Introduction
Uterine sarcomas are rare tumors, accounting for 3
to 8% of neoplasms of the uterine corpus and 1% of all tumors of
the female genital tract (1). Due
to the low incidence of these tumors their molecular biology,
including the role of epigenetic events, is poorly understood.
Uterine sarcomas are classified into smooth muscle sarcomas,
stromal sarcomas and mixed uterine tumors, i.e. carcinosarcomas.
Although the latter histological type is reclassified as a
dedifferentiated or metaplastic form of endometrial carcinoma, it
is still included in most of the studies on uterine sarcomas
(2). The standard treatment of the
uterine sarcomas comprises surgery and chemotherapy. Uterine
sarcomas are among the most lethal uterine malignancies with poorer
prognosis compared with other gynecological malignancies; 5-year
survival rates remain below 50% for early stages and do not exceed
30% in the remaining stages (3,4).
Recently, several adjuvant chemotherapy regimens
have been reported in the treatment of soft tissue sarcomas,
including sarcomas of the uterus. Some of the trials were based on
the alkylating agents, including carmustine and temozolomide (TMZ).
The regiment combining carmustine with O6-benzylguanine
did not result in objective treatment response in 12 enrolled
patients with soft tissue sarcoma (5). The initial results concerning
TMZ-based therapy as a second-line treatment in 31 patients with
advanced soft tissue sarcoma did not show activity of the drug
(6). The trial involving patients
with soft tissue sarcomas not subjected to standard chemotherapy
revealed only minimal efficacy of TMZ (7). Another trial on TMZ demonstrated a
modest activity against previously treated unresectable or
metastatic soft tissue sarcomas (8). Notably, all responding patients had
leiomyosarcoma (of uterine or non-uterine origin) (8).
Better results were obtained in 2005 by the Spanish
Group for Research on Sarcomas with the prolonged course of TMZ
which had activity in patients with pretreated soft tissue sarcomas
(9). Notably, a response was
observed in 5 of 11 patients who had gynecological leiomyosarcoma
and in one of two patients with mixed mullerian tumors. The results
of the study on the uterine leiomyosarcoma were published in the
same year, revealing therapeutic benefit of TMZ in patients with
metastatic unresectable disease (10). Of 19 patients pretreated with
doxorubicin who underwent TMZ-based therapy, two patients achieved
almost complete response and eight showed stabilization of the
disease. In a recent study of Ferriss et al (11) a clinical benefit of TMZ was achieved
in five out of six patients with advanced and recurrent uterine
leiomyosarcoma. This therapeutic benefit was associated with
silencing of the O6-methylguanine-DNA methyltransferase (MGMT)
expression as determined by immunohistochemistry. All the above
mentioned studies revealed good tolerance to TMZ (6–11).
The MGMT gene has been shown to be
epigenetically downregulated in several solid tumors. Aberrant
promoter hypermethylation of the MGMT has been associated
with the lack of its mRNA expression, the loss of MGMT protein
(12) and loss of enzymatic
activity (13). MGMT encodes
DNA repair protein, an enzyme responsible for the direct removal of
alkylating adducts from guanines. The silencing of the gene
contributes to the reduction of the genome stability and sensitizes
tumor cells to alkylating agents, including dacarbazine, carmustine
and TMZ. The main therapeutic target of these drugs are the
nitrogen bases of DNA and the most important cytotoxic derivate of
their action on the nitrogen bases is O6-methyl-guanine. Alkylation
of the guanine leads to an accumulation of the replication errors
during the S-phase of the cell cycle and, as a consequence, to the
cell cycle arrest and/or apoptosis. The high degree of removal of
alkyl adducts causes the resistance to treatment.
As has been shown in tumor cell line-based studies,
MGMT prevents TMZ-induced cell death by removing alkyl adducts from
the O6 position of guanine (14). Thus, tumor cells expressing MGMT are
resistant to alkylating agents, while those that lack the enzyme
appear to be chemosensitive. The predictive value of MGMT
epigenetic silencing is well documented for glioblastoma treatment
with TMZ. The methylation of the promoter of the gene was shown to
correlate with improved prognosis in several independent trials and
is being considered as a potential stratification marker of the
response to TMZ-based therapy (15). However, to date no formal
recommendation has been proposed as to the use of this marker in
the clinical setting. In melanomas treated with TMZ, the
MGMT methylation was associated with improved tolerance to
treatment, however not with survival (16).
This study aimed to evaluate the frequency of
MGMT promoter methylation, as well as to assess its possible
correlation with the expression levels of the gene, in
carcinosarcomas and non-epithelial malignant tumors of corpus
uteri.
Materials and methods
Patients
A total of nine patients treated for smooth muscle
uterine sarcoma, 11 for stromal uterine sarcoma and 17 for mixed
uterine tumors in the Maria Sklodowska-Curie Memorial Cancer Centre
and Institute of Oncology in Warsaw between January 2009 and
December 2010 were enrolled in the present study. The selected
patients’ characteristics are presented in Table I. The study was approved by the
Independent Ethics Committee of the Maria Sklodowska-Curie Memorial
Cancer Centre and Institute of Oncology in Warsaw and all patients
provided informed consent. Tissue specimens were divided into two
parts: one part was examined histologically, the other was frozen
in liquid nitrogen and stored at −70°C until nucleic acid
isolation. In addition to 37 tumor tissue samples, 19 samples of
normal uterine tissue were also obtained from patients enrolled in
the study.
 | Table IIndividual patient data. |
Table I
Individual patient data.
| Patient | Age (years) | MGMT
methylation status | MGMT
expression level | Tumor type | Tumor
histology/histological grade |
|---|
| Mixed uterine
tumors |
| 3 | 75.5 | Negative | 0.010 | Recurrent | Carcinosarcoma
heterologousa |
| 4 | 54.1 | Negative | 0.025 | Primary | Carcinosarcoma
heterologousb |
| 5 | 79.5 | Negative | 0.009 | Primary | Carcinosarcoma
heterologous |
| 11 | 61.4 | Positive | 0.017 | Primary | Carcinosarcoma
homologous |
| 14 | 23.2 | Positive | 0.015 | Primary | Adenosarcoma
homologous |
| 18 | 66.4 | Positive | 0.013 | Primary | Carcinosarcoma
heterologous |
| 24 | 66.6 | Negative | 0.025 | Primary | Carcinosarcoma
heterologous |
| 25 | 61.0 | Negative | 0.020 | Recurrent | Carcinosarcoma
homologous |
| 30 | 64.3 | Negative | 0.025 | Primary | Carcinosarcoma
heterologous |
| 33 | 61.6 | Negative | 0.022 | Recurrent | Carcinosarcoma
heterologous |
| 39 | 54.7 | Negative | 0.044 | Primary | Adenosarcoma
homologous |
| 44 | 68.0 | Negative | 0.030 | Primary | Adenosarcoma
heterologous |
| 47 | 65.7 | Negative | 0.014 | Primary | Carcinosarcoma
homologous |
| 51 | 56.4 | Negative | 0.005 | Primary | Mixed endometrial
stromal and smooth muscle tumor |
| 66 | 55.1 | Negative | 0.045 | Primary | Carcinosarcoma
homologous |
| 67 | 59.7 | Positive | 0.003 | Primary | Adenosarcoma
homologous |
| 71 | 55.5 | Negative | 0.126 | Recurrent | Adenosarcoma
(dediff) |
| Smooth muscle
sarcomas |
| 1 | 36.6 | Positive | 0.022 | Primary |
Rhabdomyosarcoma |
| 2 | 52.6 | Positive | 0.001 | Recurrent |
Leiomyosarcoma/G3 |
| 12 | 56.3 | Positive | 0.005 | Primary |
Leiomyosarcoma/G3 |
| 23 | 63.2 | Positive | 0.006 | Recurrent |
Leiomyosarcoma/G2 |
| 26 | 45.8 | Negative | 0.025 | Recurrent |
Leiomyosarcoma/G3 |
| 28 | 51.5 | Negative | 0.005 | Recurrent |
Leiomyosarcoma/G2 |
| 35 | 57.5 | positive | 0.018 | Recurrent |
Leiomyosarcoma/G2 |
| 37 | 25.5 | Negative | 0.016 | Recurrent | STUMP |
| 63 | 40.5 | Negative | 0.005 | Recurrent |
Leiomyosarcoma/G3 |
| Stromal uterine
sarcomas |
| 6 | 76.7 | Negative | 0.024 | Recurrent | Endometrial stromal
sarcoma, low grade |
| 8 | 59.5 | Positive | 0.019 | Primary | Undifferentiated
endometrial sarcoma |
| 13 | 60.1 | Negative | 0.025 | Recurrent | Undifferentiated
endometrial sarcoma |
| 16 | 43.3 | Negative | 0.017 | Recurrent | Endometrial stromal
sarcoma, low grade |
| 17 | 74.8 | Negative | 0.006 | Primary | Undifferentiated
endometrial sarcoma |
| 31 | 51.0 | Negative | 0.014 | Primary | Sarcoma stromale,
low grade |
| 36 | 64.6 | Negative | 0.052 | Primary | Undifferentiated
endometrial sarcoma |
| 38 | 44.8 | Negative | 0.010 | Primary | Endometrial stromal
sarcoma low grade |
| 52 | 78.1 | Negative | 0.027 | Primary | Undifferentiated
endometrial sarcoma |
| 62 | 53.4 | Negative | 0.026 | Primary | Undifferentiated
endometrial sarcoma |
| 64 | 68.9 | Negative | 0.028 | Primary | Undifferentiated
endometrial sarcoma |
DNA methylation analysis
MGMT promoter methylation analysis was
performed using the combined bisulfite restriction analysis
(COBRA).
DNA was isolated from ~50 mg of pulverized (with the
Microdismembrator II, B Braun Biotech International, Melsungen,
Germany) tumor samples using NucloSpin Tissue kit (Macherey-Nagel,
Düren, Germany), according to the manufacturer’s instructions. DNA
quantity was measured using NanoDrop 2000 (ThermoScientific,
Waltham, MA, USA). DNA (1 μg) was bisulfite converted using EpiTect
kit (Qiagen, Hilden, Germany) according to the manufacturer’s
instructions. Converted DNA was eluted with 40 μl of water.
MGMT promoter region [chr10:131155461-131155570 genome
location as determined by UCSC Genome Browser Database (http://genome.ucsc.edu/) Human March 2006 (hg18)
assembly] region was amplified using previously reported PCR
primers (17). The reaction volume
of 15 μl contained 1X PCR buffer, 2 mM MgCl2, 0.25 mM
dNTPs, 0.2 mM each primer, 0.5 U of FastStart DNA Polymerase (Roche
Applied Science, Mannheim, Germany) and 1 μl of bisulfite-treated
DNA as a template. The cycling conditions were as follows: initial
denaturation at 94°C for 3 min; followed by 38 cycles of 30 sec at
94°C, 40 sec annealing at 58°C and 50 sec at 72°C; then final
elongation for 7 min at 72°C. Subsequently, 8 μl of PCR products
were digested overnight with HpyCH4IV (TaiI)
restriction enzyme (New England Biolabs, Ipswich, MA, USA) which
cleaves sequences containing CpG dinucleotides. Restriction
fragments were electrophoresed in 10% polyacrylamide gel (acryl/bis
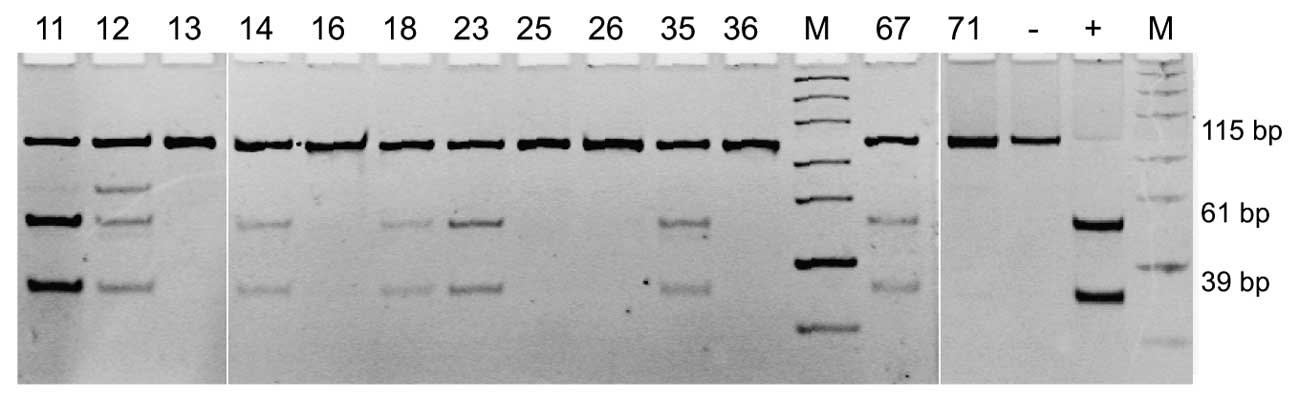
19:1) and visualized with ethidium bromide. The presence of 61- and
39-bp DNA fragments on the gel (the shortest 15-bp DNA fragment was
not visible in certain samples due to the low band intensity)
indicates the occurrence of methylated MGMT variant. The
unmethylated DNA variants, as wells as native DNA (not subjected to
bisulfite conversion), have no restriction sites for the chosen
enzyme in the analyzed region which excludes the occurrence of
false positive results.
DNA isolated from the blood sample of a healthy
donor was methylated in vitro with SssI DNA
methyltransferase (New England Biolabs) and used as a positive
(methylated) control. The same DNA sample after whole genome
amplification (GenomiPhi, GE Healthcare, Piscataway, NJ, USA) was
used as a negative (unmethylated) control.
Expression analysis
MGMT mRNA level was evaluated using the
real-time reverse transcription polymerase chain reaction
(qRT-PCR).
Total RNA from ~50 mg of pulverized (with the
Microdismembrator II, B Braun Biotech International) tumor and
normal uterine samples was extracted using RNeasy Mini kit with
on-column DNase digestion (Qiagen) according to the manufacturer’s
instructions. RNA quantity was measured using NanoDrop
(ThermoScientific), while the overall RNA quality was assessed by
electrophoresis on a denaturing agarose gel (FlashGel, Lonza,
Rockland, ME, USA). The RNA samples (1 μg each) were
reverse-transcribed using the RT2 First Strand kit (SA
Biosciences, Hilden, Germany) according to the manufacturer’s
instructions. Quantitative real-time PCR was performed in triplets
using ABI Prism 7000 Sequence Detection System (Applied Biosystems,
Carlsbad, CA, USA). The reaction mixture of 25 μl contained 2.5 μl
of 15X diluted cDNA template, 1X Power SYBR Green PCR Master Mix
(Applied Biosystems) and 0.1 mM of the forward and reverse primers.
PCR was performed as follows: precycling hold at 95°C for 10 min,
45 cycles: 95°C for 30 sec and 60°C for 60 sec. To assess the
reaction specificity, the amplification products were subjected to
melting curve analysis.
UBC was used as a reference gene, as its
stable expression in carcinosarcoma tumors and non-epithelial
malignant tumors of the corpus uteri as well as in normal
uterine tissues has been recently demonstrated (18). Primer sequences for the MGMT
and UBC were obtained from the qPrimerDepot database
(19). These were: MGMT
forward, CTCCGGACCTCCGAGAAC, and MGMT reverse,
GTCTGCACGAAATAAAGC, producing 94-bp amplicons; as well as
UBC forward, TTGCCTTGACATTCTCGATG, and UBC reverse,
ATCGCTGTGATCGTCACTTG, producing 108-bp amplicons.
Raw data were analyzed using ABI Prism 7000 SDS
Software Version 1.1 (Applied Biosystems). Relative expression
levels were calculated using the 2−ΔCt method, where ΔCt
was defined as a difference between Ct value for MGMT and
UBC reference gene.
Statistical analysis
The Chi-square test was used to compare MGMT
methylation frequencies among the three histopathological subtypes
of the analyzed tumors. The difference in the MGMT
expression levels between MGMT-methylated and unmethylated
tumors and normal uterine tissues was assessed with the use of a
two-sided Mann-Whitney U test with a significance threshold level
α=0.05. The values of the MGMT expression levels were
visualized in a plot using GraphPadPrism (La Jolla, CA, USA).
Results
MGMT promoter methylation was observed in 27%
(10/37) of tumors obtained from all the patients enrolled into our
study. In three cases that showed MGMT promoter methylation,
an additional band on the gel was observed indicating incomplete
digestion and therefore incomplete promoter methylation (Fig. 1, sample 12). When stratified by the
disease type, 55.5% (5/9) of smooth muscle sarcomas, 23.5% (4/17)
of mixed uterine tumor tissues and 9% (1/11) of stromal sarcomas
showed MGMT methylation. The difference in frequency of
MGMT promoter methylation in smooth muscle sarcomas compared
with the two other subtypes of the analyzed tumors was
statistically significant (P=0.0489).
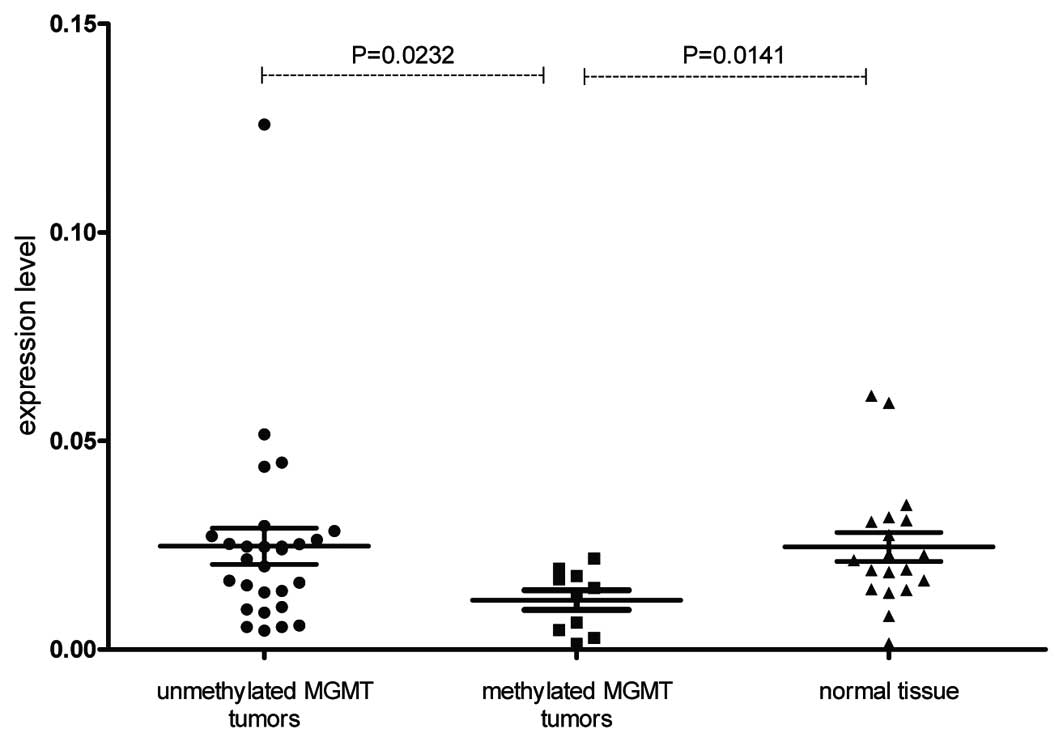
The MGMT expression level was significantly
lower in tumor samples with gene promoter methylation when compared
with unmethylated tumor tissues (P=0.0232). The MGMT
expression level was also significantly lower in tumor samples with
gene promoter methylation than in the normal uterine samples
obtained from the same patients (P=0.0141; Fig. 2). The promoter methylation status
and values of MGMT expression levels for individual uterine
sarcoma and carcinosarcoma tumors are provided in Table I.
No difference between MGMT-unmethylated
tumors and normal samples was observed.
Discussion
MGMT promoter methylation status has
previously been shown to correlate with the downregulation of the
gene expression in different types of solid tumors (20). The aim of this study was to evaluate
the frequency of MGMT promoter methylation and its
correlation with the gene expression status in carcinosarcomas and
non-epithelial malignant tumors of the corpus uteri. We
observed the methylated MGMT variant in a relatively high
fraction of tumors (27%), being the highest in smooth muscle
sarcomas where the gene was methylated in over half of the cases.
The tumors with MGMT promoter methylation showed a
significantly lower gene expression level than tumors with an
unmethylated promoter as well as normal uterine tissue samples.
A number of cell line-based experiments have
revealed the inverse correlation between the MGMT expression
and the cytotoxic effect of TMZ. These observations have been
confirmed in clinical trials. In the study by Ferriss et al
(11) on the effectiveness of TMZ
in uterine leiomyosarcomas treatment, the MGMT expression
was inversely correlated with treatment response.
Using the MGMT promoter methylation status as
a predictive biomarker has certain advantages. As cytosine
methylation is a stable covalent modification, it may be analyzed
in a wide range of tissue samples, including formalin-fixed and
paraffin-embedded (FFPE) tissues. FFPE samples are probably the
most accessible clinical tissue material for molecular analysis,
although inappropriate for the mRNA expression analysis. Currently
available laboratory techniques allow relatively fast and sensitive
MGMT methylation detection with qualitative or quantitative
results. Compared with immunohistochemical expression analysis, the
determination of MGMT methylation status is not dependent on
subjective microscopic evaluation and if a quantitative technique
is applied, more precise quantitative results may be achieved.
TMZ-based therapy is the current standard in the
treatment of glioblastoma patients and MGMT methylation
status has already been shown to be strong predictive factor of
significantly longer progression free survival and overall survival
of patients with methylation of the gene promoter (21). The systematic comparison of the
application of immunohistochemical staining with the promoter
methylation analysis in the glioblastoma patients treated with TMZ
revealed the superiority of methylation analysis as a survival
predictive factor (22).
As the group of patients enrolled in our study is
relatively small, the findings have value as preliminary results.
However, the presence of MGMT promoter methylation in a
notable proportion of patients and the observation that gene
methylation is associated with the downregulation of the gene
expression levels indicate that methylation analysis should be
included in the clinical trials on the effectiveness of TMZ in
patients with uterine sarcoma and carcinosarcoma. The results of
the present study advocate the use of TMZ in uterine leiomyosarcoma
treatment and also suggest that a smaller percentage of patients
with stromal sarcoma and carcinosarcoma may benefit from this type
of therapy. The qualitative techniques for MGMT promoter
methylation detection, including COBRA that was used in our study,
potentially allow prediction of the patients’ response to TMZ-based
treatment and thus their stratification.
To conclude, as TMZ-based chemotherapy showed
promising results in recently reported trials on the treatment of
soft tissue sarcomas, determination of MGMT promoter
methylation status may have significant clinical implications in a
fraction of patients with carcinosarcoma and non-epithelial
malignant tumors of the corpus uteri. Such studies should be
applied in the clinical practice and ultimately contribute to
future therapeutic strategies for these rare gynecological
tumors.
Acknowledgements
This study was supported by the research grant No.
NN 407 125 937 from the Ministry of Science and Higher
Education.
References
|
1
|
Brooks SE, Zhan M, Cote T and Baquet CR:
Surveillance, epidemiology, and end results analysis of 2677 cases
of uterine sarcoma 1989–1999. Gynecol Oncol. 93:204–208.
2004.PubMed/NCBI
|
|
2
|
D’Angelo E and Prat J: Uterine sarcomas: a
review. Gynecol Oncol. 116:131–139. 2010.
|
|
3
|
Major FJ, Blessing JA, Silverberg SG, et
al: Prognostic factors in early-stage uterine sarcoma. A
Gynecologic Oncology Group study. Cancer. 71(Suppl 4): 1702–1709.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zagouri F, Dimopoulos AM, Fotiou S,
Kouloulias V and Papadimitriou CA: Treatment of early uterine
sarcomas: disentangling adjuvant modalities. World J Surg Oncol.
7:382009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ryan CW, Dolan ME, Brockstein BB, et al: A
phase II trial of O6-benzylguanine and carmustine in patients with
advanced soft tissue sarcoma. Cancer Chemother Pharmacol.
58:634–639. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Woll PJ, Judson I, Lee SM, et al:
Temozolomide in adult patients with advanced soft tissue sarcoma: a
phase II study of the EORTC Soft Tissue and Bone Sarcoma Group. Eur
J Cancer. 35:410–412. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Trent JC, Beach J, Burgess MA, et al: A
two-arm phase II study of temozolomide in patients with advanced
gastrointestinal stromal tumors and other soft tissue sarcomas.
Cancer. 98:2693–2699. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Talbot SM, Keohan ML, Hesdorffer M, et al:
A phase II trial of temozolomide in patients with unresectable or
metastatic soft tissue sarcoma. Cancer. 98:1942–1946. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Garcia del Muro X, Lopez-Pousa A, Martin
J, et al; Spanish Group for Research on Sarcomas. A phase II trial
of temozolomide as a 6-week, continuous, oral schedule in patients
with advanced soft tissue sarcoma: a study by the Spanish Group for
Research on Sarcomas. Cancer. 104:1706–1712. 2005.
|
|
10
|
Anderson S and Aghajanian C: Temozolomide
in uterine leiomyosarcomas. Gynecol Oncol. 98:99–103. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ferriss JS, Atkins KA, Lachance JA,
Modesitt SC and Jazaeri AA: Temozolomide in advanced and recurrent
uterine leiomyosarcoma and correlation with o6-methylguanine DNA
methyltransferase expression: a case series. Int J Gynecol Cancer.
20:120–125. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Esteller M, Hamilton SR, Burger PC, Baylin
SB and Herman JG: Inactivation of the DNA repair gene
O6-methylguanine-DNA methyltransferase by promoter hypermethylation
is a common event in primary human neoplasia. Cancer Res.
59:793–797. 1999.PubMed/NCBI
|
|
13
|
Herfarth KK, Brent TP, Danam RP, et al: A
specific CpG methylation pattern of the MGMT promoter region
associated with reduced MGMT expression in primary colorectal
cancers. Mol Carcinog. 24:90–98. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kaina B, Fritz G, Mitra S and Coquerelle
T: Transfection and expression of human O6-methylguanine-DNA
methyltransferase (MGMT) cDNA in Chinese hamster cells: the role of
MGMT in protection against the genotoxic effects of alkylating
agents. Carcinogenesis. 12:1857–1867. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Franceschi E, Tosoni A, Pozzati E and
Brandes AA: Association between response to primary treatments and
MGMT status in glioblastoma. Expert Rev Anticancer Ther.
8:1781–1786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hassel JC, Sucker A, Edler L, et al: MGMT
gene promoter methylation correlates with tolerance of temozolomide
treatment in melanoma but not with clinical outcome. Br J Cancer.
103:820–826. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Smith E, Jones ME and Drew PA:
Quantitation of DNA methylation by melt curve analysis. BMC Cancer.
9:1232009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kowalewska M, Danska-Bidzinska A,
Bakula-Zalewska E and Bidzinski M: Identification of suitable
reference genes for gene expression measurement in uterine sarcoma
and carcinosarcoma tumors. Clin Biochem. 45:368–371. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cui W, Taub DD and Gardner K:
qPrimerDepot: a primer database for quantitative real time PCR.
Nucleic Acids Res. 35:D805–809. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jacinto FV and Esteller M: MGMT
hypermethylation: a prognostic foe, a predictive friend. DNA Repair
(Amst). 6:1155–1160. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hegi ME, Diserens AC, Gorlia T, et al:
MGMT gene silencing and benefit from temozolomide in glioblastoma.
N Engl J Med. 352:997–1003. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Preusser M, Charles Janzer R, Felsberg J,
et al: Anti-O6-methylguanine-methyltransferase (MGMT)
immunohistochemistry in glioblastoma multiforme: observer
variability and lack of association with patient survival impede
its use as clinical biomarker. Brain Pathol. 18:520–532. 2008.
|