Introduction
The anti-cancer potential of histone deacetylase
(HDAC) inhibitors has been broadly acknowledged (1,2). These
compounds block HDAC activity, resulting in profound increases in
the acetylation state of the chromatin, which in turn affects
chromatin structure and the regulation of gene expression (3). Previous studies using cDNA arrays
showed that the expression of as many as 7–10% of genes were
altered in cell lines of leukemia, multiple myeloma, and carcinomas
of the colon, bladder, kidney, prostate and breast, when cultured
for up to 48 h with butyrate, TSA, MS-275, vorinostat or FK228
(depsipeptide), using a 2-fold change as the cut-off value
(4–8). The patterns of alterations of gene
expression are similar for different HDAC inhibitors, but definite
differences were clearly induced by different agents in various
transformed cell types (9). The
development of HDAC inhibitor resistance is a major concern, as is
the case with any novel anti-tumor therapy. In preclinical studies,
resistance to HDAC inhibitor-induced transformed cell death was
noted in human bladder carcinoma cells (T24) and prostate cancer
cells (PC3) (10,11). However, the mechanisms underlying
HDAC inhibitor resistance remain to be clarified.
In this study, we evaluated the functional effects
of KBH-A42 on the growth of various cancer cell types (12,13).
Additionally, K562 (leukemia) cell lines were the most sensitive to
KBH-A42, and UM-UC-3 (bladder cancer) cells were the least
sensitive. Furthermore, in a human tumor xenograft model using
Balb/c nude mice, KBH-A42 was shown to significantly inhibit the
growth of K562 tumors, but slightly inhibited the growth of UM-UC-3
tumors. In an effort to determine the reason for the differential
response of K562 and UM-UC-3 cells to KBH-A42, cDNA microarray
analyses were conducted and confirmed by reverse
transcription-polymerase chain reaction (RT-PCR) on the two cell
types.
Materials and methods
Chemicals, cell lines and animals
All reagents were purchased from Sigma-Aldrich (St.
Louis, MO, USA) unless stated otherwise. KBH-A42 was synthesized
and supplied by Dr Gyoonhee Han at Yonsei University (Seoul,
Republic of Korea). KBH-A42 was dissolved in dimethyl sulfoxide
(DMSO) and freshly diluted in culture media for all in vitro
experiments. Female BALB/c-nu mice were purchased from SLC
(Hamahatsu, Japan) and maintained as previously described (14). All animals were permitted to
acclimate to the local environment for at least 1 week prior to
use. The cell lines CaSki, HeLa, Hep 3B, SNU709, A549, AsPC-1,
PC-3, A375, LOX-IMVI, M14 and AZ521 were cultured in RPMI-1640
(Gibco BRL, Carlsbad, CA, USA); the UM-UC-3, K562 and KB-3-1 cell
lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM;
Gibco BRL). All media were supplemented with 10% fetal bovine serum
(Hyclone, Logan, UT, USA), 2 mM L-glutamine, 100 U/ml of penicillin
and 100 μg/ml of streptomycin. The cells were maintained at 37°C in
5% CO2 humidified air.
Cell proliferation assay
Cells were plated at 0.8–1.5×104
cells/well in 96-well plates, incubated overnight and treated with
KBH-A42 for 48 h. Cell proliferation assays were performed using a
cell proliferation kit II (XTT Roche Applied Science Mannheim,
Penzberg, Upper Bavaria, Germany) in accordance with the
manufacturer’s instructions. The XTT labeling mixture was prepared
by mixing 50 volumes of 1 mg/ml sodium
3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis
(4-methoxy-6-nitro) benzenesulfonic acid hydrate with 1 volume of
0.383 mg/ml of N-methyldibenzopyrazine methyl sulfate. This XTT
labeling mixture was subsequently added to the cultures and
incubated for 2 h at 37°C. Absorbance was measured at 490 nm, with
650 nm as a reference wavelength.
Human tumor xenografts in nude mice
The K562 and UM-UC-3 cells were injected
subcutaneously into female BALB/c-nu mice. When the tumor volume
reached 100 mm3, the mice were distributed randomly and
treated with vehicle, KBH-A42 (100 mg/kg body weight, i.p., QD), or
doxorubicin (2 mg/kg body weight, i.p., Q2D) for 14 days. Following
14 days of treatment, the mice were sacrificed and all of the
tumors were removed and weighed.
Apoptosis analysis
Apoptosis was analyzed using an annexin V-FITC
apoptosis detection kit II (BD Biosciences, NJ, USA) in accordance
with the manufacturer’s instructions. In brief, the cells were
plated at 2-4×106 cells/dish in 100-mm dishes, incubated
overnight and treated for 24 h with the indicated concentrations of
KBH-A42. The cells were harvested, washed in phosphate-buffered
saline (PBS) and combined with a binding buffer containing annexin
V-FITC and propidium iodide (PI). Following 15 min of incubation in
the dark, the cells were analyzed via flow cytometry using a
FACSCalibur flow cytometer (BD Bioscience).
Caspase 3/7 assay
The activities of caspases 3 and 7 were determined
using a Caspase-Glo 3/7™ assay (Promega, Madison, WI, USA) in
accordance with the manufacturer’s instructions. In brief, the
cells were plated at 0.8–1.5×104 cells/well in 96-well
plates, incubated overnight and then treated for 24 h with the
indicated concentrations of KBH-A42. The culture supernatants were
transferred to a turbid microtiter plate and mixed with equal
volumes of Proluminescent caspase 3/7 substrate (Promega).
Following 1 h of incubation at 37°C, luminescence was measured with
a VICTOR™ light (PerkinElmer, NJ, USA).
RNA extraction
In brief, cells were plated at 2–4×106
cells/dish in 100-mm dishes, incubated overnight and treated for 24
h with KBH-A42 at the indicated concentrations. RNA was isolated
with a Qiagen RNeasy Plus Mini kit according to the manufacturer’s
instructions, and its quantity and purity were evaluated using the
A260/280 ratio (SmartSpec™ 3000, Bio-Rad, CA, USA).
cDNA microarray and data analysis
Profiling of gene expression was analyzed using a
Gene Agilent human 44K 4-plex chip (Digital Genomics, Korea) in
accordance with the manufacturer’s instructions. In brief,
approximately 2 μg of human total RNA was reverse-transcribed with
Cy3- or Cy5-conjugated dUTP (Amersham Pharmacia Biotech, Uppsala,
Sweden), respectively. After labeling the reaction for 1 h at 42°C,
the Cy3- and Cy5-labeled cDNA probes were mixed together and
hybridized to a microarray slide for 16 h at 60°C. The microarray
was scanned with a GenePix 4000B scanner (Axon Instruments, CA,
USA), and the scanned image was analyzed with GenePix v6.0 software
to determine the gene expression ratio. Raw data were normalized
via the locally weighted scatter-plot smoother (LOWESS)
normalization method. Normalized spot intensities were calculated
into gene expression ratios between the control and treatment
groups.
RT-PCR
Single-strand cDNA was synthesized from 2 μg of
total RNA, using AccuPower® RT premix (Bioneer, Korea).
PCR amplification was subsequently performed in a reaction volume
of 20 μl containing 2 μl of the appropriate cDNA, 1 μl of each set
of primers at a concentration of 10 pM and Accupower PCR premix
(Bioneer, Korea). The sequences of the primers used were:
HRK, forward: 5′-TGCTCGGCAGGC GGAACTTGTAG-3′ and reverse,
5′-GCTTCCCCAGTCCCA TTCTGTGTTT-3′; TNFRSF10B, forward:
5′-CTTGATTGT GGCTGTGTTTGTT-3′ and reverse, 5′-GCCACCTTTATC
TCATTGTCCA-3′; PYCARD, forward: 5′-CTCCTCAGTCGG CAGCCAAG-3′
and reverse, 5′-GGAGTGTTGCTGGGAA GGAG-3′; TNFRSF8, forward:
5′-CTGTGTCCCCTACCCA ATCT-3′ and reverse,
5′-CTTCTTTCCCTTCCTCTTCCA-3′. The RT-PCR products were
electrophoresed on a 1.5% agarose gel (Mupid-2) and visualized via
staining with ethidium bromide (GelDoc 1000, Bio-Rad),
respectively.
Statistical analysis
The results were expressed as the means ± SD.
One-way ANOVA followed by Dunnett’s t-test was employed for
multiple comparisons using GraphPad Prism (GraphPad Software Inc.,
CA, USA). P<0.05 was considered to be statistically
significant.
Results
Effect of KBH-A42 on human leukemia and
bladder cancer cell lines
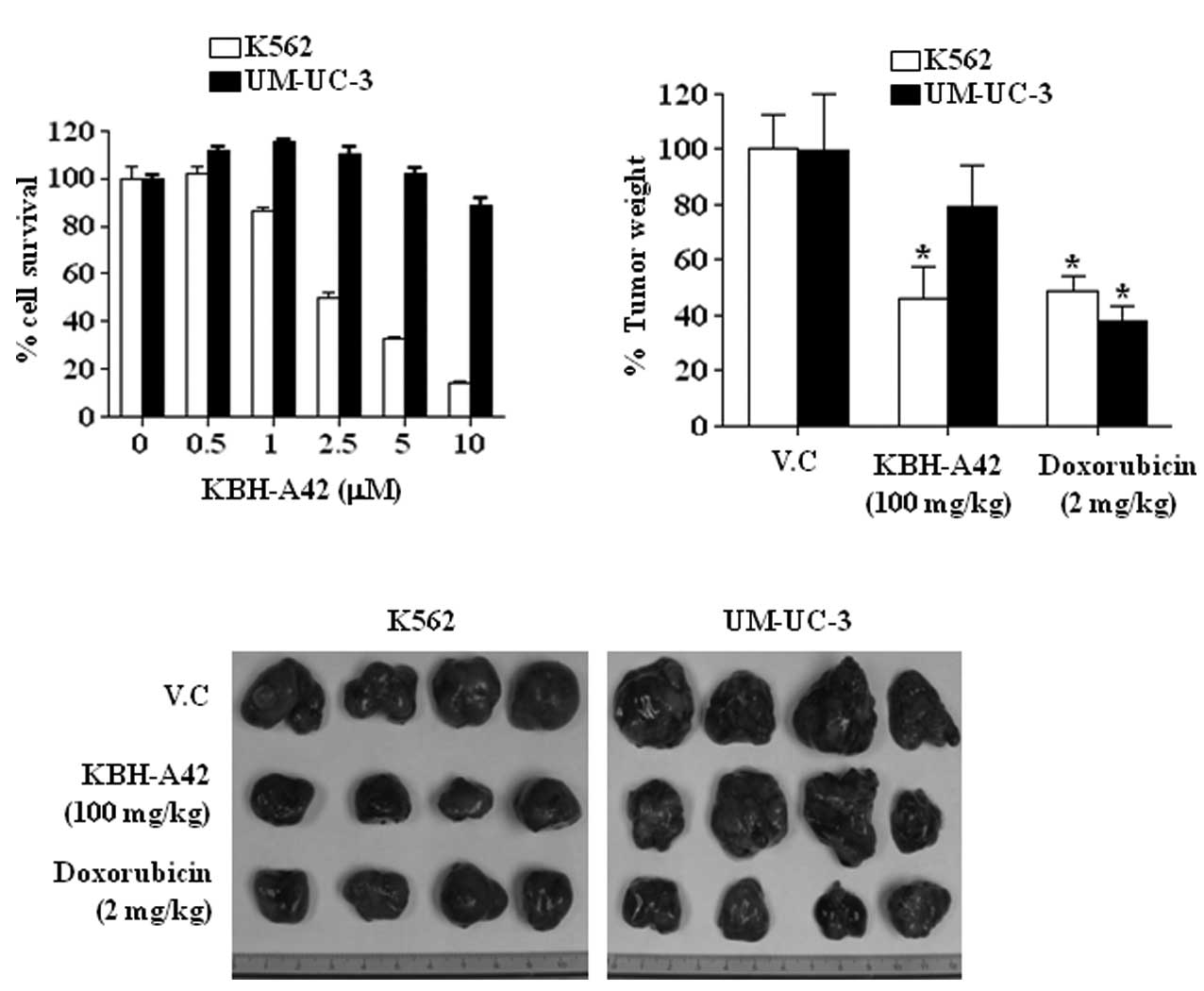
The effects of KBH-A42 on cell proliferation were
assessed in 14 human cancer cell lines obtained from 9 different
organs. Among them, the K562 human leukemia cells were the most
sensitive to KBH-A42 (GI50 = 1.41 μM), whereas the
UM-UC-3 bladder cancer cells were the least sensitive
(GI50 >10 μM) (Table
I). KBH-A42 inhibited the growth of K562 cells in a
dose-dependent manner. However, the growth of UM-UC-3 cells was not
affected at concentrations <5 μM of KBH-A42 and was inhibited by
only 11.3% at 10 μM (Fig. 1A).
Furthermore, to determine whether the in vitro effects of
KBH-A42 corresponded to the anti-tumor effects in vivo, we
investigated whether KBH-A42 affected tumor growth in a human tumor
xenograft model. As shown in Fig. 1B
and C, KBH-A42 (100 mg/kg, QD) or ADR (2 mg/kg, Q2D) induced a
37 or 40% inhibition of the growth of K562 tumors, respectively.
However, in the UM-UC-3 tumors, KBH-A42 and ADR inhibited growth by
21 or 67%, respectively (Fig. 1B and
C).
 | Table IEffect of KBH-A42 and SAHA on the
proliferation of various cell lines. |
Table I
Effect of KBH-A42 and SAHA on the
proliferation of various cell lines.
| Cell line | Origin | GI50
(μM) |
|---|
| |
|
|---|
| | KBH-A42 | ADR |
|---|
| UM-UC-3 | Bladder | >10 | 0.68 |
| K562 | Blood | 1.41 | 0.19 |
| CaSki | Cervix | 2.12 | 0.19 |
| HeLa | Cervix | 2.48 | 0.77 |
| KB-3-1 | Cervix | 3.54 | 0.51 |
| Hep 3B | Liver | 4.81 | 1.03 |
| SNU709 | Liver | 7.55 | 0.47 |
| A549 | Lung | 7.71 | 0.16 |
| AsPC-1 | Pancreas | 7.08 | 1.50 |
| PC-3 | Prostate | 9.88 | 0.76 |
| A375 | Skin | 4.44 | 0.11 |
| LOX-IMVI | Skin | 3.65 | 0.62 |
| M14 | Skin | 3.78 | 0.21 |
| AZ521 | Stomach | 7.49 | 0.21 |
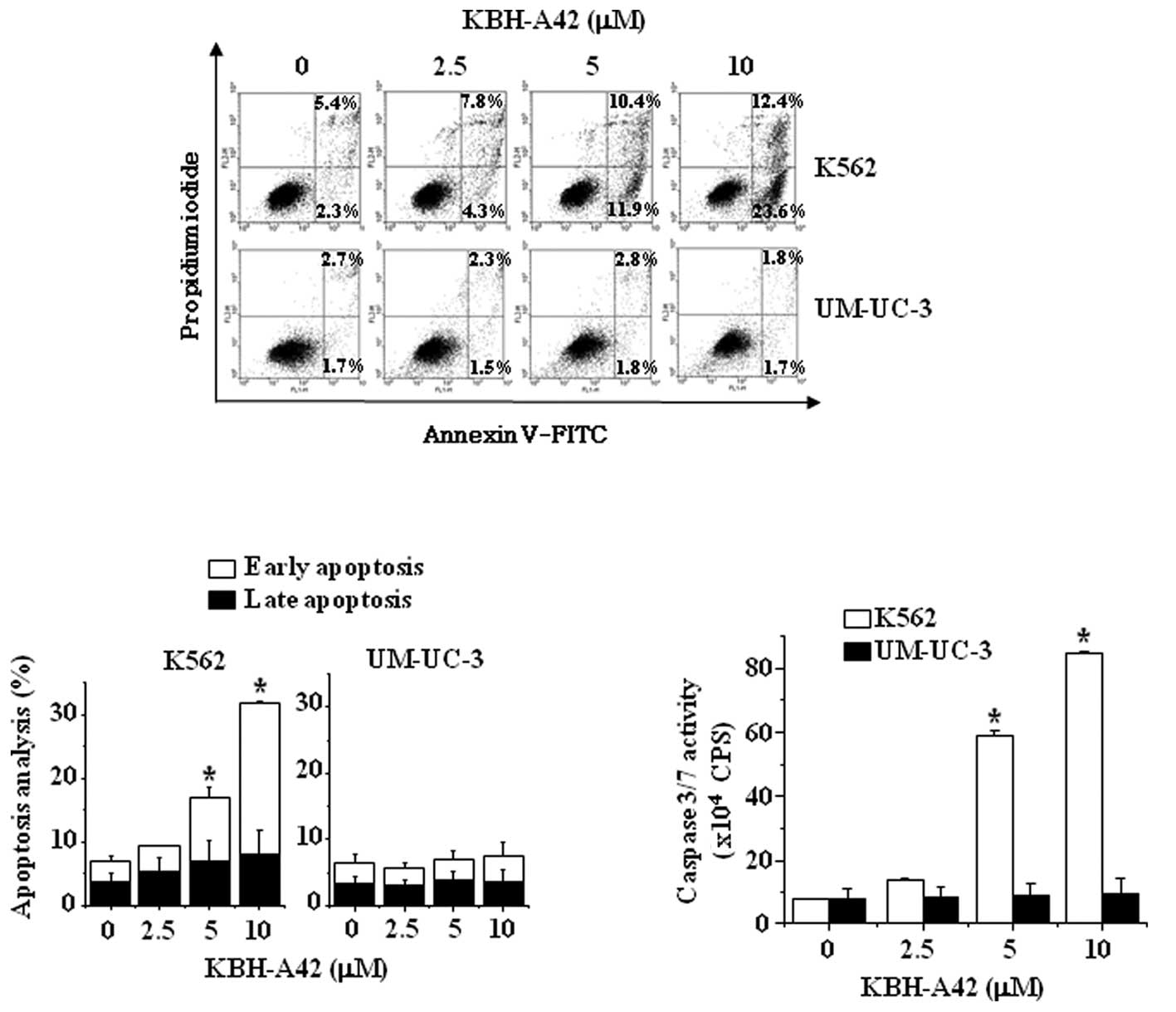
Induction of apoptosis and activation of
caspases by KBH-A42 in K562 and UM-UC-3 cells
To elucidate the mechanism underlying cell growth
inhibition by KBH-A42, the apoptosis profiles were analyzed via
flow cytometry. To detect apoptotic induction by KBH-A42, the cells
were stained with annexin V and PI. Whereas KBH-A42 induced
apoptosis by as much as 36% at 10 μM in the K562 cells, a 3.5%
induction of apoptosis was noted in the UM-UC-3 cells even at 10 μM
(Fig. 2A and B). These results were
consistent with the results shown in Fig. 2C, whereas KBH-A42 induced the
activation of caspases 3/7 in K562 cells, but not in UM-UC-3
cells.
Analysis of gene expression changes
caused by KBH-A42 treatment
To gain insight into the mechanism by which KBH-A42
inhibits cell growth, the mRNA expression profiles of K562 and
UM-UC-3 cells were analyzed in the presence and absence of KBH-A42.
Total RNAs were isolated from K562 and UM-UC-3 cells that were
either left untreated or treated with 5 μM KBH-A42 for 24 h,
labeled and hybridized to arrays (Agilent human 44K 4-plex chip),
and at least a 2-fold difference was detected between the untreated
and treated groups with regard to apoptosis-related genes. Among
the 139 apoptosis-related genes assessed, 14 (10.07%) and 17
(12.23%) genes were up-regulated (ratio >2.0) in the K562 and
UM-UC-3 cells, and 34 (24.46%) and 42 (30.22%) genes were
down-regulated (ratio <0.5) in the K562 and UM-UC-3 cells. The
most prominently up-regulated genes showing at least a two-fold
difference between the K562 and UM-UC-3 cells treated with KBH-A42
were: activity-regulated cytoskeleton-associated protein
(ARC), baculoviral IAP repeat-containing 3 (BIRC3),
CD70 molecule (CD70), CASP2 and RIPK1 domain-containing
adaptor with death domain (CRADD), harakiri (HRK),
myeloid cell leukemia sequence 1 (MCL1), tumor necrosis
factor receptor superfamily, member 10b (TNFRSF10B) and
tumor necrosis factor receptor superfamily, member 25
(TNFRSF25) (Table II). The
most profoundly down-regulated genes that also evidenced at least a
2-fold difference between K562 and UM-UC-3 cells were: B-cell
CLL/lymphoma2 (BCL2), BCL2-like 13 (BCL2L13), caspase
recruitment domain family, member 6 (CARD6), insulin-like
growth factor 1 receptor (IGF1R), PYD and CARD domain
containing protein gene (PYCARD), RPA interacting protein,
transcript variant 2 (RIP), TRAF family member-associated
NF-κB activator (TANK), tumor necrosis factor receptor
superfamily, member 8 (TNFRSF8) and tumor protein p53
binding protein 2 (TP53BP2) (Table II).
 | Table IIUp- and down-regulated genes by
KBH-A42 treatment in K562 and UM-UC-3 cells. |
Table II
Up- and down-regulated genes by
KBH-A42 treatment in K562 and UM-UC-3 cells.
| GeneBank
number | Gene symbol | Fold change vs.
control |
|---|
| |
|
|---|
| | K562 | UM-UC-3 |
|---|
| Up-regulated
genes |
| NM_015193 | ARC | 7.71 | 2.49 |
| NM_001165 | BIRC3 | 12.25 | 2.23 |
| NM_001252 | CD70 | 5.06 | 1.13 |
| NM_003805 | CRADD | 2.52 | 1.07 |
| NM_003806 | HRK | 5.69 | 0.40 |
| NM_021960 | MCL1 | 3.43 | 0.91 |
| NM_003842 |
TNFRSF10B | 5.30 | 2.65 |
| NM_148968 |
TNFRSF25 | 10.62 | 7.99 |
| Down-regulated
genes |
| NM_000633 | BCL2 | 0.35 | 1.39 |
| NM_015367 | BCL2L13 | 0.45 | 1.27 |
| NM_032587 | CARD6 | 0.18 | 0.79 |
| BC010607 | IGF1R | 0.47 | 1.26 |
| NM_013258 | PYCARD | 0.20 | 1.42 |
| NM_032308 | RIP | 0.44 | 1.60 |
| NM_133484 | TANK | 0.41 | 1.38 |
| NM_001243 | TNFRSF8 | 0.14 | 0.76 |
| NM_005426 | TP53BP2 | 0.44 | 1.23 |
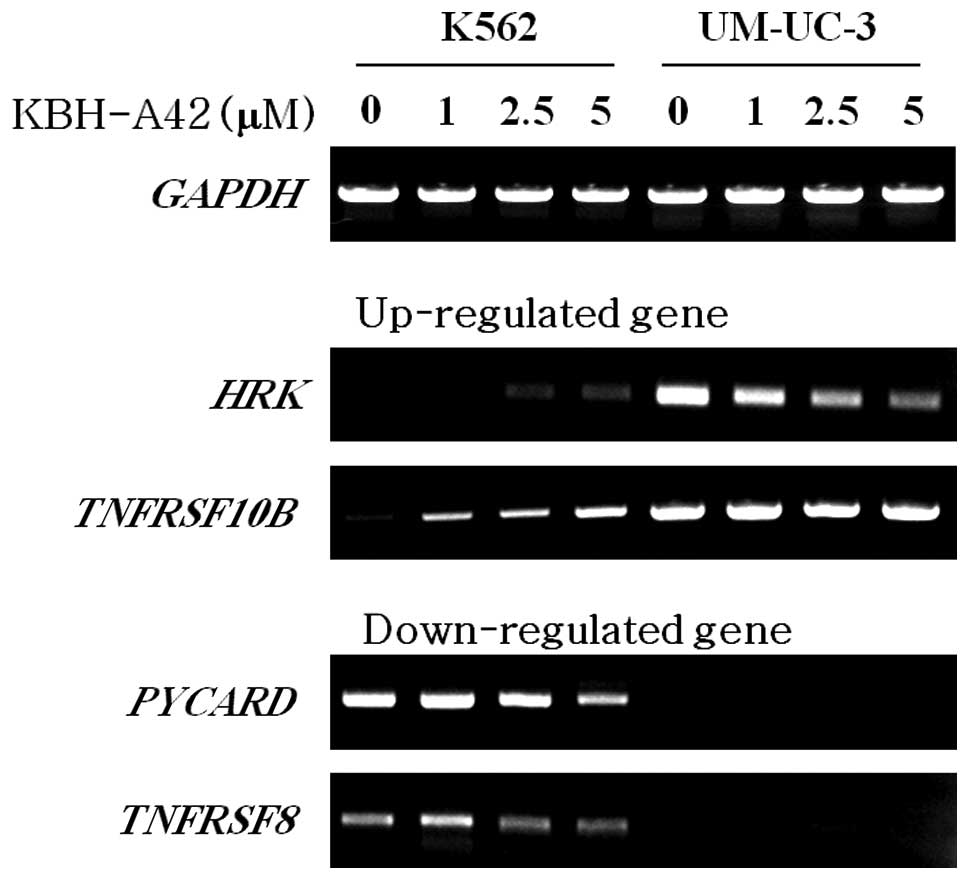
Effect of KBH-A42 on the expression of
apoptosis-related genes
Since KBH-A42 induced apoptosis in K562 cells but
not in UM-UC-3 cells, apoptosis-related genes that were altered by
KBH-A42 were selected from the cDNA microarray results and
subjected to RT-PCR for confirmation. Two genes, HRK and
TNFRSF10B, were up-regulated by KBH-A42, as demonstrated by
the results of the microarray analysis. The expression of
HRK mRNA was reduced in the UM-UC-3 cells, but increased
slightly in the K562 cells. The expression of the TNFRSF10B
mRNA was greatly increased as a result of KBH-A42 treatment in the
K562 cells, but was unaltered in the UM-UC-3 cells (Fig. 3). The mRNA expression of
PYCARD and TNFRSF8, which was down-regulated by
KBH-A42 in the cDNA microarray, was re-examined and confirmed via
RT-PCR. PYCARD and TNFRSF10B mRNA showed a reduced
expression as a result of KBH-A42 treatment in the K562 cells, but
was not detected in the UM-UC-3 cells (Fig. 3).
Discussion
In this study, we demonstrated that a novel
δ-lactam-based HDAC inhibitor, KBH-A42, inhibited the growth of 14
selected cancer cell lines. The findings described have shown that
KBH-A42 significantly suppressed the growth of almost all of the
tested cancer cell lines, but certain cell types were more
susceptible to the compound than others. Whereas the leukemia cell
lines were the most sensitive to KBH-A42, the glioma and prostate
cancer cell lines were weakly sensitive, and the bladder cancer
cell line was the least sensitive. Our findings demonstrate the
cell type-specific growth inhibitory effect of KBH-A42. In a human
tumor xenograft model using Balb/c nude mice, KBH-A42 has been
shown to inhibit the growth of K562 tumors significantly, but only
slightly inhibited the growth of UM-UC-3 tumors.
Since HDAC inhibitors have been reported to induce
apoptosis in a variety of cancer cell lines (15–17),
we evaluated the effect of KBH-A42 on the apoptosis of K562 and
UM-UC-3 cells. The results of the apoptosis analysis demonstrate
that the growth inhibition of K562 cells by KBH-A42 was mediated,
at least in part, by the induction of apoptosis, but that of
UM-UC-3 cells was not. The in vitro caspase assays, which
measured the involvement of the major effectors of apoptosis,
showed a significant difference between the activities of caspase
3/7.
A cDNA microarray was conducted to screen for any
changes in the expression of apoptosis-associated genes. The DNA
microarray technique is a powerful method, allowing for the
simultaneous analysis of the expression levels of multiple genes.
In this study, gene expression was profiled using cDNA microarrays
on KBH-A42-treated cells. A number of apoptosis-related genes were
identified from the list of the genes modulated by KBH-A42 on the
cDNA microarray assays. Among them, four genes that were up- or
down-regulated by KBH-A42 were selected and their expression
patterns were confirmed via RT-PCR.
In the present study, 139 apoptosis-related genes
whose expression was altered by KBH-A42 treatment in K562 and
UM-UC-3 cells were screened out. Eight genes that were up-regulated
(ratio >2.0) and showed at least a 2-fold difference between the
K562 and UM-UC-3 cells were apoptosis-associated genes. In
mammalian cells, the pro-survival members (Bcl-2, Bcl-xL, Bcl-w,
Mcl-1 and A1) oppose the pro-apoptotic members, which consist of
the Bax group (Bax, Bak and Bok) and the BH3-only proteins (Bim,
Bad, Bid, Bik, Bmf, Puma, Noxa and HRK). The BH3-only proteins
monitor cellular well-being and, when activated by cytotoxic
signals, engage pro-survival relatives by inserting the BH3 domain
into a hydrophobic groove on their surfaces (18). The expression of HRK induced rapid
cell death, which was repressed by Bcl-2 and Bcl-xL. It was
determined that the deletion of the BH3 region eliminated the
ability of HRK to interact with Bcl-2 or Bcl-xL and eliminated or
profoundly attenuated HRK killing activity (19). In this study, HRK was
slightly up-regulated in K562 cells but significantly
down-regulated in UM-UC-3 cells as a result of KBH-A42 treatment.
In addition, Takimoto et al previously asserted that
TNFRSF10B was a viable tumor suppressor candidate, and the
overexpression of TNFRSF10B induced apoptosis in tumor cells
in a p53-dependent manner (20).
The results of this study have shown that KBH-A42 up-regulated the
expression of TNFRSF10B in K562 cells in a
concentration-dependent manner, but caused no such changes in
UM-UC-3 cells.
In addition, nine genes were found to be
down-regulated (ratio <0.5) and exhibited more than a 2-fold
difference between the K562 and UM-UC-3 cells; these cells were
also associated with apoptosis. Apoptosis-associated speck-like
protein containing a C-terminal caspase-recruitment domain (CARD)
is a protein encoded by the PYCARD gene in humans. This gene
encodes for an adaptor protein composed of a PYD and a CARD domain
(21). PYCARD mediates the assembly
of large signaling complexes in the inflammatory and apoptotic
signaling pathways, via caspase activation. In K562 cells,
PYCARD was reduced in a dose-dependent manner and was not
detected in UM-UC-3 cells. TNFRSF8, which is involved in apoptosis,
is a member of the TNF receptor superfamily, which is capable of
inducing apoptosis by activating the caspase pathway (22). In K562 cells, TNFRSF8 was
reduced in a dose-dependent manner and was not detected in UM-UC-3
cells. Thus, KBH-A42 may inhibit cell growth by regulating the
expression of apoptosis-related genes, and may have better efficacy
in leukemia.
In conclusion, the results of this study have
demonstrated that KBH-A42 inhibited the growth of cancer cells such
as K562 cells under in vitro and in vivo conditions,
and also that the growth inhibitory effects of KBH-A42 appear to be
mediated by apoptosis. In the microarray assays, apoptosis-related
genes containing ARC, BIRC3, CD70,
CRADD, HRK, MCL1, TNFRSF10B and
TNFRSF25 were up-regulated and BCL2, BCL2L13,
CARD6, IGF1R, PYCARD, RIP, TANK,
TNFRSF8 and TP53BP2 were down-regulated. Among these
genes, changes in the expression of HRK, TNFRSF10B,
PYCARD and TNFRSF8 mRNAs was confirmed via RT-PCR.
Collectively, our results have shown that KBHA42 possesses
anti-cancer properties and has a strong potential as a therapeutic
candidate with improved efficacy on leukemia under in vitro
and in vivo conditions. However, different cells may be
regulated differentially by KBH-A42.
Acknowledgements
This study was supported by the Bio R&D program
through the Korea Science and Engineering Foundation funded by the
Ministry of Education, Science and Technology (2008-0029594) and
the KRIBB Research Initiative program.
References
|
1
|
Saito A, Yamashita T, Mariko Y, et al: A
synthetic inhibitor of histone deacetylase, MS-27-275, with marked
in vivo antitumor activity against human tumors. Proc Natl Acad Sci
USA. 96:4592–4597. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Saunders NA, Popa C, Serewko MM, Jones SJ,
Dicker AJ and Dahler AL: Histone deacetylase inhibitors: novel
anticancer agents. Expert Opin Investig Drugs. 8:1611–1621. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grunstein M: Histone acetylation in
chromatin structure and transcription. Nature. 389:349–352. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chambers AE, Banerjee S, Chaplin T, et al:
Histone acetylation-mediated regulation of genes in leukaemic
cells. Eur J Cancer. 39:1165–1175. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Glaser KB, Staver MJ, Waring JF, Stender
J, Ulrich RG and Davidsen SK: Gene expression profiling of multiple
histone deacetylase (HDAC) inhibitors: defining a common gene set
produced by HDAC inhibition in T24 and MDA carcinoma cell lines.
Mol Cancer Ther. 2:151–163. 2003.PubMed/NCBI
|
|
6
|
Mitsiades CS, Mitsiades NS, McMullan CJ,
et al: Transcriptional signature of histone deacetylase inhibition
in multiple myeloma: biological and clinical implications. Proc
Natl Acad Sci USA. 101:540–545. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peart MJ, Smyth GK, van Laar RK, et al:
Identification and functional significance of genes regulated by
structurally different histone deacetylase inhibitors. Proc Natl
Acad Sci USA. 102:3697–3702. 2005. View Article : Google Scholar
|
|
8
|
Sasakawa Y, Naoe Y, Sogo N, et al: Marker
genes to predict sensitivity to FK228, a histone deacetylase
inhibitor. Biochem Pharmacol. 69:603–616. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gray SG, Qian CN, Furge K, Guo X and Teh
BT: Microarray profiling of the effects of histone deacetylase
inhibitors on gene expression in cancer cell lines. Int J Oncol.
24:773–795. 2004.PubMed/NCBI
|
|
10
|
Butler LM, Agus DB, Scher HI, et al:
Suberoylanilide hydroxamic acid, an inhibitor of histone
deacetylase, suppresses the growth of prostate cancer cells in
vitro and in vivo. Cancer Res. 60:5165–5170. 2000.PubMed/NCBI
|
|
11
|
Richon VM, Sandhoff TW, Rifkind RA and
Marks PA: Histone deacetylase inhibitor selectively induces p21WAF1
expression and gene-associated histone acetylation. Proc Natl Acad
Sci USA. 97:10014–10019. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kang MR, Kang JS, Han SB, et al: A novel
delta-lactam-based histone deacetylase inhibitor, KBH-A42, induces
cell cycle arrest and apoptosis in colon cancer cells. Biochem
Pharmacol. 78:486–494. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kang MR, Lee K, Kang JS, et al: KBH-A42, a
histone deacetylase inhibitor, inhibits the growth of
doxorubicin-resistant leukemia cells expressing P-glycoprotein.
Oncol Rep. 23:801–809. 2010.PubMed/NCBI
|
|
14
|
Lee SD, Park SK, Lee ES, et al: A
lipid-soluble red ginseng extract inhibits the growth of human lung
tumor xenografts in nude mice. J Med Food. 13:1–5. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Louis M, Rosato RR, Brault L, et al: The
histone deacetylase inhibitor sodium butyrate induces breast cancer
cell apoptosis through diverse cytotoxic actions including
glutathione depletion and oxidative stress. Int J Oncol.
25:1701–1711. 2004.
|
|
16
|
Doi S, Soda H, Oka M, et al: The histone
deacetylase inhibitor FR901228 induces caspase-dependent apoptosis
via the mitochondrial pathway in small cell lung cancer cells. Mol
Cancer Ther. 3:1397–1402. 2004.PubMed/NCBI
|
|
17
|
Roh MS, Kim CW, Park BS, et al: Mechanism
of histone deacetylase inhibitor Trichostatin A induced apoptosis
in human osteosarcoma cells. Apoptosis. 9:583–589. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Willis SN and Adams JM: Life in the
balance: how BH3-only proteins induce apoptosis. Curr Opin Cell
Biol. 17:617–625. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Inohara N, Ding L, Chen S and Nunez G:
Harakiri, a novel regulator of cell death, encodes a protein that
activates apoptosis and interacts selectively with
survival-promoting proteins Bcl-2 and Bcl-X(L). EMBO J.
16:1686–1694. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takimoto R and El-Deiry WS: Wild-type p53
transactivates the KILLER/DR5 gene through an intronic
sequence-specific DNA-binding site. Oncogene. 19:1735–1743. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stehlik C, Lee SH, Dorfleutner A,
Stassinopoulos A, Sagara J and Reed JC: Apoptosis-associated
speck-like protein containing a caspase recruitment domain is a
regulator of procaspase-1 activation. J Immunol. 171:6154–6163.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Staber PB, Noehammer C, Durkop H, et al:
mRNA expression patterns indicate CD30 mediated activation of
different apoptosis pathways in anaplastic large cell lymphoma but
not in Hodgkin’s lymphoma. Leuk Res. 30:343–348. 2006.PubMed/NCBI
|