Introduction
The P3/NS1/1-Ag4-1 (NS-1) cell line, which is
derived from a BALB/c mouse with myeloma, synthesizes the κ light
chain but not the heavy chain and is a non-Ig-secreting subclone of
P3X27 (1,2). It was widely used in hybridoma
technology for the production of monoclonal antibodies (McAbs) due
to its high proliferation and cell-fusion rate (3). The property of synthesizing the κ
light chain of Immunoglobulin G (IgG1) with non-secretion causes
certain problems in antibody engineering, for example the
humanization of the murine-derived McAbs. To the best of our
knowledge, there have been no studies performed concerning the
sequence of the variable region gene of NS-1 cells. In this study,
we synthesized 17 primers, 4 pairs of heavy chain primers and 9
light chain primers, to clone and sequence the genes encoding the
variable region of the NS-1 cells using reverse transcription PCR
(RT-PCR).
Materials and methods
Cell line, plasmids and main
reagents
The NS-1 cell line was purchased from CellBank
Australia (Wentworthville, Australia). The mouse hybridoma cell
line ZCH-7-2F9 (2F9), which generated anti-human CD14 McAb, was
established by our laboratory (4).
E. coli DH5α, the restriction enzyme EcoRI, Taq DNA
polymerase, M-MLV reverse transcriptase, RNasin®, RQ1
RNase-free DNase, pGEM®-T easy Vector system, X-gal,
IPTG and TRIzol total RNA extract reagent were purchased from
Invitrogen Life Technologies (Carlsbad, CA, USA). T4 DNA ligase,
RPMI-1640 medium and fetal bovine serum were purchased from
Gibco-BRL (Carlsbad, CA, USA) and the DL2000 marker was purchased
from Takara Bio, Inc. (Shiga, Japan). The QIAquick Gel Extraction
kit was purchased from Qiagen (Hilden, Germany).
Olig(dT)12–18 primers were purchased from Promega
(Mannheim, Germany).
Primer synthesis
The primers are cited by Chiang et al
(5) and were synthesized by Sangon
Biotech (Shanghai) Co,. Ltd. (Shanghai, China). The sequences of
the primers are listed in Table
I.
 | Table IDNA sequences of the primers used in
reverse transcription PCR analysis. |
Table I
DNA sequences of the primers used in
reverse transcription PCR analysis.
| Primer number | Primer sequence
(5′-3′) |
|---|
| Primers for the
variable region of the murine heavy chain |
| Forward primers
complementary to the leading sequence |
| P1 |
atggaatgcagctgggtcatcctctt |
| P2 |
atgggatggagctgtgtaatgctctt |
| P3 |
atgaacttcgggctgagcttgatttt |
| P4 |
atggctgtcttggggctgctcttct |
| Reverse primers
complementary to the J fragment |
| P5 |
tgaggagacggtgaccatggtccc |
| P6 |
tgaggagactgtgagagtggtgcc |
| P7 |
tgcagagacagtgaccagactccc |
| P8 |
tgaggagacggtgactgaggtccc |
| Primers for the
variable region of the murine light chain |
| Forward primers
complementary to the leading sequence |
| P9 |
atggagacagacacactcctgctat |
| P10 |
atggattttcaagtgcagattttcag |
| P11 |
atggagtcacagactcaggtctttata |
| P12 |
atggccccaactcagctcctggt |
| P13 |
atgaagttgcctgttaggctgttg |
| Reverse primers
complementary to the Cκ and J fragment |
| P14 |
ccgttttatttccaacttggtccc |
| P15 |
ccgtttgatttcctgcttggtgcc |
| P16 |
ccgtttcagctccagcttggtccc |
| P17 |
ggatacagttggtgcagcatcagcccgttt |
Cell culture
The NS-1 mouse myeloma and 2F9 mouse hybridoma cell
lines were maintained in RPMI-1640 medium supplemented with
antibiotics and 10% fetal bovine serum at 37°C, 5% CO2
and saturated humidity.
Extraction of total RNA
For the isolation of total RNA from the NS-1 and 2F9
cells, TRIzol reagent was used according to the manufacturer's
instructions. Prior to reverse transcription, the total RNA was
digested with RNase-free DNase and the quality was determined by
agarose gel electrophoresis and ultraviolet spectrophotometer
analysis. The cDNA coding for the variable chains was synthesized
from the total RNA template using murine leukemia virus reverse
transcriptase and Olig(dT)12–18 primers.
Cloning and sequencing
All DNA manipulation and bacterial transformations
were based on the methods described by Sambrook et al
(6). The conditions for PCR
amplification were as follows: pre-denaturation at 94°C for 2 min,
30 cycles of denaturation at 94°C for 30 sec, annealing at 57°C for
30 sec, extension at 72°C for 30 sec and, following the final
cycle, an additional extension at 72°C for 5 min. The PCR products
were purified according to the QIAquick DNA reagent kit
instructions. The concentration was determined using an ultraviolet
spectrophotometer. At a molar ratio of 3:1, the DNA fragment of
interest and the pGEM®-T easy vector were combined by T4
ligase in a 16°C water bath overnight. The product (5 μl) was then
transferred to competent DH5α bacteria. Positive recombinants were
selected on a Luria-Bertain (LB) plate with X-gal, IPTG and 100
μg/ml Amp. The white bacterial colonies were amplified and plasmids
were extracted and purified using the QIAquick DNA reagent kit.
Following further determination with the EcoRI restriction
enzyme and 1% agarose gel electrophoresis, the DNA of the positive
recombinants was sequenced.
The genetic sequencing was performed using the
dideoxynucleotide chain termination method with an automatic
sequencer (ABI PRISM377). The sequence was determined using BLAST
analysis (IMGT, the international ImMunoGeneTics information
system® http://www.imgt.org).
The study was approved by the Ethics Committee of
The Children's Hospital of Zhejiang University School of
Medicine.
Results
Extraction of total RNA
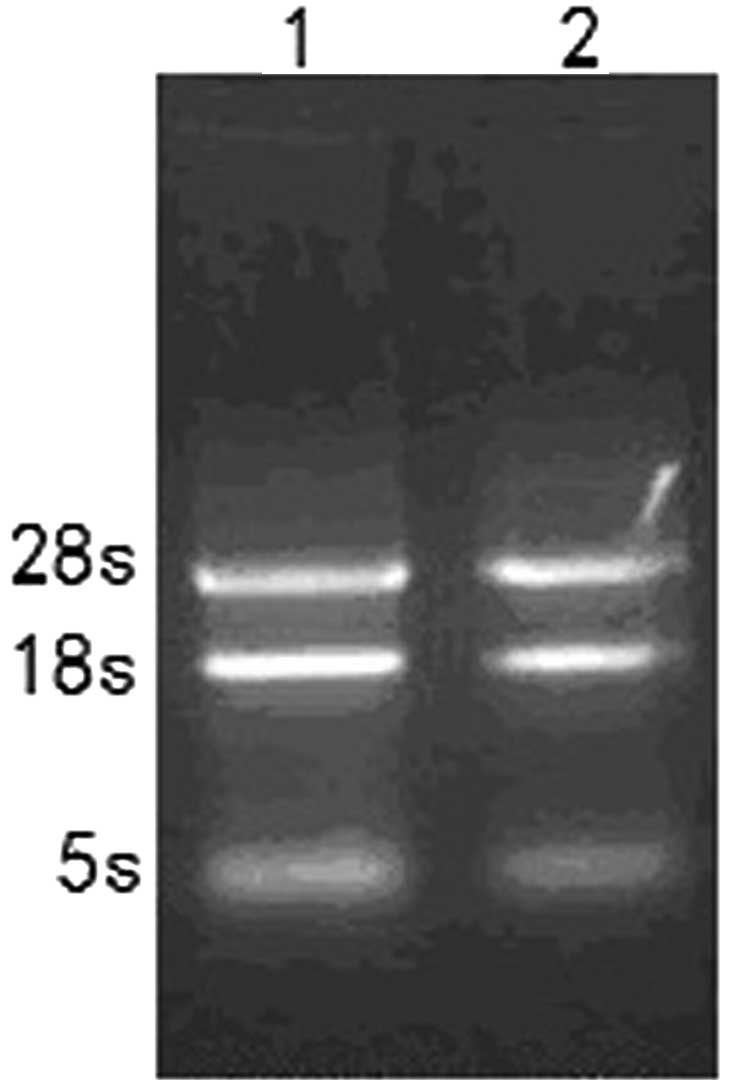
The total RNA was quantified using agarose gel
electrophoresis. On the gel there were 3 clear bands of 28S, 18S
and 5S ribosomal RNAs (Fig. 1). The
A260/A280 ratio of the total RNA preparations was between 1.8 and
2.0. The yields of the total RNA were ~40 and 55 μg/107
cells for NS-1 and 2F9, respectively.
Cloning and sequencing
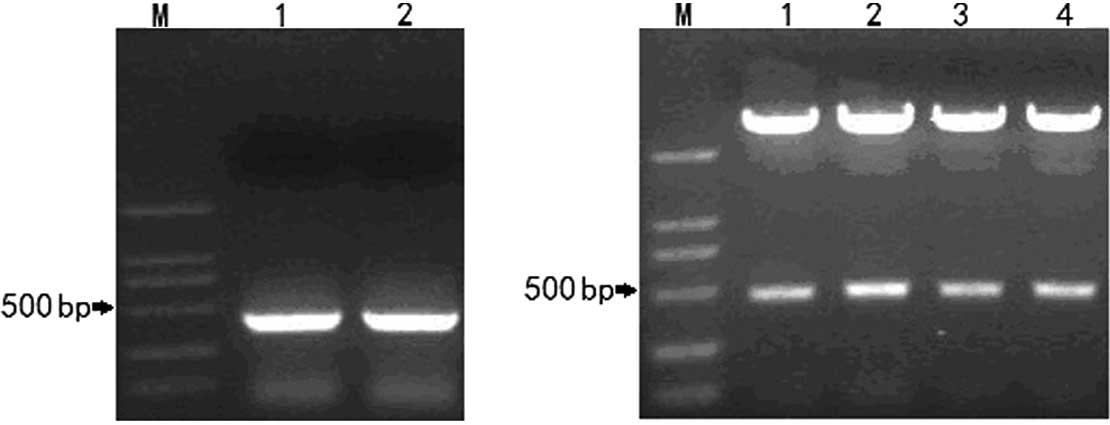
Following PCR screening of the cDNA templates of the
NS-1 and 2F9 cells, the light chain primers P9 and P14 amplified a
specific band with a size of 392 bp in the NS-1 and 2F9 cells
(Fig. 2A), whereas all the heavy
chain primers were negative in the NS-1 cells. The positive
recombinants contained the band of interest with a size of ~400 bp
according to the results of the EcoRI digestion and agarose
gel electrophoresis (Fig. 2B).
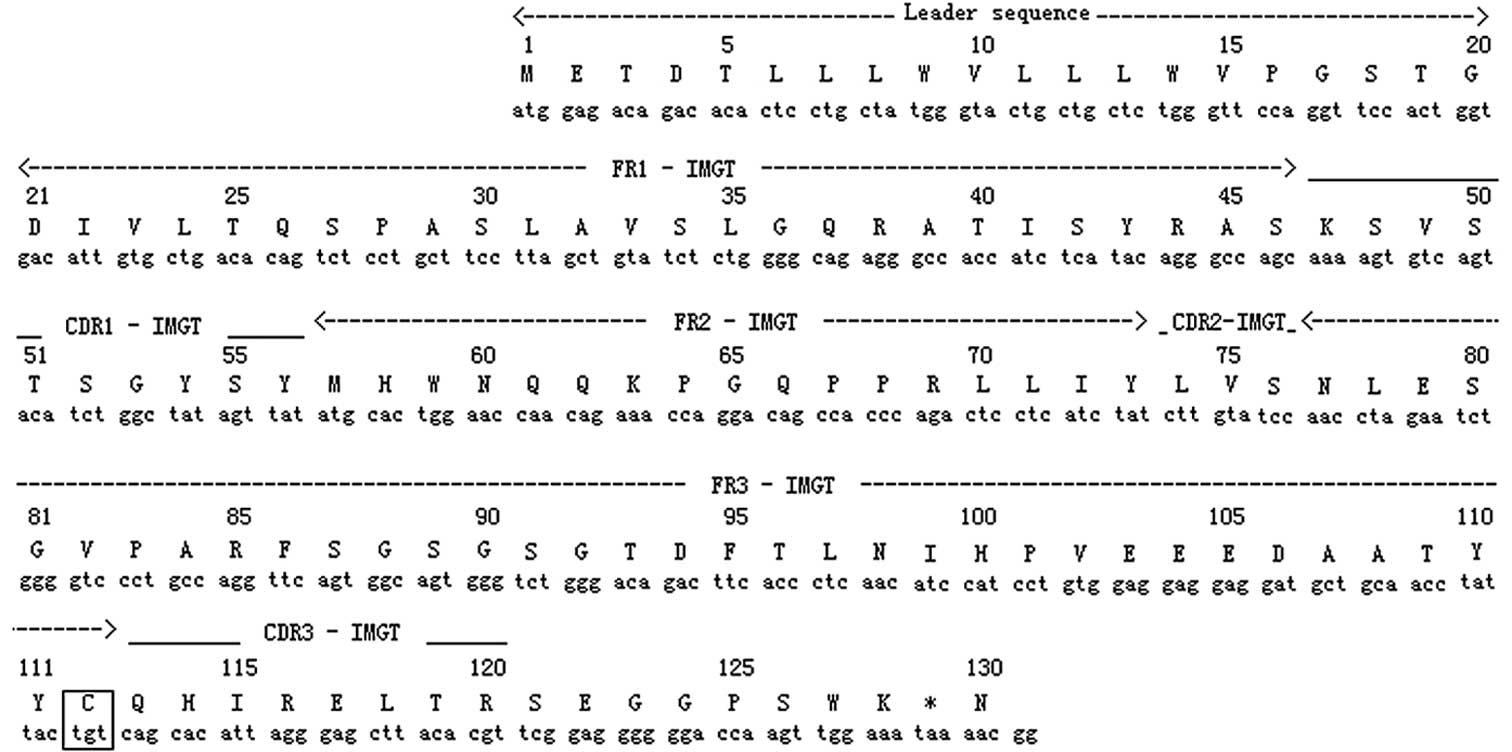
The results of the DNA sequencing revealed that the
amplified light chain variable region (VL) from the NS-1
cells was identical to that of the 2F9 cells. The nucleotide and
deduced amino acid (AA) sequences of the NS-1 VL are
shown in Fig. 3. The sequence was
387 bp long, encoded 128 AA and included a leader sequence of 60
bp. There was a TAA stop codon at 385–387 bp and only one
cysteine was found, at 112AA/128AA, as shown in the box in Fig. 3. The leader sequence, frame regions
(FRs) and complementarity determining regions (CDRs) 1–3 of the
VL were positioned as shown in Fig. 3. The gene segment family of the NS-1
VL was identified by a search for similarities against
the IMGT/V-QUEST database. The V- and J-segments were identified as
Musmus IGκKV3-12*01 and Musmus IGκKJ2*01,
respectively. Accordingly, the NS-1 VL gene belongs to
the Igκ gene family V3 subgroup. The results were
analyzed using the IMGT/V-QUEST program (version 3.2.21) and are
summarized in Table II.
 | Table IIDetailed results of the IMGT/V-QUEST
analysis of the NS-1 VL sequence. |
Table II
Detailed results of the IMGT/V-QUEST
analysis of the NS-1 VL sequence.
| Segment of NS-1
VL | Gene and allele | Scorea | Identity, %b (nt)c |
|---|
| V | Musmus
IGκV3-12*01 F | 1405 | 98.28 (286/291) |
| J | Musmus
IGκJ2*01 F | 171 | 97.22 (35/36) |
Discussion
When Köhler and Milstein first described hybridoma
technology in 1975, it appeared to have the potential to develop
treatments for a variety of human diseases (1). The technique involves forming
hybridomas by fusing a specific antibody-producing B cell (from a
murine spleen) with a murine myeloma cell (for example NS-1 or SP
2/0) that is selected for its ability to grow in tissue culture and
for an absence of antibody chain secretion (8). The antibodies produced by the
hybridoma are of a high specificity and are therefore McAbs. The
fused hybridomas, being cancer cells, multiply rapidly and
indefinitely and produce large amounts of the desired
antibodies.
The uses of McAbs are numerous and include the
prevention, diagnosis and treatment of disease, vaccine production
and antigenic characterization of pathogens. However, in hybridoma
technology the majority of McAbs are derived from mice and the
clinical application of murine antibodies has been greatly
restricted due to the occurrence of severe serum disease and the
presence of human anti-mouse antibody (HAMA) in patients during
therapy (9). Therefore, it is
necessary to reduce the immunogenicity of the mouse antibody in
order to be able to administer large doses of antibody repeatedly
to patients. The most commonly used method is to humanize the
murine-derived McAbs by gene cloning (10).
NS-1 and SP 2/0 are the two most widely used murine
myeloma cell lines. Myeloma lines, including SP2/0 and X63.6.5.3,
do not synthesize the heavy or light chains of immunoglobulins;
therefore, hybridomas established with these myeloma lines secrete
homogeneous McAbs with heavy and light chains derived only from
spleen cells. However, myeloma lines such as NS-1 and P3U1
synthesize κ light chains, although they are not secreted, meaning
that the NS-1 VL is encoded by mRNA (11,12).
This causes certain problems for the humanization of murine McAbs,
including interrupting the sequencing of the McAb variable region
genes. In this study, in order to resolve this problem, we
successfully cloned and sequenced the NS-1 VL gene.
Following screening with 4 pairs of heavy chain
primers by RT-PCR, no heavy chain variable region (VH)
gene was observed; with 9 pairs of light chain primers, there was a
387-bp VL gene amplified with the P9 and P14 primers in
the NS-1 and 2F9 cells (Fig. 2A).
In order to avoid the influence of the residual genomic DNA, the
total RNA was treated with RNase-free DNase prior to RT-PCR.
Following the purification of the amplified NS-1 VL
gene, we inserted it into the pGEM®-T easy vector using
TA cloning and the recombinants were checked with EcoRI
digestion and agarose gel electrophoresis. These steps ensured that
the gene sequencing was reliable and accurate. The resulting
sequence was identical in the NS-1 and 2F9 cells.
According to the analysis from IMGT (Table II and Fig. 3), the sequence contained 3 CDRs and
4 FRs, a TAA stop codon at the end of the cDNA and only one
cysteine in the AA sequence. The NS-1 VL gene was a
nonproductive IGκ rearranged sequence and considered to be a
pseudogene due to the stop codon and out-of-frame junction.
This study successfully cloned and sequenced the
VL gene of the NS-1 cell line and determined that it was
a pseudogene. The results of this study may prevent the selection
of the wrong VL gene from the fused partner NS-1 cells
during McAb humanization.
Acknowledgements
The study was supported by the National Natural
Science Foundation of China (no. 30901327) and Zhejiang Provincial
Natural Science Foundation of China (no. Y2100070).
References
|
1
|
Köhler G and Milstein C: Continuous
cultures of fused cells secreting antibody of predefined
specificity. Nature. 256:495–497. 1975.
|
|
2
|
Köhler G, Howe SC and Milstein C: Fusion
between immunoglobulin-secreting and nonsecreting myeloma cell
lines. Eur J Immunol. 6:292–295. 1976.PubMed/NCBI
|
|
3
|
Tomita M and Tsumoto K: Hybridoma
technologies for antibody production. Immunotherapy. 3:371–380.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tang YM, Ning BT, Cao J, et al:
Construction and expression of single-chain antibody derived from a
new clone of monoclonal antibody against human CD14 in CHO cells.
Immunopharmacol Immunotoxicol. 29:375–386. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chiang YL, Sheng-Dong R, Brow MA, et al:
Direct cDNA cloning of the rearranged immunoglobulin variable
region. Biotechniques. 7:360–366. 1989.PubMed/NCBI
|
|
6
|
Sambrook J and Russell DW: Molecular
Cloning: A Laboratory Manual. 3rd edition. Cold Spring Harbor
Laboratory Press; New York: 2001
|
|
7
|
Siddiqui MZ: Monoclonal antibodies as
diagnostics; an appraisal. Indian J Pharm Sci. 72:12–17. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kennett RH: Hybridomas: a new dimension in
biological analyses. In Vitro. 17:1036–1050. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Presta LG: Engineering of therapeutic
antibodies to minimize immunogenicity and optimize function. Adv
Drug Deliv Rev. 58:640–656. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bernett MJ, Karki S, Moore GL, et al:
Engineering fully human monoclonal antibodies from murine variable
regions. J Mol Biol. 396:1474–1490. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Abe N and Inouye K: Purification of
monoclonal antibodies with light-chain heterogeneity produced by
mouse hybridomas raised with NS-1 myelomas: application of
hydrophobic interaction high-performance liquid chromatography. J
Biochem Biophys Methods. 27:215–227. 1993. View Article : Google Scholar
|
|
12
|
Cowan NJ, Secher DS and Milstein C:
Intracellular immunoglobulin chain synthesis in non-secreting
variants of a mouse myeloma: detection of inactive light-chain
messenger RNA. J Mol Biol. 90:691–701. 1974. View Article : Google Scholar
|