Introduction
Oral squamous cell carcinoma (OSCC) is one of the
most common malignant tumors of the oral and maxillofacial region.
The occurrence and development of OSCC are multi-stage processes
involving a variety of changes at the gene level, including the
activation of oncogenes and the inactivation of tumor suppressor
genes (1). Apoptosis is a mechanism
that is responsible for the physiological deletion of cells and
appears to be intrinsically programmed in certain physiological or
pathological conditions. The inactivation of apoptosis-related
tumor suppressor genes may lead to abnormal cell proliferation and
eventually to tumorigenesis.
Apoptosis protease activating factor-1 (Apaf-1) and
death-associated protein kinase (DAPK) are apoptosis-related tumor
suppressor genes that have generated interest due to their
downregulated expression in various tumors. Studies have shown that
their reduced expression is associated with promoter
hypermethylation (2–5). DAPK is able to activate the
p19ARF/p53-dependent apoptotic pathway through phosphorylation of
p19ARF (6). p53 then triggers the
mitochondrial apoptotic pathway, leading to DNA fragmentation and
apoptosis by inducing the expression of certain specific apoptotic
genes, including Bax, PUMA, Noxa and Apaf-1. The significance of
p53 in initiating the early stages of apoptosis has been widely
confirmed (7). Therefore, studies
with regard to the expression of Apaf-1 and DAPK in OSCC may
contribute to exploring the anti-apoptotic pathway of tumor cells
and may also provide guidance for future treatment.
The expression and methylation levels of Apaf-1 and
DAPK in OSCC tissues or cells have rarely been reported. The
present study detected the expression and methylation of Apaf-1 and
DAPK in OSCC tissues and Tca8113 tongue squamous cell carcinoma
cell lines. Demethylation was also observed in the transcriptional
regulation of Apaf-1 and DAPK in the Tca8113 cell line.
Materials and methods
Patients, diagnosis and samples
A total of 33 male and 20 female patients with OSCC,
with a median age of 55 years (range, 40–72 years), were selected
for the study. A control group comprising 23 cases of normal oral
mucosa was used. No patients were administered antitumoral
treatment prior to the tumor samples being taken. The diagnosis of
OSCC was based on standard criteria. A total of 19 patients (35.8%)
were classified with stage I tumors, 25 (47.2%) with stage II and 9
(17%) with stage III. All the samples were obtained using aseptic
techniques in the operating theatre and placed into liquid nitrogen
immediately. No patients were administered chemotherapy or
radiotherapy prior to surgery. All the samples were obtained
according to the agreement of the ethics committee of the Medical
Department of Jilin University (Changchun, Jilin, China).
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total RNA was extracted from 53 OSCC samples, 23
normal oral mucosa samples and from the Tca8113 cell line using
TRIzol (Invitrogen, Carlsbad, CA, USA), according to the
manufacturer’s instructions, and stored at −80°C. All RNA samples
with an A260/280 ratio >1.9 were selected for the RT procedure
using a Quantscript RT kit (Tiangen, Beijing, China). cDNA
amplification and the detection of specific products were performed
using PCR. Apaf-1 and DAPK expression was determined on the basis
of the endogenous control, β-actin. The specific primer sequences
are given in Table I. PCR was
performed in a thermal cycler using the following cycling
conditions: 94°C for 5 min, 30 cycles at 94°C for 30 sec, 55°C for
30 sec, 72°C for 30 sec and a final extension of 10 min at 72°C.
The PCR mixture contained 2 μl cDNA, 1 μl of each
primer (10 μM), 12.5 μl 2X Taq MasterMix in a final
volume of 25 μl. The 10 μl PCR products were then
loaded onto 1.5% (m/v) agarose gel, electrophoresed and visualized
under ultraviolet light subsequent to being stained with ethidium
bromide.
 | Table I.Specific primers used in PCR. |
Table I.
Specific primers used in PCR.
| Gene name | Primer sequence | Length (bp) | Product size
(bp) |
|---|
| Apaf-1 | | | |
| F | 5′-TTG CTG CCC TTC
TCC ATG AT-3′ | 20 | 318 |
| R | 5′-TCC CAA CTG AAA
CCC AAT GC-3′ | 20 | |
| DAPK | | | |
| F | 5′-GAT AGA AAT GTC
CCC AAA-3′ | 18 | 343 |
| R | 5′-TCT TCT TTG GAT
CCT TGA-3′ | 18 | |
| β-actin | | | |
| F | 5′-GTG GGG CGC CCC
AGG CAC CA-3′ | 20 | 540 |
| R | 5′-CTC CTT AAT GTC
ACG CAC GAT TTC-′ | 24 | |
Methylation-specific PCR (MSP)
Genomic DNA was extracted from 53 OSCC samples, 23
normal oral mucosa samples and from the Tca8113 cell line using the
TIANcombi DNA Lyse&Amp PCR kit (Tiangen). MSP was performed on
the Apaf-1 and DAPK promoter regions. The DNA was modified and
purified using the EpiTect Bisulfite kit (Qiagen, Hilden, Germany).
The primer sequences that were used to detect the methylated and
unmethylated promoters in the Apaf-1 and DAPK genes are shown in
Table II. MSP was performed in a
thermal cycler using the following cycling profile: Apaf-1, 94°C
for 5 min, 39 cycles at 94°C for 30 sec, 61°C for 90 sec, 72°C for
60 sec and a final extension of 5 min at 72°C; DAPK, 95°C for 5
min, 40 cycles at 95°C for 30 sec, 58°C for 30 sec, 72°C for 30 sec
and a final extension of 5 min at 72°C. The MSP mixture contained
50 ng of bisulphite-treated DNA, 0.2 mM dNTPs, 2 mM
MgCl2, 10 pmol of each primer, 1X PCR buffer and 1 unit
Taq polymerase at a final volume of 50 μl. The 10 μl
PCR products were then loaded onto 1.5% (m/v) agarose gel,
electrophoresed and visualized under ultraviolet light subsequent
to being stained with ethidium bromide.
 | Table II.Specific primers used in MSP. |
Table II.
Specific primers used in MSP.
| Gene name | Primer sequence | Length (bp) | Product size
(bp) | Reference |
|---|
| Apaf-1 | | | | |
| Methylation | | | | |
| F | 5′-GAG GTG TCG TAG
CGG TAT TC-3′ | 20 | 212 | (3) |
| R | 5′-CGA AAA TTA ACG
AAA TAA ACG TC-3′ | 23 | | |
| Unmethylation | | | | |
| F | 5′-ATT TGA GGT GTT
GTA GTG GTA TTT G-3′ | 25 | 221 | |
| R | 5′-ACC TCC AAA AAT
TAA CAA AAT AAA CAT-3′ | 27 | | |
| DAPK | | | | |
| Methylation | | | | |
| F | 5′-GGA TAG TCG GAT
CGA GTT AAC GTC-3′ | 24 | 98 | (8) |
| R | 5′-CCC TCC CAA ACG
CCG -3′ | 15 | | |
| Unmethylation | | | | |
| F | 5′-GGA GGA TAG TTG
GAT TGA GTT AAT GTT-3′ | 27 | 106 | |
| R | 5′-CAA ATC CCT CCC
AAA CAC CAA-3′ | 21 | | |
5-Aza-deoxycytidine (DAC) treatment of
the Tca8113 cell line
The Tca8113 cells were cultured in Iscove’s modified
Dulbecco’s medium (Invitrogen Gibco) supplemented with 10% (v/v)
newborn calf serum. The cells in the logarithmic proliferative
phase were seeded in a 96-well plate at a density of
5×103 cells/well and cultured in an incubator at 37°C
with 5% CO2 saturated humidity for 24 h. The cells were
then treated with 1, 3 or 5 μM DAC (Sigma-Aldrich, St.
Louis, MO, USA) for 72 h, with a fresh medium containing DAC that
was replenished every 24 h. The cells in the treatment and control
groups were harvested. Cell viability was detected using MTT and
the cell morphology was observed under a contrast phase microscope.
Apoptosis of the Tca8113 cells was studied using an Annexin V-PI
dual staining assay and TUNEL. Demethylation of the Apaf-1 and DAPK
promoter regions was detected by MSP and the mRNA expression of
each gene was detected by RT-PCR.
Statistical analysis
Quantitative data were expressed as arithmetic mean
± standard deviation (SD) and analyzed using the SPSS program
(SPSS® release 16.0, Chicago, IL, USA). mRNA expression
in the normal oral mucosa and OSCC samples was analyzed by a
t-test. The results were analyzed using the relative optical
density of the target gene ratio to β-actin (mRNA index). mRNA
expression was considered to be decreased in tumor tissues if the
mRNA index was <50% than that in normal tissues and was
considered normal if the mRNA index was ≥50% that in normal
tissues. Pearson correlation was performed to assess Apaf-1 and
DAPK gene expression in OSCC tissues. The association between mRNA
expression and other clinical parameters, including the
pathological grade, age and gender of patients, was studied using
the χ2 test. Gene methylation was compared between the
normal oral mucosa and OSCC groups using the χ2 test.
The χ2 test was also performed to study the association
between gene methylation and mRNA expression. A one-way analysis of
variance was used to evaluate the statistical significance in the
cell experiment. The P-value was evaluated according to Tukey’s
method. All P-values were two-sided and P=0.05 was considered to
indicate a statistically significant difference.
Results
RT-PCR
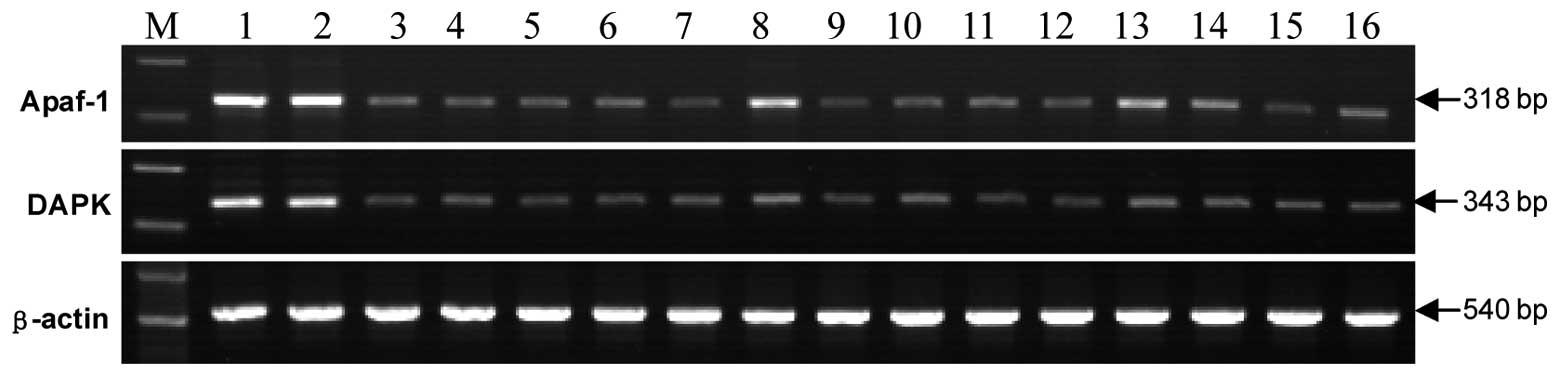
Apaf-1 and DAPK mRNA expression was detected in all
23 cases of normal oral mucosa and the positive rate was 100%, with
no gene reduction or deletion. The mRNA index of the Apaf-1 and
DAPK genes was 50% greater in two (3.77%) and three (5.66%) OSCC
tissues compared to that of normal tissues, while in the remaining
51 (96.23%) and 50 (94.34%) cases, the mRNA index of the two genes
showed a decreased expression (Fig.
1; Table III).
 | Table III.Statistical analysis of Apaf-1 and
DAPK mRNA expression. |
Table III.
Statistical analysis of Apaf-1 and
DAPK mRNA expression.
| Gene | Tumor (n=53) | Control (n=23) | P-value |
|---|
| Apaf-1/β-actin | 0.11±0.02 | 0.52±0.02 | P<0.01 |
| DAPK/β-actin | 0.10±0.03 | 0.80±0.01 | P<0.01 |
MSP
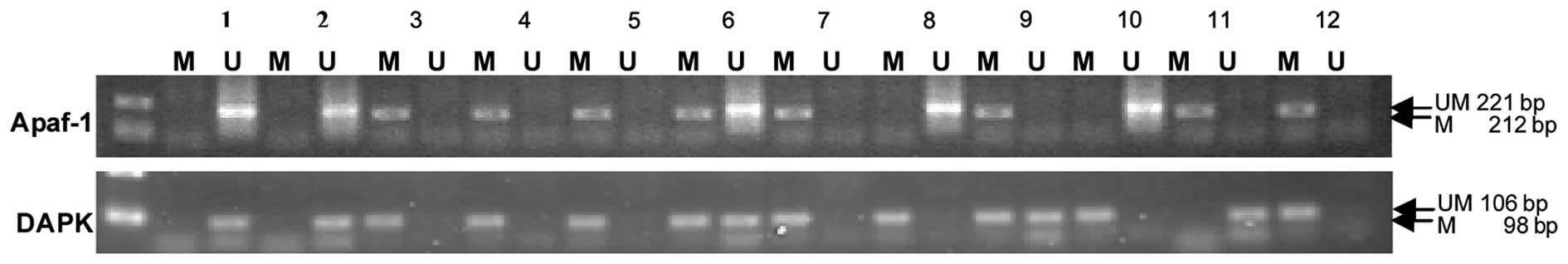
Methylation was not detected in the Apaf-1 and DAPK
promoter regions in the normal oral mucosa samples. However,
exhaustive methylation in the promoter region of the Apaf-1 gene
was detected in 41 cases (77.36%) and partial methylation in 5
cases (9.43%) of OSCC samples. Exhaustive methylation in the
promoter region of the DAPK gene was observed in 30 cases (56.60%)
and partial methylation in 8 cases (15.09%) of OSCC samples. Apaf-1
and DAPK promoter methylation correlated with a decreased mRNA
expression in the OSCC tissues (Fig.
2; Table IV).
 | Table IV.Association between the methylation
status and mRNA expression downregulation of Apaf-1 and DAPK. |
Table IV.
Association between the methylation
status and mRNA expression downregulation of Apaf-1 and DAPK.
| Gene | mRNA decrease
(n) | mRNA normal
(n) | χ2 | P-value |
|---|
| Apaf-1 | | | | |
| Methylated | 46 | 0 | 13.658 | P<0.01 |
| Unmethylated | 5 | 2 | | |
| DAPK | | | | |
| Methylated | 38 | 0 | 8.256 | P<0.01 |
| Unmethylated | 12 | 3 | | |
DAC treatment of the Tca8113 cell
line
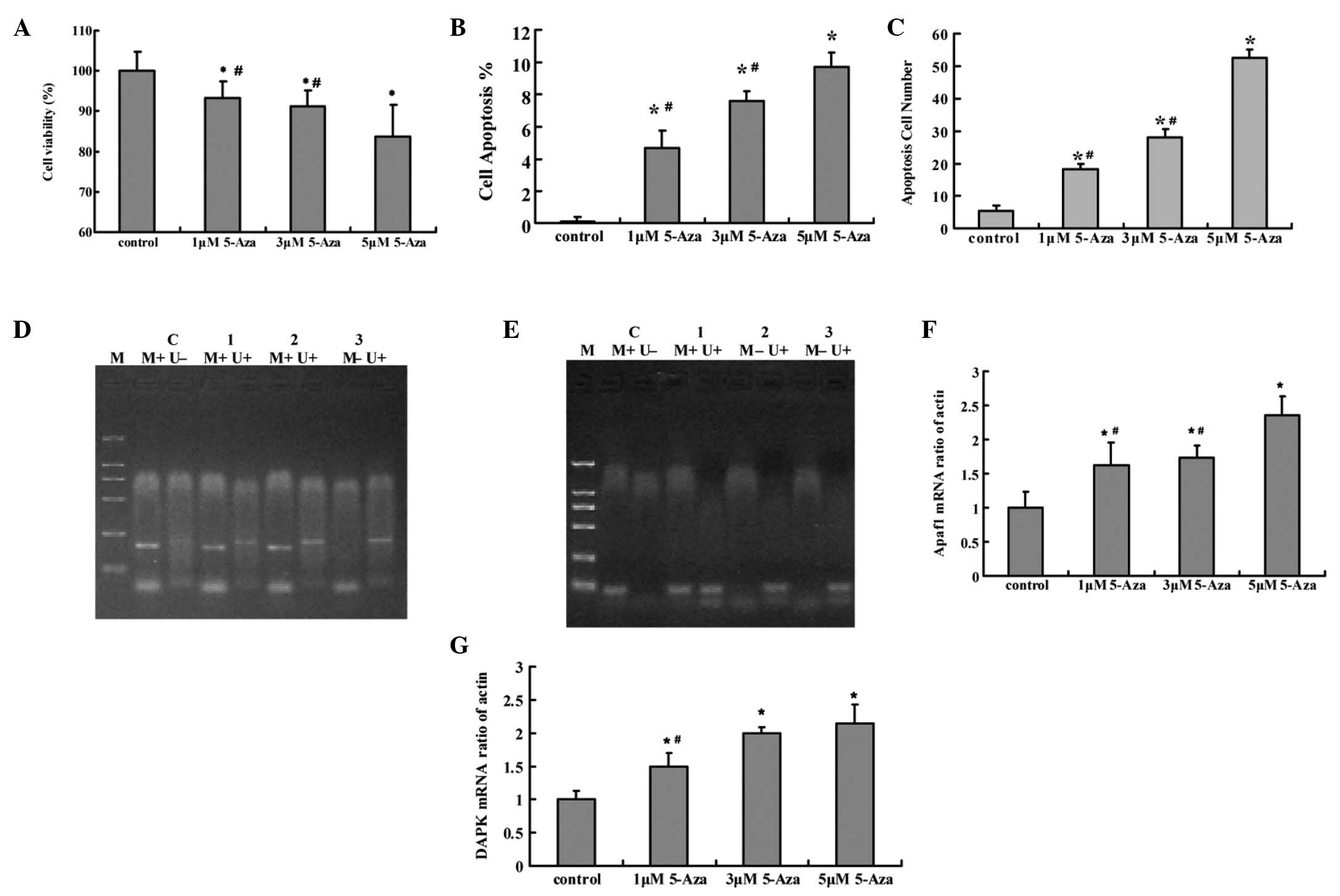
Following treatment with DAC, the Tca8113 cell
viability rate was significantly decreased compared with that of
the control group (P<0.05). Apoptosis of the Tca8113 cells was
induced by DAC and was also significantly different compared with
that of the control group (P<0.05). A dose-dependent effect was
observed in the cell viability and apoptosis. The two genes were
demethylated following treatment with DAC and the most significant
effect was observed in the 5 μM DAC group. Apaf-1 and DAPK
mRNA expression in the Tca8113 cells was significantly increased
following treatment with DAC compared with the control group
(P<0.05). A dose-dependent effect was observed in the mRNA
expression. (Fig. 3)
Statistical analysis
Apaf-1 and DAPK mRNA expression was significantly
decreased in OSCC tissues compared with the normal oral mucosa
samples (P<0.01). The Pearson correlation coefficient was 0.950
(P<0.01), which indicated that a correlation existed between the
expression of the two genes in the OSCC tissues (Table V). The total methylation rate of the
Apaf-1 and DAPK genes was 86.79% (46/53) and 71.69% (38/53),
respectively in the OSCC tissues, which is significantly different
compared with the normal oral mucosa samples (P<0.01). Apaf-1
and DAPK mRNA expression and promoter methylation were not shown to
be significantly correlated with the pathological grade, age and
gender of the patients (P>0.05; Tables VI and VII).
 | Table V.Pearson correlation analysis between
Apaf-1 and DAPK. |
Table V.
Pearson correlation analysis between
Apaf-1 and DAPK.
| Gene | n | Mean ± SD | P-value |
|---|
| Apaf-1 | 53 | 0.5113±0.2131 | P<0.01 |
| DAPK | 53 | 0.2944±0.2922 | |
 | Table VI.Association between the mRNA
expression of Apaf-1 and DAPK and pathological grade, age and
gender of the patients. |
Table VI.
Association between the mRNA
expression of Apaf-1 and DAPK and pathological grade, age and
gender of the patients.
| Variable | n | Apaf-1 mRNA
| χ2 | P-value | DAPK mRNA
| χ2 | P-value |
|---|
| Decreased | Normal | Decreased | Normal |
|---|
| Grade, n | | | | 3.719 | >0.05 | | | 1.512 | >0.05 |
| I | 19 | 17 | 2 | | | 17 | 2 | | |
| II | 25 | 25 | 0 | | | 24 | 1 | | |
| III | 9 | 9 | 0 | | | 9 | 0 | | |
| Gender, n | | | | 0.019 | >0.05 | | | 0.183 | >0.05 |
| Male | 29 | 28 | 1 | | | 27 | 2 | | |
| Female | 24 | 23 | 1 | | | 23 | 1 | | |
| Age (years), n | | | | 2.711 | >0.05 | | | 0.701 | >0.05 |
| <55 | 23 | 21 | 2 | | | 21 | 2 | | |
| ≥55 | 30 | 30 | 0 | | | 29 | 1 | | |
 | Table VII.Association between the Apaf-1 and
DAPK methylation status and the pathological grade, age and gender
of patients. |
Table VII.
Association between the Apaf-1 and
DAPK methylation status and the pathological grade, age and gender
of patients.
| Variable | n | Apaf-1
| χ2 | P-value | DAPK
| χ2 | P-value |
|---|
| + | − | + | − |
|---|
| Grade | | | | 2.421 | >0.05 | | | 0.266 | >0.05 |
| I | 19 | 15 | 4 | | | 13 | 6 | | |
| II | 25 | 22 | 3 | | | 18 | 7 | | >0.05 |
| III | 9 | 9 | 0 | | | 7 | 2 | | |
| Gender | | | | 0.458 | >0.05 | | | 0.547 | >0.05 |
| Male | 29 | 26 | 3 | | | 22 | 7 | | |
| Female | 24 | 20 | 4 | | | 16 | 8 | | |
| Age (years) | | | | 0.620 | >0.05 | | | 0.359 | >0.05 |
| <55 | 23 | 19 | 4 | | | 15 | 8 | | |
| ≥55 | 30 | 27 | 3 | | | 23 | 7 | | |
Discussion
Epigenetics is a study of heritable changes that
occur in gene function without changes in the sequence of nuclear
DNA (9). The understanding of
epigenetic mechanisms, including DNA methylation and changes in
chromatin structure, has shown rapid progress (10). These changes may induce gene
silencing, imprinting, paramutation and RNA interference.
Therefore, abnormal modification may lead to tumorigenesis
(11). Results of studies have
demonstrated that Apaf-1 and DAPK are tumor suppressor genes, which
rarely mutate. Therefore, the mechanism underlying the loss of
function in the genes may be closely associated with promoter
hyper-methylation (12–14).
The functional significance of Apaf-1 and DAPK
methylation in OSCC is uncertain. Apaf-1 and DAPK mRNA expression
and promoter hypermethylation status in OSCC have never been
reported previously. The present study demonstrated that the
expression of the Apaf-1 and DAPK mRNA indices was decreased (96.23
and 94.34%, respectively) and the gene promoter region was
hypermethylated (86.79 and 71.69%, respectively) in OSCC. Further
analysis showed that hypermethylation of Apaf-1 (P<0.05) and
DAPK (P<0.01) promoter regions correlated with a decreased mRNA
expression in the tumor tissues. The results indicate that
epigenetic alterations of the Apaf-1 and DAPK genes exist and may
play a role in OSCC.
As the core of the apoptosome complex, the Apaf-1
gene is a significant pro-apoptotic factor in the mitochondrial
apoptosis pathway (15) and may be
directly or indirectly associated with a number of diseases
(16,17). Since the Apaf-1 gene is rarely
mutated, promoter methylation appears to be more frequent in
tumors. (18–20). As a positive regulator of apoptosis
(21), the DAPK gene participates
in numerous transduction pathways that are mediated by p53, TNF-α,
Fas, TNF-β and other apoptotic factors, which also undergo promoter
hyper-methylation in various types of tumors. An analysis of DNA
hypermethylation in the serum of patients with non-small-cell lung
carcinoma has shown that the DAPK hypermethylation frequency was
68.4% (22). DAPK is frequently
methylated in malignant mesothelioma, which is associated with gene
silencing and may be of prognostic significance (23). Studies have shown the DAPK gene to
be hypermethylated in head and neck squamous cell carcinoma
(24), bladder cancer (25), B-cell lymphoma (26) and cervical cancer (27).
In the present study, following statistical
analysis, the expression and hypermethylation of Apaf-1 and DAPK
was not significantly correlated with the pathological grade, age
and gender of the patients (P>0.05). The Pearson correlation
coefficient was 0.950 (P<0.01), indicating that the expression
of the two genes in the OSCC tissues were correlated. Since they
are the significant genes in the DAP kinase/p53/Apaf-1 apoptotic
pathway, obtaining information in future studies may provide novel
information for the treatment of OSCC.
The results have shown that Apaf-1 and DAPK promoter
hypermethylation correlated with a decreased mRNA expression,
indicating that the two genes may regain expression following
demethylation. A dynamic pattern of methylation requires the
presence of methylating and demethylating activities. The most
controversial issue in the DNA methylation field is the question of
whether DNA methylation is reversible (28). The DNA methylation reaction is
catalyzed by DNA methyltransferase (DNMT). The consensus is that
DNMT inhibitors are only active in replicating cells, through the
passive inhibition of DNA methylation by blocking DNMT1 during DNA
synthesis (29). As a type of DNMT1
inhibitor, DAC has been shown to reactivate tumor suppressor genes
that have been silenced by promoter DNA methylation, in order to
restore their function of inducing apoptosis, inhibiting tumor cell
growth and providing therapeutics for chemotherapy-resistance
tumors (30). Studies have
indicated that DAC may be able to restore the function of the
Apaf-1 gene in acute myeloid leukemia (31) and induce apoptosis of bladder cancer
cells by reversing the unmethylated status of the DAPK promoter
(32). The mechanism of function is
associated with the induction of terminal differentiation,
senescence or apoptosis, resulting in an irreversible loss of
proliferative potential (33).
Since Apaf-1 and DAPK promoter hypermethylation has been detected
in OSCC, the present study aimed to confirm the demethylation role
of DAC on the two genes in OSCC cells. The results revealed that
apoptosis of the Tca8113 cells was induced by DAC. Furthermore, the
cell viability rate was significantly decreased and the cell
apoptotic rate was significantly increased in the Tca8113 cells
that were treated by DAC. Demethylation of the Apaf-1 and DAPK
genes correlated with the upregulation of the mRNA, indicating that
methylation is responsible for the inactivation of Apaf-1 and DAPK
in OSCC. A dose-dependent effect was observed in cell viability,
apoptosis and mRNA expression. In the present study, the mRNA
expression of Apaf-1 and DAPK was upregulated following gene
demethylation and the genes were able to exert their pro-apoptotic
roles.
In conclusion, the present study is consistent with
numerous observations in carcinogenesis that have identified the
loss of Apaf-1 and DAPK as a key feature in tumor progression. The
occurrence and development of OSCC may be closely associated with a
significant decrease in Apaf-1 and DAPK mRNA expression. In OSCC
tissues, the Apaf-1 and DAPK promoter regions are usually
methylated. Gene hypermethylation frequently leads to a decrease in
Apaf-1 and DAPK mRNA expression. The Apaf-1 and DAPK gene
expression was increased following demethylation, which promoted
apoptosis of the tumor cells. Therefore, Apaf-1 and DAPK may serve
as significant diagnostic markers or potential targets for OSCC
treatment. Further research should clarify the DAPK/p53/Apaf-1
apoptosis pathway and its association with other pathways. Since
5-Aza-2′-deoxycytidine has limitations in clinical application,
drug combination studies are also a significant aspect in
additional investigations.
Acknowledgements
This study was financially supported
by the Jilin Provincal Science & Technology Department of China
(Grant no. 200705349). The authors would like to thank Ling Zhang,
Guangxiang Zang and Chunhua Zhou for their expert technical
assistance and Liwei Wang for linguistic advice.
References
|
1.
|
Kodama M, Murakami M and Kodama T:
Topological evidence of differential oncogene activation-tumor
suppressor gene inactivation features in 10 human neoplasias, as
revealed by sequential regression analysis of world cancer
incidence data. Anticancer Res. 17:3809–3816. 1997.
|
|
2.
|
Christoph F, Kempkensteffen C, Weikert S,
Köllermann J, Krause H, Miller K, et al: Methylation of tumour
suppressor genes APAF-1 and DAPK-1 and in vitro effects of
demethylating agents in bladder and kidney cancer. Br J Cancer.
95:1701–1707. 2006. View Article : Google Scholar
|
|
3.
|
Wang HL, Bai H, Li Y, Sun J and Wang XQ:
Rationales for expression and altered expression of apoptotic
protease activating factor-1 gene in gastric cancer. World J
Gastroenterol. 13:5060–5064. 2007.PubMed/NCBI
|
|
4.
|
Leung RC, Liu SS, Chan KY, Tam KF, Chan
KL, Wong LC, et al: Promoter methylation of death-associated
protein kinase and its role in irradiation response in cervical
cancer. Oncol Rep. 19:1339–1345. 2008.
|
|
5.
|
Kato K, Iida S, Uetake H, Takagi Y,
Yamashita T, Inokuchi M, et al: Methylated TMS1 and DAPK genes
predict prognosis and response to chemotherapy in gastric cancer.
Int J Cancer. 122:603–608. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Raveh T, Droguett G, Horwitz MS, DePinho
RA and Kimchi A: DAP kinase activates a p19ARF/p53-mediated
apoptotic checkpoint to suppress oncogenic transformation. Nat Cell
Biol. 3:1–7. 2001.PubMed/NCBI
|
|
7.
|
Pruschy M, Rocha S, Zaugg K, Tenzer A,
Hess C, Fisher DE, et al: Key targets for the execution of
radiation-induced tumor cell apoptosis: the role of p53 and
caspases. Int J Radiat Oncol Biol Phys. 49:561–567. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Soria JC, Rodriguez M, Liu DD, Lee JJ,
Hong WK and Mao L: Aberrant promoter methylation of multiple genes
in bronchial brush samples from former cigarette smokers. Cancer
Res. 62:351–355. 2002.
|
|
9.
|
Conerly M and Grady WM: Insights into the
role of DNA methylation in disease through the use of mouse models.
Dis Model Mech. 3:290–297
|
|
10.
|
Mitsiades CS and Anderson KC: Epigenetic
modulation in hematologic malignancies: challenges and progress. J
Natl Compr Canc Netw. 7(Suppl 8): S1–S12; quiz. S14–S16.
2009.PubMed/NCBI
|
|
11.
|
Lopez-Serra L and Esteller M: Proteins
that bind methylated DNA and human cancer: reading the wrong words.
Br J Cancer. 98:1881–1885. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Tost J: DNA methylation: an introduction
to the biology and the disease-associated changes of a promising
biomarker. Methods Mol Biol. 507:3–20. 2009. View Article : Google Scholar
|
|
13.
|
Suzuki H, Toyota M, Sato H, Sonoda T,
Sakauchi F and Mori M: Roles and causes of abnormal DNA methylation
in gastrointestinal cancers. Asian Pac J Cancer Prev. 7:177–185.
2006.PubMed/NCBI
|
|
14.
|
Suzuki E, Imoto I, Pimkhaokham A, Nakagawa
T, Kamata N, Kozaki KI, et al: PRTFDC1, a possible tumor-suppressor
gene, is frequently silenced in oral squamous-cell carcinomas by
aberrant promoter hypermethylation. Oncogene. 26:7921–7932. 2007.
View Article : Google Scholar
|
|
15.
|
Lindholm D and Arumäe U: Cell
differentiation: reciprocal regulation of Apaf-1 and the inhibitor
of apoptosis proteins. J Cell Biol. 167:193–195. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Sánchez I, Xu CJ, Juo P, Kakizaka A,
Blenis J and Yuan J: Caspase-8 is required for cell death induced
by expanded polyglutamine repeats. Neuron. 22:623–633.
1999.PubMed/NCBI
|
|
17.
|
Gervais FG, Xu D, Robertson GS,
Vaillancourt JP, Zhu Y, Huang J, et al: Involvement of caspases in
proteolytic cleavage of Alzheimer’s amyloid-beta precursor protein
and amyloidogenic A beta peptide formation. Cell. 97:395–406.
1999.
|
|
18.
|
Johnson CE, Huang YY, Parrish AB, Smith
MI, Vaughn AE, Zhang Q, et al: Differential Apaf-1 levels allow
cytochrome c to induce apoptosis in brain tumors but not in normal
neural tissues. Proc Natl Acad Sci USA. 104:20820–20825. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Hinz S, Kempkensteffen C, Weikert S,
Schostak M, Schrader M, Miller K, et al: EZH2 polycomb
transcriptional repressor expression correlates with methylation of
the APAF-1 gene in superficial transitional cell carcinoma of the
bladder. Tumour Biol. 28:151–157. 2007. View Article : Google Scholar
|
|
20.
|
Christoph F, Weikert S, Kempkensteffen C,
Krause H, Schostak M, Köllermann J, et al: Promoter
hypermethylation profile of kidney cancer with new proapoptotic p53
target genes and clinical implications. Clin Cancer Res.
12:5040–5046. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Cohen O, Inbal B, Kissil JL, Raveh T,
Berissi H, Spivak-Kroizaman T, et al: DAP-kinase participates in
TNF-alpha- and Fas-induced apoptosis and its function requires the
death domain. J Cell Biol. 146:141–148. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Hoffmann AC, Kaifi JT, Vallböhmer D,
Yekebas E, Grimminger P, Leers JM, et al: Lack of prognostic
significance of serum DNA methylation of DAPK, MGMT, and GSTPI in
patients with non-small cell lung cancer. J Surg Oncol.
100:414–417. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Chim CS, Liang R, Fung TK, Choi CL and
Kwong YL: Epigenetic dysregulation of the death-associated protein
kinase/p14/HDM2/p53/Apaf-1 apoptosis pathway in multiple myeloma. J
Clin Pathol. 60:664–669. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
De Schutter H, Geeraerts H, Verbeken E and
Nuyts S: Promoter methylation of TIMP3 and CDH1 predicts better
outcome in head and neck squamous cell carcinoma treated by
radiotherapy only. Oncol Rep. 21:507–513. 2009.PubMed/NCBI
|
|
25.
|
Jarmalaite S, Jankevicius F, Kurgonaite K,
Suziedelis K, Mutanen P and Husgafvel-Pursiainen K: Promoter
hypermethylation in tumour suppressor genes shows association with
stage, grade and invasiveness of bladder cancer. Oncology.
75:145–151. 2008. View Article : Google Scholar
|
|
26.
|
Takino H, Li C, Hu S, Kuo TT, Geissinger
E, Muller-Hermelink HK, et al: Primary cutaneous marginal zone
B-cell lymphoma: a molecular and clinicopathological study of cases
from Asia, Germany, and the United States. Mod Pathol.
21:1517–1526. 2008. View Article : Google Scholar
|
|
27.
|
Zhao XL, Meng ZY, Qiao YH and Zhang HL:
Promoter methylation of DAPK gene in cervical carcinoma. Ai Zheng.
27:919–923. 2008.(In Chinese).
|
|
28.
|
Ramchandani S, Bhattacharya SK, Cervoni N
and Szyf M: DNA methylation is a reversible biological signal. Proc
Natl Acad Sci USA. 96:6107–6112. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Fridman AL, Tang L, Kulaeva OI, Ye B, Li
Q, Nahhas F, et al: Expression profiling identifies three pathways
altered in cellular immortalization: interferon, cell cycle, and
cytoskeleton. J Gerontol A Biol Sci Med Sci. 61:879–889. 2006.
View Article : Google Scholar
|
|
30.
|
Wang X, Tryndyak V, Apostolov EO, Yin X,
Shah SV, Pogribny IP, et al: Sensitivity of human prostate cancer
cells to chemotherapeutic drugs depends on EndoG expression
regulated by promoter methylation. Cancer Lett. 270:132–143. 2008.
View Article : Google Scholar
|
|
31.
|
Del Poeta G, Bruno A, Del Principe MI,
Venditti A, Maurillo L, Buccisano F, et al: Deregulation of the
mitochondrial apoptotic machinery and development of molecular
targeted drugs in acute myeloid leukemia. Curr Cancer Drug Targets.
8:207–222. 2008.PubMed/NCBI
|
|
32.
|
Xu NR, Liu CX, Zheng SB, Li HL, Xu YW and
Xu K: Reversion transcriptional expression of DAPK in bladder
cancer T24 cells 5-aza-2′-deoxycytidine. Nan Fang Yi Ke Da Xue Xue
Bao. 29:1882–1886. 2009.(In Chinese).
|
|
33.
|
Lemaire M, Chabot GG, Raynal NJ, Momparler
LF, Hurtubise A, Bernstein ML and Momparler RL: Importance of
dose-schedule of 5-aza-2′-deoxycytidine for epigenetic therapy of
cancer. BMC Cancer. 8:1282008.PubMed/NCBI
|