Introduction
We recently identified F10, the hydatidiform
mole-related gene, using screening of suppression subtractive
hybridization cDNA libraries of normal and hydatidiform villi
(GenBank accession number, AB196290) (1). Previous studies have suggested that
F10 is involved in the malignant transformation of hydatidiform
moles, as well as the development of gynecological cancer (2,3). F10
is expressed at low levels in human lung cancer A549 cells. The
gene promotes cell proliferation by upregulating proliferating cell
nuclear antigen and cyclin D1 (4),
suggesting that F10 plays a pro-proliferative role in accelerating
cancer development. Since excessive proliferation and inhibited
apoptosis are involved in cancer occurrence and development
(5,6), this study aimed to examine whether F10
also exerts anti-apoptotic roles. We induced the overexpression of
F10 in A549 cells, examined the apoptosis level and compared the
expression of apoptosis-associated genes, including BCL2-associated
X protein (BAX) and caspase-3.
Materials and methods
Cells and reagents
The human lung cancer cell line A549 was maintained
in RPMI-1640 medium with 10% fetal bovine serum in a 37°C, 5%
CO2 incubator. Mouse anti-human BAX and mouse anti-human
β-actin monoclonal antibodies were purchased from Boster Biological
Technology, Ltd. (Wuhan, China). Mouse anti-human caspase-3
monoclonal antibody was obtained from Cell Signaling Technology,
Inc. (Danvers, MA, USA). Rabbit anti-mouse secondary antibody was
purchased from Dako Company (Glostrup, Denmark).
Transfection
F10 was inserted into the pEGFP-N1 vector (Takara
Bio, Inc., Shiga, Japan) as an EcoRI-Kpn I fragment and the
recombinant plasmid was confirmed by sequencing. The plasmid
pEGFP-N1-F10 or pEGFP-N1 empty vector was transfected into A549
cells using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The
single clones were selected with G418 (Sigma, St. Louis, MO, USA).
The expression of F10 was confirmed by RT-PCR analysis following
the manufacturer’s instructions (Takara Bio, Inc.). The recombinant
cell lines were named A549-F10 and A549-empty, respectively.
MTT cell proliferation assay
Untransfected A549, A549-F10 and A549-empty cells
were seeded at 1×104 cells/well in 96-well plates in 200
μl of medium. The cells were cultured for 0.5, 1, 6, 12, 24 or 48 h
(sextuplicate per time point) before 20 μl of 5 mg/ml MTT (Sigma)
was added to each well. Following a 4-h incubation, the cell
supernatant was discarded and 150 μl/well of DMSO (Sigma) was
added. After 5 min of mixing, the OD value at 490 nm was measured
using a Bio-Tek microplate reader (Bio-Rad, Hercules, CA, USA),
then the tumor cell growth curve was drawn.
TUNEL-FITC/Hoechst 33258 apoptosis
detection assay
Apoptosis was detected using the Annexin V-FITC
Apoptosis Detection kit and the Hoechst Staining kit (KeyGEN
Biotech., Nanjing, China). Untransfected A549, A549-F10 and
A549-empty cells were seeded on coverslips, washed three times in
PBS for 5 min each, fixed in 4% formaldehyde for 20 min and
incubated in 70% ethanol at −20°C for 30 min. The coverslips were
washed a further 3 times and the cells were permeabilized. The
permeabilization was performed in 0.1% Triton X-100/0.1% sodium
citrate at room temperature for 10 min. After three 5-min washes in
PBS, the cells were incubated with 3% H2O2 at
room temperature for 10 min. After another three 5-min washes in
PBS, the cells were incubated with TdT enzyme at 37°C for 90 min,
which was protected from light. After two 2-min washes in PBS, the
nuclei were stained with Hoechst 33258 at room temperature for 20
min in the dark. The cells were finally washed in the dark three
times in PBS containing 0.5% Tween 20, 2 min each, and mounted in
glycerol. Images were captured using a fluorescence microscope
(Nikon, Tokyo, Japan).
Western blot analysis
Lysates from untransfected A549, A549-F10 and
A549-empty cells were separated on gels, transferred to membranes,
first stained with anti-BAX, anti-caspase-3 or anti-β-actin
antibody, then stained with HRP-labeled secondary antibodies and
developed with an ECL kit. The quantification was analysed using
the SensiAnsys software (Shanghai Peiqing Science & Technology,
Co., Ltd., Shanghai, China).
Statistical analysis
Data were analyzed using the SPSS 13.0 software
(SPSS, Inc., Chicago, IL, USA) and expressed as mean ± standard
deviation. RT-PCR results were analyzed using the two-sample
t-test. For the MTT cell proliferation assay, the factorial design
analysis of variance (ANOVA) was used to compare inter-group
differences. For the TUNEL-FITC/Hoechst 33258 assay and western
blot analysis, the results were analyzed by one-way ANOVA followed
by Fisher’s LSD post hoc tests if variance homogenenity was
assumed, or by Welch and Dunnett T3 tests if homogeneity was not
assumed. P<0.05 was considered to indicate a statistically
significant result.
Results
Stable transfection of F10 in A549
cells
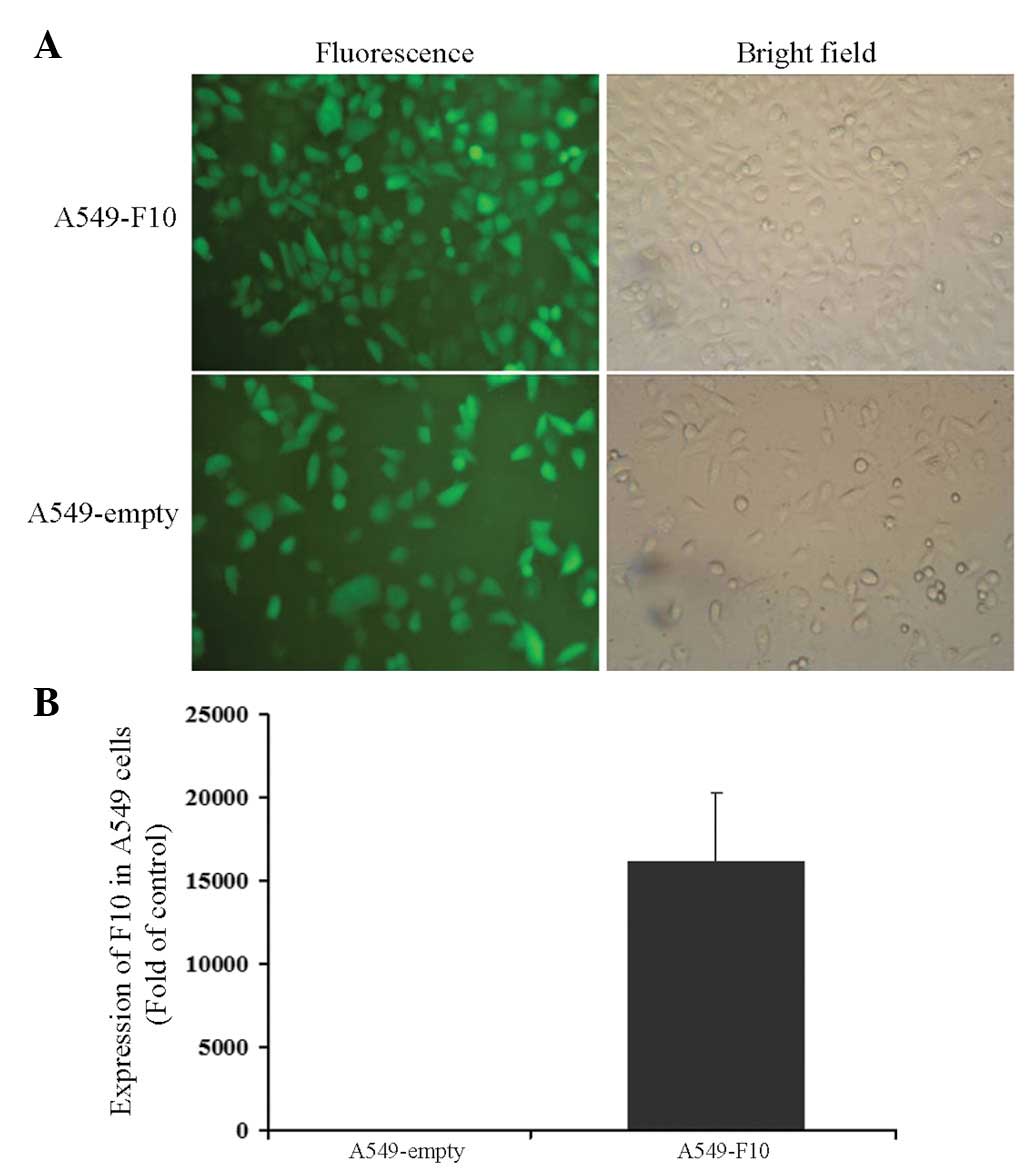
A549 cells were transfected with pEGFP-N1-F10 or the
pEGFP-N1 empty vector and selected for 4 weeks with G418. After
another 1-week culture, cells from the two groups were green under
a microscope, showing that transfection efficiency was close to
100% (Fig. 1A). The expression of
F10 was confirmed by RT-PCR (A549-F10 vs. A549-empty, t=−6.904,
P=0.002) (Fig. 1B).
F10 transfection accelerates A549
proliferation
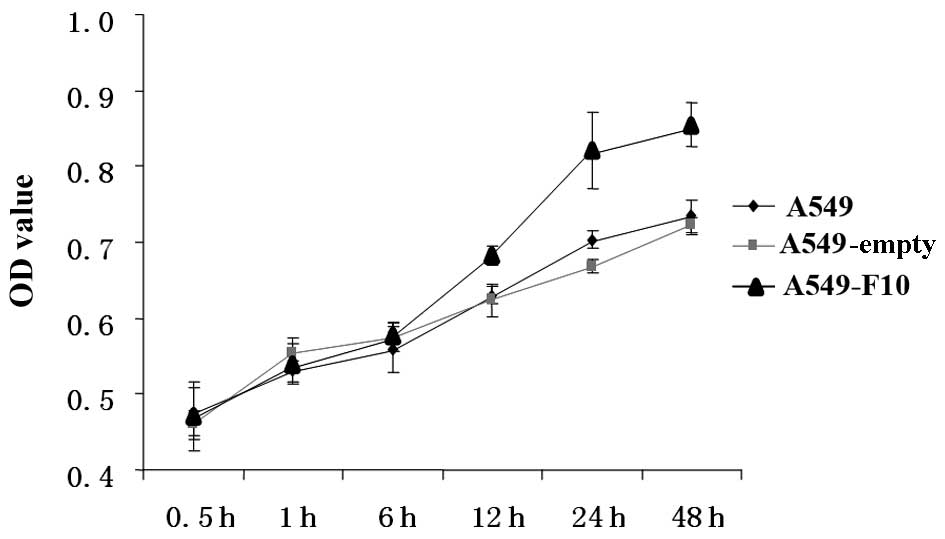
We compared proliferation among untransfected A549,
A549-F10 and A549-empty cells using the MTT assay (Fig. 2). The difference between the groups
was significant (F=48.039, P=0.000). The effect of time and the
correlation between group and time were significant (F=323.264,
P=0.000 and F=11.442, P=0.000, respectively). There was no
difference between the three groups at 0, 0.5 and 1 h. After 12 h,
A549-F10 cells proliferated markedly faster than A549-empty and
untransfected A549 cells (P<0.05). No difference in
proliferation was observed between the A549-empty and untransfected
A549 cells (P>0.05).
F10 transfection inhibits apoptosis in
A549 cells
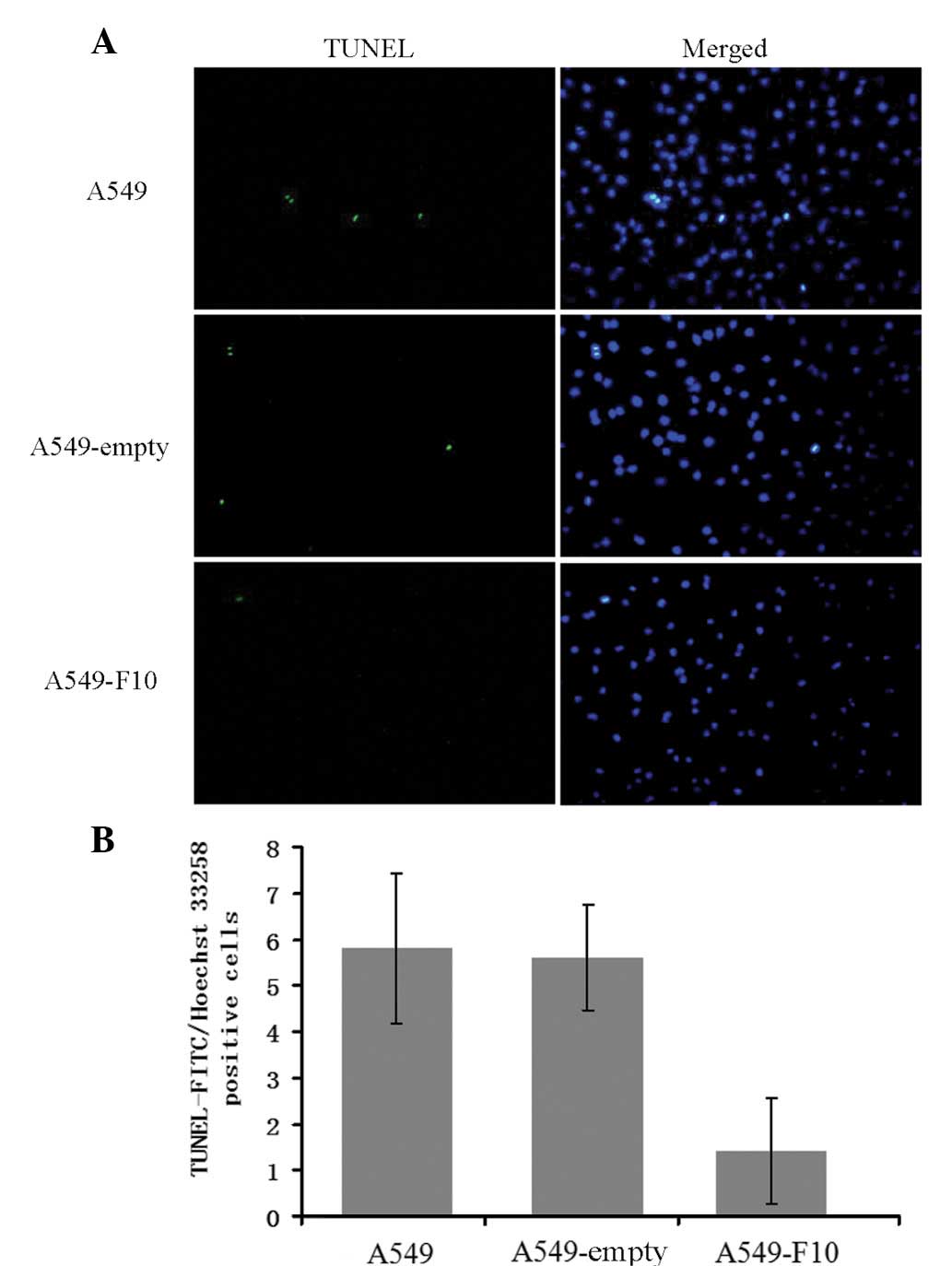
To examine the effect of F10 overexpression on
apoptosis, untransfected A549, A549-F10 and A549-empty cells were
double-stained with TUNEL and Hoechst 33258 (Fig. 3). Apoptotic cells were
TUNEL-positive and their nuclei exhibited strong Hoechst blue
staining. By contrast, normal cells were TUNEL-negative and showed
weak blue nulcei Hoechst staining. TUNEL and Hoechst 33258
double-positive cells were counted and the number of
double-positive (apoptotic) cells differed significantly among the
three cell lines (F=17.472, P=0.000). There were markedly fewer
apoptotic A549-F10 cells than untransfected A549 and A549-empty
cells (P<0.001), suggesting that F10 overexpression inhibits
apoptosis in A549 cells. No difference in the apoptotic level was
observed between the untransfected A549 cells and A549-empty cells
(P=0.816).
F10 transfection reduces BAX and
caspase-3 protein levels in A549 cells
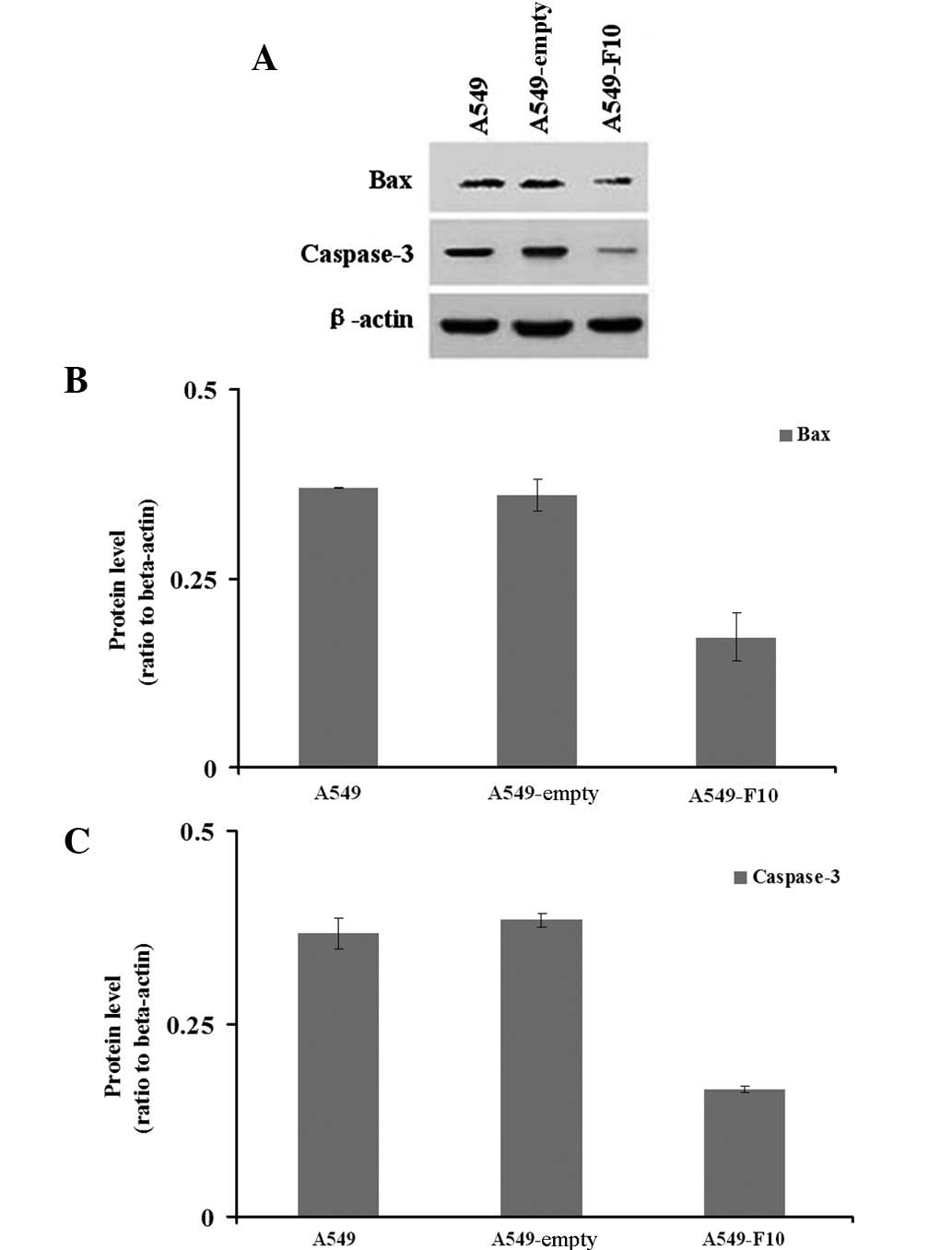
We next examined how F10 transfection affects the
expression of pro-apoptotic genes, BAX and caspase-3 (Fig. 4). Our western blotting results
showed significant difference among the three cell lines in the
expression of BAX (Welch=45.966, P=0.008) and caspase-3 (F=268.790,
P=0.000). The Dunnett’s T3 test demonstrated that A549-F10 cells
exhibited lower BAX protein expression than untransfected A549
cells and A549-empty cells (P<0.05). Similarly, the LSD test
revealed that the caspase-3 expression in A549-F10 cells was
markedly lower than that in untransfected A549 and A549-empty cells
(P=0.000). No difference in the expression of BAX (P=0.833) or
caspase-3 (P=0.155) was observed between the untransfected A549 and
A549-empty cells.
Discussion
F10 has been suggested to participate in the
malignant transformation of hydatidiform moles and the development
of the gynecological cancer. Therefore, it is imperative to study
the potential role of F10 to improve diagnosis and treatment of
cancer. One promising strategy is to establish a cell system
overexpressing F10. We previously screened F10 mRNA expression by
RT-PCR in eight different cell lines (Bel7402, HIC, HepG2, PC,
A549, MGC, 16HBE and 293 cells) (7). The human lung cancer cell line A549
was identified as expressing low levels of F10 and thus served as a
model cell system for studies using F10 overexpression. In this
study, we transfected A549 cells with pEGFP-N1-F10 plasmid stably
and selected single positive clones using G418. F10 mRNA expression
was confirmed by RT-PCR.
The occurrence and development of cancer often
involves two aspects: excessive proliferation and inhibited
apoptosis (8). F10 has been shown
previously to promote tumor cell proliferation (4). In our study this observation was
confirmed by the MTT in vitro proliferation assay, which
showed that from 12 h, A549 cells overexpressing F10 proliferated
markedly faster than untransfected cells or cells transfected with
the empty expressing vector. We then examined whether F10 also
contributes to cancer development by inhibiting apoptosis. The
TUNEL-FITC/Hoechst 33258 staining demonstrated that the level of
apoptosis was significantly lower in F10 overexpressing cells.
Apoptosis, first identified in 1972 by Kerr and
colleagues, is different from necrosis. It is an actively
controlled cell suicide regulated by multiple genes and a series of
signal transductions. Pro-apoptotic genes, including caspase-3 and
BAX, play important roles in this progress. The caspase family
participates in multiple apoptosis-associated physiological and
pathological processes (9–11). caspase-3, a member of the caspase
family, exerts its pro-apoptotic function through the death
receptor (12) and
mitochondrial-mediated pathways (13). caspase-3, as an apoptotic effector,
may be used as an indicator of apoptosis: upregulation of caspase-3
expression indicates an increase in apoptosis, whereas
downregulation indicates decrease (14). BAX, a pivotal BCL-2 family member,
is located in the cytoplasm under normal circumstances. During
apoptosis, due to conformational changes, BAX is translocated into
the mitochondria to form homodimers, which target and open the
mitochondrial intermembrane contact sites and subsequently release
cytochrome C and apoptosis-inducing factors, which in turn promote
protein hydrolysis and activate caspases, leading to apoptosis
(15,16).
To examine the potential mechanisms underlying the
apoptosis inhibition role of F10, we further examined the levels of
BAX and caspase-3 in the presence or absence of F10 overexpression.
Western blot analysis demonstrated that F10 overexpression markedly
decreases the expression of BAX and caspase-3 in A549 cells,
suggesting that F10 inhibits apoptosis by targeting BAX and
caspase-3.
In conclusion, our study established a
F10-overexpression model in the human lung cancer cell A549 and
demonstrated that F10 overexpression promotes cell proliferation
and inhibits apoptosis, probably through downregulating the
pro-apoptotic genes caspase-3 and BAX. Since apoptosis involves
numerous participants and its mechanism may be different in
different cell types, further studies are required to examine the
mechanism by which F10 reduces the levels of BAX and caspase-3 and
whether other molecules also contribute to the role of F10 in
cancer occurrence and development.
Acknowledgements
The authors would like to thank Yanguo Cui and
Xiaomin Cao for their technical assistance. This study was
supported by the National Natural Science Foundation of China (no.
30672234).
References
|
1
|
Li GT, Pang ZJ, Zhou J, et al: Cloning of
new genes associated with the pathogenesis of hydatidiform mole.
Guangdong Medical Journal. 27:22–24. 2006.(In Chinese).
|
|
2
|
Zhou J, Chen SL, Xing FQ, et al:
Association of the novel hydatidiform mole-related gene F10 with
the invasiveness of trophoblastic tumor. Di Yi Jun Yi Da Xue Xue
Bao. 25:171–173. 2005.(In Chinese).
|
|
3
|
Zhou J, Liang W, Li B, et al: The
expression of hydatidiform mole associated new gene F10 in
different tumor tissues. Guangdong Medical Journal. 26:596–597.
2005.(In Chinese).
|
|
4
|
Cao XM, Pang ZJ, Quan S and Xing FQ:
Effect of F10 gene on expression of proliferating cell nuclear
antigen and Cyclin D1. Journal of Sun Yat-Sen University (Medical
Sciences). 30:6–9. 2009.(In Chinese).
|
|
5
|
Valásková Z, Kinová S, Danihel L, et al:
The complexity of interactions of the tumour growth process. Vnitr
Lek. 55:1145–1158. 2009.(In Slovak).
|
|
6
|
Caroppi P, Sinibaldi F, Fiorucci L and
Santucci R: Apoptosis and human diseases: mitochondrion damage and
lethal role of released cytochrome C as proapoptotic protein. Curr
Med Chem. 16:4058–4065. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cao XM, Pang ZJ and Quan S: Construction
and identification of a stable eukaryotic expression system for F10
gene. Nan Fang Yi Ke Da Xue Xue Bao. 28:57–59. 2008.(In
Chinese).
|
|
8
|
Sarcević B: Apoptosis in tumors. Acta Med
Croatica. 63(Suppl 2): 43–47. 2009.(In Croatian).
|
|
9
|
Kumar S and Dorstyn L: Analysing caspase
activation and caspase activity in apoptotic cells. Methods Mol
Biol. 559:3–17. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vaculova A and Zhivotovsky B: Caspases:
determination of their activities in apoptotic cells. Methods
Enzymol. 442:157–181. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Denault JB and Salvesen GS: Apoptotic
caspase activation and activity. Methods Mol Biol. 414:191–220.
2008.PubMed/NCBI
|
|
12
|
Wang Y, Sun LG and Xia CH:
Caspase-mediated Fas apoptosis pathway. World Chinese Journal of
Digestology. 14:3439–3442. 2006.(In Chinese).
|
|
13
|
Jin LF and Chen TY: Proteinum family of
Bcl-2 gene and apoptosis. Med Recapitul. 11:446–447. 2005.(In
Chinese).
|
|
14
|
Mazumder S, Plesca D and Almasan A:
Caspase-3 activation is a critical determinant of genotoxic
stress-induced apoptosis. Methods Mol Biol. 414:13–21.
2008.PubMed/NCBI
|
|
15
|
Pan G, O’Rourke K and Dixit VM: Caspase-9,
Bcl-XL, and Apaf-1 form a ternary complex. J Biol Chem.
273:5841–5845. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li P, Nijhawan D, Budihardjo I, et al:
Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9
complex initiates an apoptotic protease cascade. Cell. 91:479–489.
1997. View Article : Google Scholar : PubMed/NCBI
|