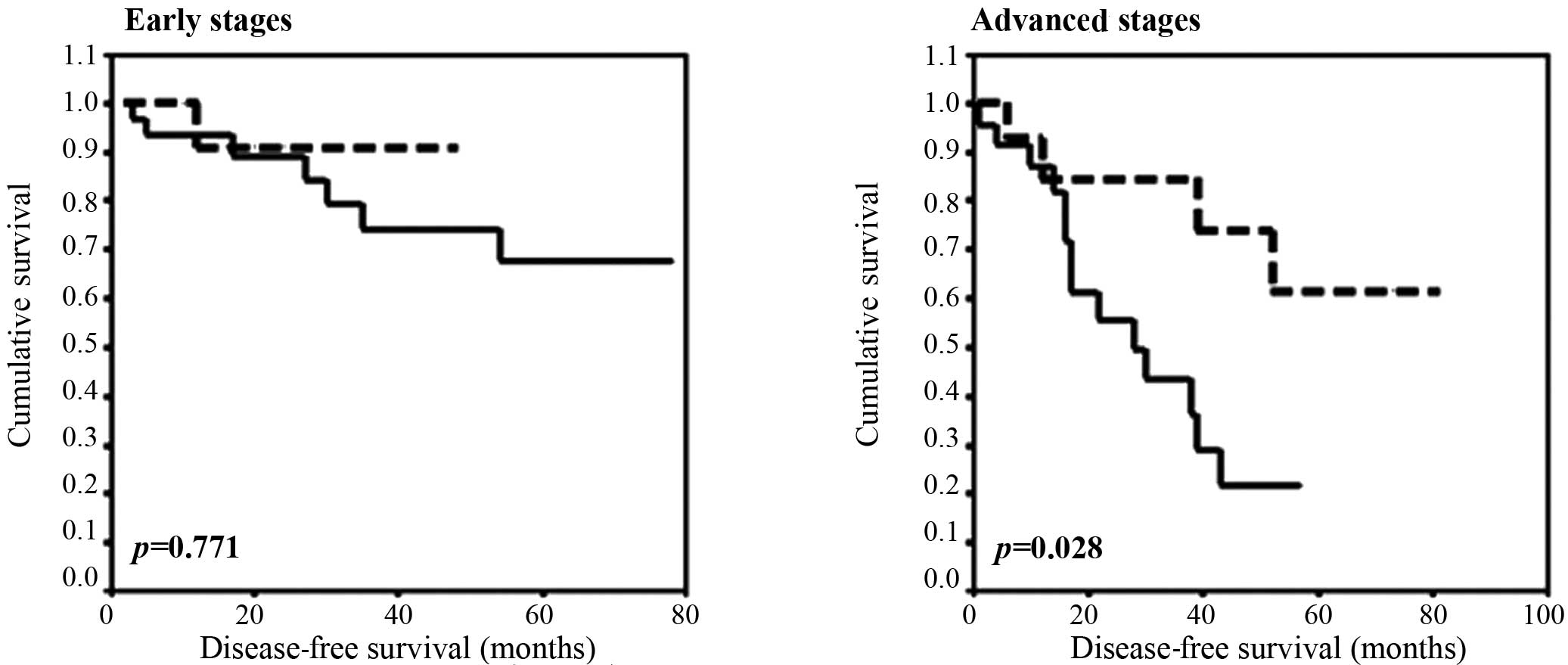
Disease-free survival
No apparent differences were observed regarding
KIAA0101, EPAS1 and UBE2D3 mRNA levels in the
plasma and DFS following the Kaplan-Meier analysis. However, when
patients divided into tertiles on the basis of their EPAS1
mRNA levels were sub-grouped as early (I + II) and advanced (III)
stages, a trend towards statistical association in the advanced
stages (III) was observed (p=0.082, Kaplan-Meier test). As
mentioned above, patients with medium and high levels of
EPAS1 mRNA levels in the plasma were grouped together and
only two categories were analyzed: patients with low or high
EPAS1 levels. Therefore, statistical significance was
evident for the mRNA levels of EPAS1 in advanced stages of
disease; patients with high levels of mRNA showed a 4-year DFS rate
of 21.7% (95% CI 0.9–42.5), while patients with low levels showed a
rate of 73.9% (95% CI 47.8–99.9) (p=0.028, Kaplan-Meier test,
Fig. 1).
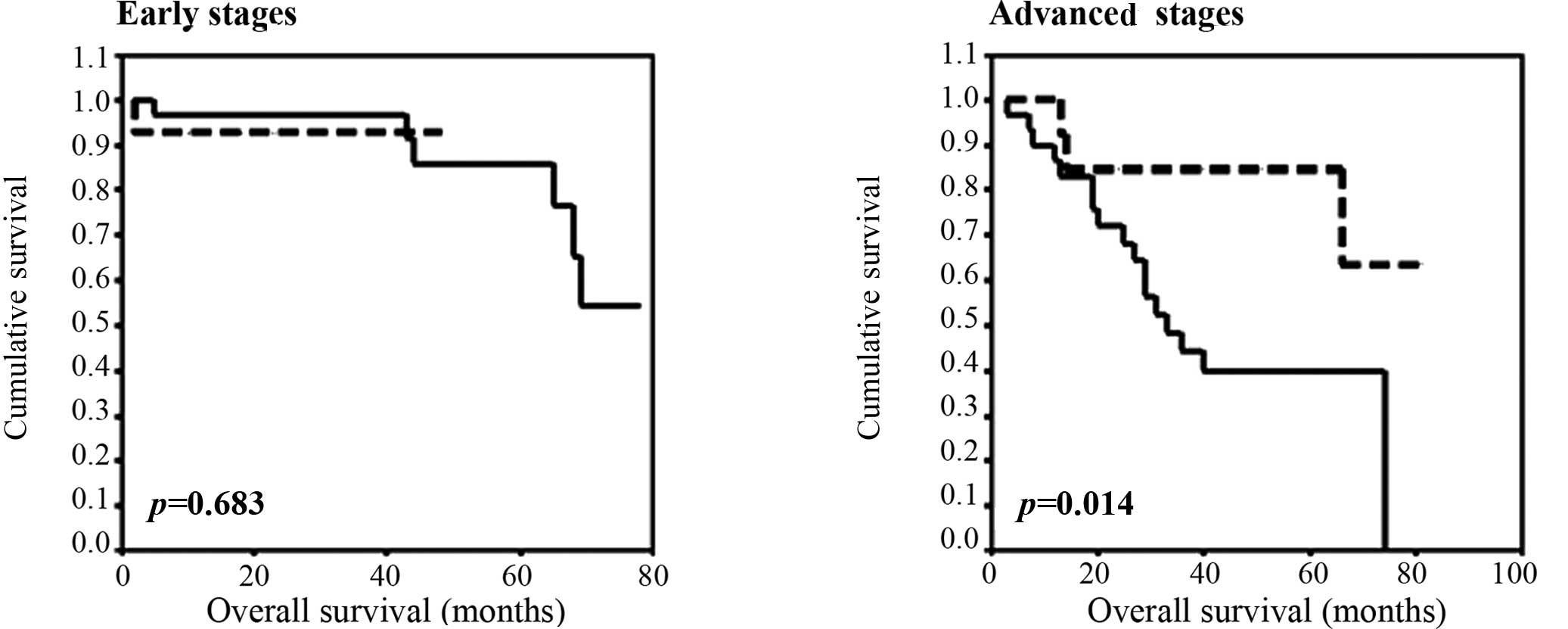
Overall survival
No differences were observed in OS for
KIAA0101 and UBE2D3 mRNA levels in the plasma.
However, a statistical association was found between high levels of
EPAS1 mRNA and shorter OS in patients at advanced stages
(III + IV) (p=0.047, Kaplan-Meier test). As with DFS, patients with
medium and high levels of EPAS1 mRNA levels in the plasma
were grouped together. The statistical significance was evident for
the high mRNA levels of EPAS1 at advanced stages of disease.
Thus, at 48 months follow-up for the advanced stage patients, OS
for patients with high levels was 39.8% (95% CI 20.8–58.9), while
that of patients with low levels was 84.6% (95% CI 65–100)
(p=0.014, Kaplan-Meier test, Fig.
2).
Discussion
Since genetic alterations are associated with an
altered expression of numerous genes at the mRNA level, plasma RNA
is a useful source of molecular information associated with tumor
development. The presence of RNA released from tumor cells in the
plasma of cancer patients has been proven. Plasma RNA in cancer
patients circulates highly protected by tumor-specific
microvesicle-like structures (18).
Moreover, these released microvesicles efficiently transfer content
to other cell types and have various effects on immune responses,
cell growth, angiogenesis and metastasis (19–21).
Plasma RNAs may provide prognostic information similar to RNA from
primary tumors. The potential use of plasma RNA in this field has
been reported (2,7,11,22,23).
In this sense, the application of genomic profiling of plasma RNA
may allow the unbiased selection of a cancer marker in the plasma
(19). In the context of cancer
research, DNA microarrays have been widely used to identify changes
that are common to various types of cancer, as well as signature
profiles unique to a sub-category of cancer. Furthermore, this
technology may be useful in the detection of early stages of the
disease, aiding prognosis, predicting response to specific
therapies and monitoring the treatment of disease (24).
In a previous study, the feasibility of a genomic
approach to studying plasma RNA was analyzed (12), as well as its diagnostic value. A
set of four genes (EPAS1, KIAA0101, UBE2D3 and
DDX46) was selected from a list of 40 differentially
expressed genes identified after profiling plasma RNA from CRC and
from healthy donors by cDNA microarray hybridization. The four
genes were analyzed by quantitative RT-PCR in new plasma samples,
which confirmed higher mRNA levels of EPAS1, KIAA0101
and UBE2D3 in the plasma of CRC patients as compared to
normal subjects. The levels of EPAS1 and UBE2D3 mRNA
were significantly lower in plasma samples obtained after surgery,
returning to normal levels, which shows a favorable correlation
between these indicators and the tumor condition. Moreover, class
prediction using support vector machines with a training set
composed of real-time PCR data for EPAS1 classified
pre-surgery samples as the CRC group and post-surgery samples as
the normal group (12).
In the present study, the mRNA level of
EPAS1, KIAA0101 and UBE2D3 in plasma was
analyzed to evaluate the prognostic value of the four genes.
Although the mRNA levels of KIAA0101 and UBE2D3 in
plasma may have diagnostic value (12), we found no prognostic value of these
indicators due to associations with clinicopathological parameters
and patient survival. However, high mRNA levels of EPAS1 in
plasma were associated with relapse of disease and patient
mortality, as well as with shorter DFS and OS in advanced stages.
EPAS1 is a factor induced under hypoxia and is an essential
mediator of cell adaptation to hypoxia, regulating the expression
of genes involved in tumor angiogenesis, glucose metabolism and
resistance to oxidative stress (25–27).
The overexpression of EPAS1 has been characterized in the
tumor tissue of various types of human cancer, including CRC, and
has shown a close correlation with metastatic tumor activity
(28–30). However, previous data on
EPAS1 and clinical outcome in CRC have been inconclusive,
showing both associations and no associations with poor survival
(30–32). Our results indicate that the results
reported by other investigators, who have shown that EPAS1
is an indicator of poor prognosis in tumors, a reasonable
assumption given the roles of this gene, may also be reflected in
the plasma of patients in advanced stages.
Acknowledgements
This study was supported by grants SAF2007-60214
from the Fundación Científica de la AECC, and CM:S-GEN/0266/2006,
ISCIII-RTICC-RD06/0020/0020 from the Fundación Banco Santander. The
study concept and design were by M.N. and S.J. M.N., R.M. and H.M
were involved in the acquisition of date, while G.V., G.J.M., D.G.,
P.C. and R.M carried out the analysis and interpretation of data.
M.N. and G.V. drafted the manuscript, and statistical analysis was
carried out by G.I., D.R., S.B. and H.A. B.F. was responsible for
obtaining funding for the study. Finally, S.J. and B.F. were
involved in the supervision of the study.
References
|
1
|
Schoen RE: The case for population-based
screening for colorectal cancer. Nat Rev Cancer. 2:65–70. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fleischhacker M and Schmidt B: Circulating
nucleic acids (CNAs) and cancer – a survey. Biochim Biophys Acta.
1775:181–232. 2007.
|
|
3
|
Leon SA, Shapiro B, Sklaroff DM and Yaros
MJ: Free DNA in the serum of cancer patients and the effect of
therapy. Cancer Res. 37:646–650. 1977.PubMed/NCBI
|
|
4
|
Shapiro B, Chakrabarty M, Cohn EM and Leon
SA: Determination of circulating DNA levels in patients with benign
or malignant gastrointestinal disease. Cancer. 51:2116–2120. 1983.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Silva JM, Rodriguez R, Garcia JM, Muñoz C,
Silva J, Dominguez G, Provencio M, España P and Bonilla F:
Detection of epithelial tumour RNA in the plasma of colon cancer
patients is associated with advanced stages and circulating tumour
cells. Gut. 50:530–534. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Silva J, Silva JM, Garcia JM, Dominguez G
and Bonilla F: RNA is more sensitive than DNA in identification of
breast cancer patients bearing tumor nucleic acids in plasma. Gene
Chromosome Cancer. 3:375–376. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Silva JM, Dominguez G, Silva J, Garcia JM,
Sanchez A, Rodriguez O, Provencio M and Bonilla F: Detection of
epithelial messenger RNA in the plasma of breast cancer patients is
associated with poor prognosis tumor characteristics. Clin Cancer
Res. 7:2821–2825. 2001.PubMed/NCBI
|
|
8
|
Dasi F, Lledo S, Garcia-Granero E, Ripoll
R, Marugan M, Tormo M, Garcia-Conde J and Aliño SF: Real-time
quantification in plasma of human telomerase reverse transcriptase
(hTERT) mRNA: a simple blood test to monitor disease in cancer
patients. Lab Invest. 81:767–769. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wong SC, Lo SF, Cheung MT, Ng KO, Tse CW,
Lai BS, Lee KC and Lo YM: Quantification of plasma catenin mRNA in
colorectal cancer and adenoma patients. Clin Cancer Res.
10:1613–1617. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Garcia V, Garcia JM, Peña C, Silva J,
Dominguez G, Lorenzo Y, Diaz R, Espinosa P, Garcia de Sola J,
Cantos B and Bonilla F: Free circulating mRNA in plasma from breast
cancer patients and clinical outcome. Cancer Lett. 263:312–320.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Garcia V, Garcia JM, Peña C, Silva J,
Dominguez G, Hurtado A, Alonso I, Rodriguez R, Provencio M and
Bonilla F: Thymidylate synthase mRNA expression in plasma from
colon cancer patients. Clin Cancer Res. 12:2095–2100. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Collado M, Garcia V, Garcia JM, Alonso I,
Lombardia L, Diaz-Uriarte R, Fernandez Luis Lopez, Zaballos A,
Bonilla F and Serrano M: Genomic profiling of circulating plasma
RNA for the analysis of cancer. Clin Chem. 53:1860–1863. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pilarsky C, Wenzig M, Specht T, Saeger HD
and Grutzmann R: Identification and validation of commonly
overexpressed genes in solid tumors by comparison of microarray
data. Neoplasia. 6:744–750. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
Large-scale meta-analysis of cancer microarray data identifies
common transcriptional profiles of neoplastic transformation and
progression. Proc Natl Acad Sci USA. 101:9309–9314. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yoshimura H, Dhar DK, Kohno H, Kubota H,
Fujii T, Ueda S, Kinugasa S, Tachibana M and Nagasue N: Prognostic
impact of hypoxia-inducible factors 1alpha and 2alpha in colorectal
cancer patients: correlation with tumor angiogenesis and
cyclooxygenase-2 expression. Clin Cancer Res. 10:8554–8560. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Saville MK, Sparks A, Xirodimas DP,
Wardrop J, Stevenson LF, Bourdon JC, Woods YL and Lane DP:
Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in
vivo. J Biol Chem. 279:42169–42181. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gonen H, Bercovich B, Orian A, Carrano A,
Takizawa C, Yamanaka K, Pagano M, Iwai K and Ciechanover A:
Identification of the ubiquitin carrier proteins, E2s, involved in
signal-induced conjugation and subsequent degradation of
IkappaBalpha. J Biol Chem. 74:14823–14830. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Garcia JM, Garcia V, Peña C, Dominguez G,
Silva J, Diaz R, Espinosa P, Citores MJ, Collado M and Bonilla F:
Extracellular plasma RNA from colon cancer patients is confined in
a vesicle-like structure and is mRNA-enriched. RNA. 14:1424–1432.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Al-Nedawi K, Meehan B, Micallef J, Lhotak
V, May L, Guha A and Rak J: Intercellular transfer of the oncogenic
receptor EGFRvIII by microvesicles derived from tumor cells. Nat
Cell Biol. 10:619–624. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Skog J, Würdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky
AM and Breakefield XO: Gliobastoma microvesicles transport RNA and
proteins that promote tumor growth and provide diagnostic
biomarkers. Nat Cell Biol. 10:1470–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hasselmann DO, Rappl G, Rossler M, Ugurel
S, Tilgen W and Reinhold U: Detection of tumor-associated
circulating mRNA in serum, plasma and blood cells from patients
with disseminated malignant melanoma. Oncol Res. 8:115–118.
2001.PubMed/NCBI
|
|
23
|
Kopreski MS, Benko FA and Gocke CD:
Circulating RNA as a tumor messenger in the serum of patient of
malignant melanoma. Clin Cancer Res. 5:1961–1965. 1999.
|
|
24
|
Duffy MJ, van Dalen A, Haglund C, Hansson
L and Kalapdor R: Clinical utility of biochemical markers in
colorectal cancer: European Group on tumor markers (EGTM). Eur J
Cancer. 39:718–727. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bertout JA, Patel SA and Simon MC: The
impact of O2 availability on human cancer. Nat Rev
Cancer. 8:967–975. 2008.
|
|
26
|
Wouters BG and Koritzinsky M: Hypoxia
signaling through mTOR and the unfolded protein response in cancer.
Nat Rev Cancer. 8:851–864. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Erler JT, Bennewith KL, Nicolau M,
Dornhofer N, Kong C, Le QT, Chi JT, Jeffrey SS and Giaccia AJ:
Lysyl oxidase is essential for hypoxia-induced metastasis. Nature.
440:1222–1226. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Flamme I, Krieg M and Plate KH:
Up-regulation of vascular endothelial growth factor in stromal
cells of hemangioblastomas is correlated with up-regulation of the
transcription factor HRF/HIF-2alpha. Am J Pathol. 153:25–29. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Talks KL, Turley H, Gatter KC, Maxwell PH,
Pugh CW, Ratcliffe PJ and Harris AL: The expression and
distribution of the hypoxia-inducible factors HIF-1alpha and
HIF-2alpha in normal human tissues, cancers, and tumor-associated
macrophages. Am J Pathol. 157:411–421. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Baba Y, Nosho K, Shima K, Irahara N, Chan
AT, Meyerhardt JA, Chung DC, Giovannucci EL, Fuchs CS and Ogino S:
HIF1A overexpression is associated with poor prognosis in a cohort
of 731 colorectal cancers. Am J Pathol. 176:2292–2301. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rasheed S, Harris AL, Tekkis PP, Turley H,
Silver A, McDonald PJ, Talbot IC, Glynne-Jones R, Northover JM and
Guenther T: Hypoxia-inducible factor-1α and -2α are expressed in
most rectal cancers but only hypoxia-inducible factor-1 α is
associated with prognosis. Br J Cancer. 100:1666–1673. 2009.
|
|
32
|
Yoshimura H, Dhar DK, Kohno H, Kubota H,
Fujii T, Ueda S, Kinugasa S, Tachibana M and Nagasue N: Prognostic
impact of hypoxia-inducible factors 1α and 2α in colorectal cancer
patients: correlation with tumor angiogenesis and cyclooxygenase-2
expression. Clin Cancer Res. 10:8554–8560. 2004.
|