Introduction
Colorectal cancer (CRC) is one of the most common
types of malignant tumor worldwide. In recent years, due to the
early diagnosis and treatment of CRC, the mortality rates have
decreased, however, the prognosis remains poor. Therefore, it is
crucial to identify an effective means for the chemoprevention of
CRC to reduce morbidity and mortality. Indomethacin (IN) belongs to
a group of non-steroidal anti-inflammatory drugs (NSAIDs), which
are associated with the decreased incidence of CRC and familial
adenomatous polyposis tumor formation (1,2).
Previously, it has also been shown that these drugs may inhibit
cell proliferation of esophageal, stomach, liver, pancreatic and
other types of cancer (3–6). In addition, previous studies have
reported a 40–50% decrease in mortality from CRC with the prolonged
use of NSAIDs (7,8). The well-documented pharmacological
action of NSAIDs is the inhibition of cyclooxygenase (COX)-2
(9–11), but NSAIDs in general have numerous
targets, other than COX-2, which may inhibit tumor cell growth and
induce apoptosis, including activation of the transcription factor
nuclear factor κB and extracellular signal-regulated kinase
(ERK)1/2 and reactive oxygen species (ROS) generation (12–14).
However, the molecular mechanisms by which IN exerts its effects
are not well understood. Functional proteomics provide a
high-throughput method to study the complexity of life. Proteome
technology is a useful tool for the identification of new cancer
markers and treatment-related changes in cancer. The comparative
analysis of protein alterations between pre-treated and treated
cells or tissues using high-throughput proteome technology has
allowed for the identification of special treatment-related
proteins and the development of new molecular-based therapies. In
the current study, the total proteins from IN-treated and untreated
groups were separated by immobilized pH gradient-based
two-dimensional gel electrophoresis (2-DE). The various expression
proteins were identified by peptide mass fingerprint (PMF), based
on matrix-assisted laser desorption/ionization time of flight mass
spectrometry (MALDI-TOF-MS). The PMF maps were searched in the
SWISS-PROT/TrEMBL database using PeptIdent software (http://www.expasy.ch/sprot). The purpose of the
current study was to use functional proteomics to identify the
correlated proteins of IN-treated CRC via the non-COX-dependent
pathway.
Materials and methods
Cell culture and materials
The HCT116 human CRC cell line was purchased from
the American Type Culture Collection (ATCC, Manassas, VA, USA). The
cells were cultured in RPMI-1640 medium supplemented with 10% fetal
calf serum in a 37°C, 5% CO2 environment.
IN, thiourea, iodoacetamide, dithiothreitol (DTT)
and the second-dimension sodium dodecyl sulfate (SDS)-PAGE standard
proteins, TPCK-Trypsin, K3Fe(CN)6,
trifluoroacetic acid (TFA), α-cyano-4-hydroxycinnamic acid (CCA),
DTT and acetonitrile (ACN), were obtained from Sigma-Aldrich (St.
Louis, MO, USA). RPMI-1640 and 10% FBS were purchased from the
Medical College of Xiangya (Changsha, China). The BCA Protein Assay
kit, Immobiline pH-gradient DryStrips (pH 3–10; 24 cm), Acrylamide,
SDS, Tris,
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS),
IPGphor isoelectric focusing (IPG-IEF) cell, ImageMaster 2D Elite
4.01 analysis software and LabScan software on Imagescanner were
obtained from Amersham Pharmacia Biotech (Amersham, UK). The
ProTEAN II electrophoresis apparatus was obtained from Bio-Rad
(Hercules, CA, USA) and the Applied Biosystems Voyager System 4307
MALDI-TOF-MS was bought from Applied Biosystems Inc. (Foster City,
CA, USA).
Sample preparation
HCT116 cells were seeded in 75-cm2 tissue
culture flasks and grown for 1–2 days prior to use. When 50%
confluent growth had been reached, the media was changed to fresh
standard media or media containing 316 μmol IN (IC50)
(15). The cells were harvested 48
h after treatment, rinsed with PBS (0.8 g/l NaCl, 0.2 g/l KCl, 1.44
g/l NaH2PO4 and 0.24 g/l
KH2PO4) and trypsinized with a solution of
2.5 g/l trypsin and 0.2 g/l EDTA. After 1 min, media containing FBS
was added to terminate the action of the trypsin. The resulting
suspension was centrifuged at 1,000 rpm for 7 min at 4°C and the
supernatant was discarded. Next, the cells were resuspended in
ice-cold PBS and centrifuged at 1,500 rpm for 10 min at 4°C and the
supernatant was removed. This wash step was repeated three times
and the cells were stored at −80°C until further use. Protein
extraction from the untreated and IN-treated cells was performed
with lysis buffer. The harvested IN-treated and untreated HCT116
cells were left in lysis buffer (7 mol/l urea, 2 mol/l thiourea, 4%
CHAPS, 40 mmol/l Tris and 1 mmol/l PMSF) for 30 min in ice. The
resulting cell lysate was then vortexed and the sample was
incubated at room temperature for 30 min. Following centrifugation
at 15,000 rpm at 4°C for removal of particulate material, the
protein solution was collected and stored at −80°C until use.
Protein concentrations were determined using the Bio-Rad Protein
Assay kit (Bio-Rad) with BSA (Sigma-Aldrich) as the standard.
IPG-2D-PAGE
2-DE was performed mainly according to the
laboratory instructions (15).
IPG-IEF was run on an IEF system (Amersham Pharmacia Biotech).
Immobilized pH gradient strips (24 cm long; pH 3–10) were
rehydrated for 14 h with 450 μl of 2-D solubilizing solution (8
mol/l urea, 2% CHAPS, 0.5% IPG buffer, pH 3–10L, 3% DTT and a trace
of bromophenol blue) containing 260 μg total proteins for
analytical runs and mixed with a rehydration solution to a total
volume of 450 μl. Following rehydration for 14 h, IEF was performed
with a low initial voltage (500–1,000 V) during the initial 2 h and
then a voltage gradient of up to 8,000 V was applied with a
limiting current of 15 μA/strip. The total product time × voltage
applied was 69,920 vh for the analytical runs. The temperature was
maintained at 20°C following IEF separation, and the gel strips
were equilibrated for 2×15 min in an equilibration buffer
containing 50 mmol/l Tris-HCl (pH 8.8), 6 M urea, 30% glycerol, 2%
SDS and a trace of bromophenol blue. DTT (1%) was added to the
first equilibration buffer and was replaced with 2.5% iodoacetamide
in the second equilibration buffer. The equilibrated gel strips
were then applied onto 1-mm thick 12.5% SDS linear polyacrylamide
gradient vertical slab gels and sealed with 0.5% agarose. SDS-PAGE
was run using Bio-Rad Protean II electrophoresis apparatus for 30
min at a constant current of 10 mA/gel and then switched to 25
mA/gel until the bromophenol blue frontier had reached the bottom
of the gels. During the whole run, the temperature was set at 15°C.
To determine the isoelectric point (pI) and molecular weight (Mr)
of the separated proteins, 2-D standards were added to the protein
samples as internal markers. Following 2-DE, the protein spots were
visualized by a silver-based staining technique with the protein
silver stain kit (Amersham Pharmacia Biosciences).
Image analysis
The stained 2-DE gels were scanned using LabScan
software on Imagescanner (Amersham Pharmacia Biotech). The spot
intensity calibration, spot detection, background abstraction,
matching, 1-D calibration and establishment of an average gel were
performed using the ImageMaster 2D Elite 4.01 analysis software
(Amersham Pharmacia Biotech). The intensity of each spot was
quantified by calculating the spot volume following normalization
of the image using the total spot volume normalization method,
multiplied by the total area of all spots. The reproducibility of
the spot position was calculated according to Gorbett’s method
(16). The statistical analysis was
performed with SPSS for Windows 10.0 and Excel (SPSS, Inc.,
Chicago, IL, USA).
Enzymatic digestion of protein spots and
MALDI-TOF-MS analysis of tryptic peptides
In-gel digestion was performed mainly according to
the laboratory instructions. A total of 15 differential spots were
excised from preparative gels using biopsy punches and then
transferred to a 1.5-ml siliconized Eppendorf tube. One
protein-free gel piece was treated in parallel as a negative
control. The gel-spots were destained in a destaining solution
consisting of 100 mmol/l Na2S2O3
and 30 mmol/l K3Fe(CN)6 (V/V, 1:1). The
protein-containing gel-spots were reduced in a reduction buffer
(100 mmol/l NH4HCO3 and 10 mmol/l DTT) for 1
h at 57°C and then in alkylation buffer (100 mmol/l
NH4HCO3 and 55 mmol/l indoacetamide) in the
dark for 30 min at room temperature. The gel pieces were dried in a
vacuum centrifuge and then the dried gel-pieces were incubated in a
digestion solution containing 50 mmol/l
NH4HCO3, 5 Mm/l CaCl2 and 0.1 g/l
TPCK-trypsin for 24 h at 37°C. The digest buffer was removed and
saved. The gel pieces were then extracted with 100% ACN/5% TFA for
1 h at 37°C and the supernatant was removed. The extracts plus the
first saved digest buffer were finally pooled and concentrated to
10 μl. The tryptic peptide mixture (1 μl) was mixed with 1 μl CCA
matrix solution and vortexed gently. A volume (2 μl) of the mixture
containing CCA matrix was loaded onto a stainless steel plate and
air-dried, then 1 μl 0.1% TFA was added and removed after 30 sec,
then air-dried. The sample was analyzed by Applied Biosystems
Voyager System 4307 MALDI-TOF-MS (Applied Biosystems Inc.). The
parameters were set up as follows: Positive ion-reflector mode;
accelerating voltage, 20 KV; grid voltage, 64.5%; mirror voltage
ratio, 1.12; N2 laser wavelength, 337 nm; pulse width, 3
nsec; number of laser shots, 50; acquisition mass range,
1,000–3,000 Da; delay, 100 nsec; and vacuum degree,
4×10−7 Torr. A trypsin-fragment peak served as an
internal standard for mass calibration. A list of the corrected
mass peaks formed the PMF.
Database searching and identification of
proteins
Proteins were identified using the PMF results by
searching the SWISS-PROT/TrEMBL database (http://www.expasy.ch/sprot) using the PeptIdent
software. The search parameters were set as follows: Mass
tolerance, ±0.5 Da; number of missed cleavage sites, ≤1; cysteine
residue modified as carbamidomethyl-cys; number of matched
peptides, ≥4; species selected, Homo sapiens (human);
peptide ion, [M+H]+; isotope masses were used; and the
search range was within the experimental pI value of ±0.5 pH unit
and the experimental Mr of ±20%.
Data analysis
Data are expressed as the mean ± SD with the
exception of the MTT and 2-DE data, which were expressed as the
mean only. Data were analyzed using Student’s t-test. Statistical
analysis was performed using SPSS for Windows 10.0 and Excel (SPSS,
Inc.). P≤0.05 was considered to indicate a statistically
significant difference.
Results
Result of 2-DE and ImageMast
analysis
Using the same conditions and parameters, the
experiment was repeated four times from cell culture to 2-DE,
respectively. For the IN-treated and untreated HCT116 cells, a
clear background and well-resolved and reproducible 2-DE patterns
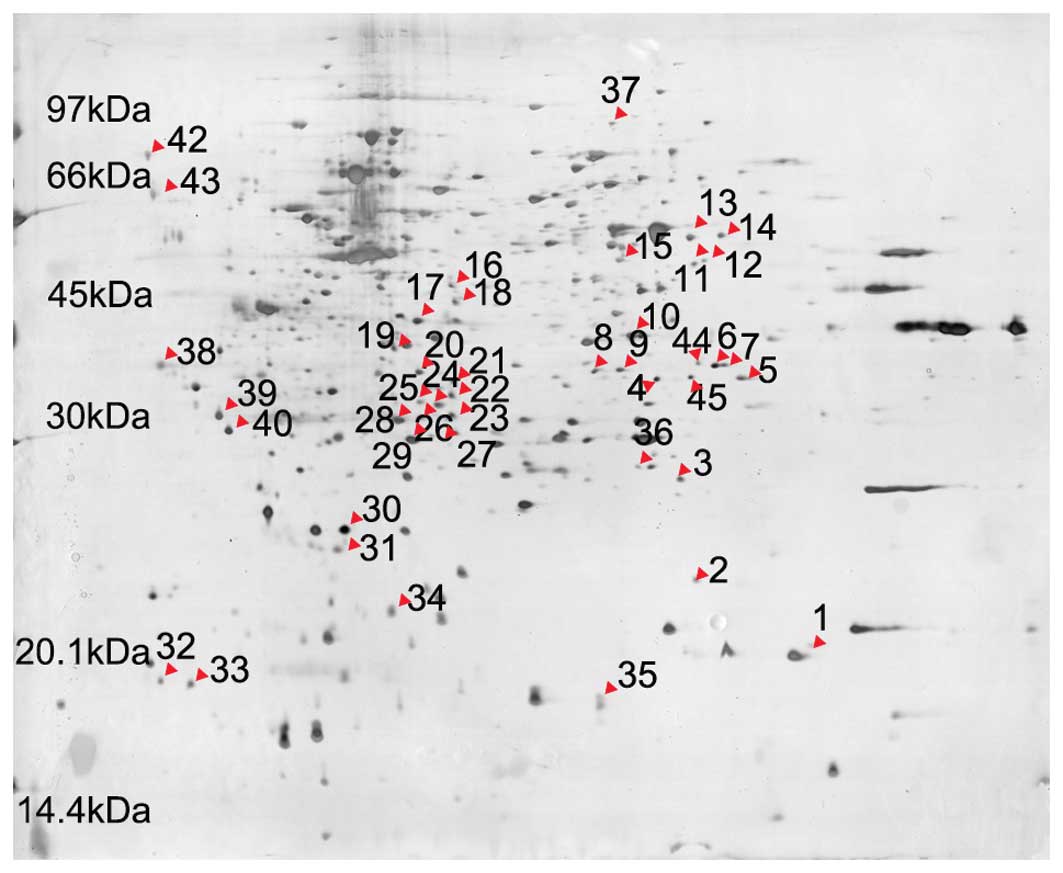
were attained. Fig. 1 shows 2-DE
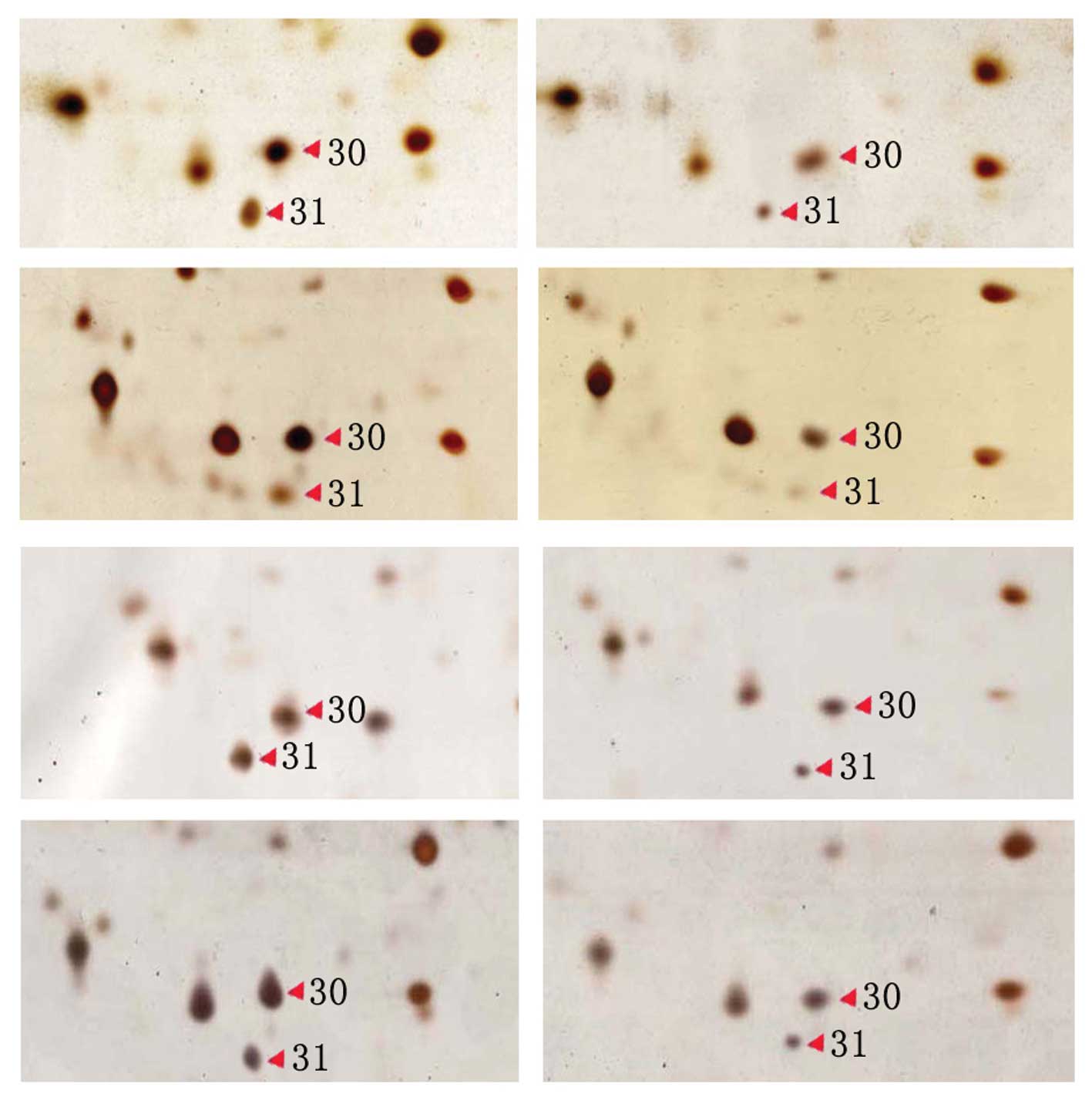
profiles of the untreated cells and Fig. 2 shows the partial 2-DE profiles of
the IN-treated and untreated HCT116 cells at four different times.
The results show that the levels of differentially-expressed
protein spots 30 and 31 were decreased in the IN-treated group.
Compared with the untreated maps, the average number of spots
decreased by 7% in the IN-treated group (P<0.05). Furthermore,
the differentially-detected protein spots between the IN-treated
and untreated groups in the four experiments were consistent with a
significant difference in relative volume (P<0.05) (Table I). Forty-five differential protein
spots were identified between the IN-treated and untreated groups.
Table I shows the relative volumes
of the partial differential proteins.
 | Table ICharacterized differential expression
of IN-treated and untreated HCT116 cells. |
Table I
Characterized differential expression
of IN-treated and untreated HCT116 cells.
| Protein spot no. | Peptide matches,
n | AC | Protein
description | Coverage, % | Theoretical molar
mass, KDa/pI | Experimental molar
mass, KDa/pI | Relative volume of
untreated, % | Relative volume of
IN-treated, % | P-value |
|---|
| 3 | 5/50 | Q14964 | Ras-related protein
Rab-39 | 30.0 | 24.87/6.90 | 27.82/7.61 | 0.238±0.043 | 0.136±0.041 | 0.043 |
| 4 | 9/30 | Q92782 | Zinc finger protein
neuro-d4 | 45.5 | 38.91/6.28 | 38.14/7.44 | 0.236±0.019 | 0.077±0.027 | 0.027 |
| 5 | 6/37 | Q9BQ16–2 | Splice isoform 2 of
testican-3 precursor | 41.2 | 35.59/8.79 | 38.30/8.08 | 0.319±0.029 | 0.122±0.035 | 0.034 |
| 6 | 8/57 | P00747 | Angiostatin | 40.2 | 41.64/7.74 | 40.13/7.87 | 0.334±0.037 | 0.193±0.007 | 0.026 |
| 8 | 9/53 | O14753 | Putative
transcription factor Ovo-like1 | 37.4 | Undefined | 39.32/6.98 | 0.236±0.014 | 0.071±0.012 | 0.017 |
| 9 | 11/60 | P48745 | Chain1:NOV protein
homolog | 35.8 | 36.09/7.95 | 39.54/7.17 | 0.233±0.023 | 0.070±0.014 | 0.010 |
| 11 | 20/76 | Q9Y297–2 | Splice insoform 2
of F-box/ WD-repeat protein 1A | 30.1 | 65.05/8.24 | 56.08/7.72 | 0.227±0.040 | 0.065±0.027 | 0.006 |
| 14 | 11/59 | Q9H7B4 | Set and Mynd domain
containing protein 3 | 31.5 | 49.11/6.75 | 58.02/7.92 | 0.225±0.006 | 0.017±0.001 | 0.001 |
| 20 | 8/40 | P27361 | p44MAPK | 26.8 | 43.74/6.28 | 39.16/5.66 | 0.234±0.054 | 0.080±0.029 | 0.017 |
| 24 | 12/34 | O95388 | WISP-1 | 36.5 | 38.01/6.47 | 34.34/5.80 | 0.320±0.011 | 0.128±0.015 | 0.001 |
| 28 | 9/20 | Q03405 | uPAR | 83.2 | 31.46/5.95 | 32.04/5.51 | 0.404±0.021 | 0.249±0.036 | 0.049 |
| 30 | 4/35 | Q16548 | Bfl-1 | 32.6 | 20.13/5.32 | 25.37/5.10 | 0.516±0.107 | 0.093±0.072 | 0.039 |
| 31 | 5/51 | P12004 | PCNA | 25.9 | 28.77/4.57 | 24.43/5.04 | 0.293±0.009 | 0.077±0.016 | 0.002 |
| 36 | 4/34 | O43609 | spry-1 | 40.3 | Undefined | 28.44/7.29 | 0.070±0.003 | 0.197±0.012 | 0.007 |
| 39 | 5/36 | P17034 | Zinc finger protein
KOX23 | 87.5 | Undefined | 32.79/4.19 | 0.343±0.042 | 0.512±0.063 | 0.029 |
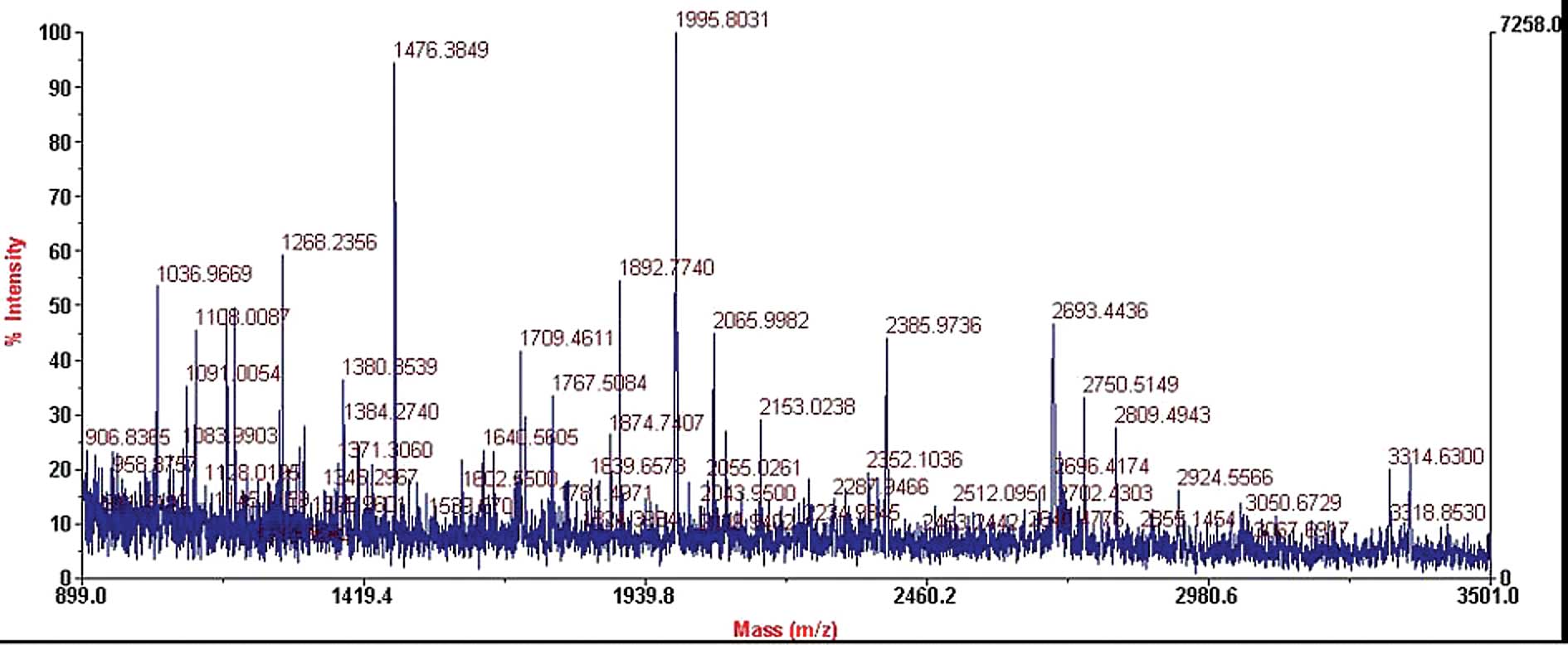
MALDI-TOF-MS PMF analysis of the
differential protein spots
The differential protein spots between the
IN-treated and untreated groups were detected by 2-DE gel image
analysis software. To determine the accuracy of the matched result,
two matched spots (16 differential protein spots in the IN-treated
and untreated groups) were identified by MALDI-TOF-MS. The results
showed that the two matched spots were the same protein [AC O95388,
Wnt1-inducible signaling pathway protein 1 (WISP-1); Fig. 3]. Between ≥3 paired, IN-treated and
untreated HCT116 cells, 15 differential protein spots were
detected. These differential protein spots were excised from the
silver stained gels and digested in-gel with trypsin. The PMF maps
were obtained by MALDI-TOF-MS and calibrated with the TPCK-trypsin
auto-degraded peak (m/z, 1,993.9772 Da). These PMF results were
used to search the SWISS-PROT and TrEMBL databases using the
PeptIdent software. The resulting proteins were determined by
comprehensively considering the corresponding experimental pI, Mr,
number of matched peptides and sequence coverage (Table I). Table II shows the matching of the
differential protein spot 28 PMF results with protein Q03405 in the
database, which identified a signaling pathway protein, WISP-1. It
was first identified that HCT116 cells may be induced to undergo
apoptosis by IN via the Wnt1 signaling pathway. Through the use of
PMF, 15 differential proteins were identified, a number of which
are the products of oncogenes and other molecules involved in the
regulation of the cell cycle, apoptosis and signal
transduction.
 | Table IIMatching of differential protein spot
28 PMF results with protein Q03405 in the database. |
Table II
Matching of differential protein spot
28 PMF results with protein Q03405 in the database.
| User mass(Da) | Matching mass
(Da) | Δ mass (Da) | MC | Modification | Position | Peptide |
|---|
| 1274.2457 | 1274.4745 | 0.2288 | 1 | 1xMSO | 25–35 | CMQCKTNGDCR |
| 1445.2916 | 1445.5389 | 0.2473 | 1 | 3xCys_CAM,
1xMSO | 25–35 | CMQCKTNGDCR |
| 1995.8514 | 1995.7915 | −0.0598 | 0 | 3xCys_PAM | 114–129 |
YLECISCGSSDMSCER |
| 1995.8514 | 1995.8027 | −0.0486 | 1 | | 114–131 |
YLECISCGSSDMSCERGR |
| 2679.0663 | 2679.2324 | 0.1661 | 1 | 1xCys_PAM | 198–220 |
CNEGPILELENLPQNGRQCYSCK |
| 2692.4470 | 2692.3393 | −0.1076 | 1 | 2xCys_CAM | 81–105 |
TGLKITSLTEVVCGLDLCNQGNSGR |
| 2750.4127 | 2750.2695 | −0.1431 | 1 | 2xCys_PAM | 198–220 |
CNEGPILELENLPQNGRQCYSCK |
| 2810.3884 | 2810.1637 | −0.2246 | 1 | 2xCys_PAM | 215–238 |
QCYSCKGNSTHGCSSEETFLIDCR |
| 3123.8803 | 3123.4436 | −0.4366 | 1 | Cys_PAM:144 | 139–164 |
SPEEQCLDVVTHWIQEGEEGRPKDDR |
| 3124.9551 | 3125.2825 | 0.3274 | 0 | 1xCys_PAM,
1xMSO | 262–290 |
GCATASMCQHAHLGDAFSMNHIDVSCCTK |
Discussion
Compared with the genome, the proteome is dynamic,
showing the various expression levels of proteins in various
tissues and cells, the stages of up-growth and the physiology or
pathology. The basic challenges of proteomics are identifying the
proteins and predicting their functions. This is likely to lead to
a new understanding of human biology, as well as to the design of
new molecular structures as potential novel diagnostic or drug
discovery targets (17). 2-DE is
regarded as the most powerful separation method for resolving
complex mixtures of proteins. Combined with mass spectrometry and
ever-growing protein databases, powerful tools have become
available that have made ‘functional proteomics’ feasible.
Functional proteomics is defined as the use of proteomic methods to
monitor and analyze molecular networks and fluxes within the living
cell and to identify the molecular species that participate in
these networks upon perturbation of the cellular environment. In
order to further explore the molecular mechanisms of IN on CRC, a
high-throughput proteomics technique was used in the current study
to address the molecular basis of this effect by the study of the
protein expression profiles of HCT116 cells prior to and following
IN treatment. Analysis of the 2-DE profiles of the IN-treated and
untreated cells was repeated four times. Quantitative expression
changes were exhibited in 45 differential protein spots following
IN-treatment, 34 of which decreased in abundance, 10 of which
showed higher expression and 2 of which were expressed in untreated
cells only. Differential protein spots (n=15) were selected to
perform in-gel trypsin digestion and MALDI-TOF-MS-based PMF
analysis, which identified a new signaling pathway protein, WISP-1.
It was first identified that HCT116 cells may be induced to undergo
apoptosis by IN via the Wnt1 signaling pathway. Specific proteins
were shown to be the products of oncogenes and other molecules were
involved in the regulation of the cell cycle, apoptosis and signal
transduction.
The present study identified that IN induced
apoptosis and inhibited the proliferation of HCT116 cells through
various independent-COX methods. In addition, it was identified
that HCT116 cells may be induced to undergo apoptosis by IN via the
Wnt1 signaling pathway. Wnt family members are critical to a number
of developmental processes, and components of the Wnt signaling
pathway have been associated with tumorigenesis in familial and
sporadic colon carcinomas (18).
Wnt-1 is a member of an expanding family of cysteine-rich and
glycosylated signaling proteins that mediate diverse developmental
processes, including the control of cell proliferation, adhesion,
cell polarity and the establishment of cell fates. It has been
previously shown that WISP-1 activates the antiapoptotic Akt/PKB
signaling pathway. It has also been demonstrated that WISP-1
prevents cells from undergoing apoptosis following DNA damage
through the inhibition of the mitochondrial release of cytochrome
c and the upregulation of antiapoptotic
Bcl-XL(19). In the
current study, the 24th differential protein spot was
identified as WISP-1, which decreased in expression in the
IN-treated HCT116 cells. This implies that IN induces apoptosis and
inhibits the proliferation of HCT116 cells.
In addition, the present study identified that the
30th differential protein spot decreased in expression
in the IN-treated cells. This spot was identified as Bcl-2-related
protein A1 (BfL-1). The decrease in BfL-1 expression is likely to
be an important mechanism to promote apoptosis in HCT116 cells.
BfL-1 is a new member of the Bcl-2-related proteins, and previous
studies have shown that the expression and regulation of Bcl-2
family members is one of the key factors of apoptosis. The Bcl-2
protein family is divided into antiapoptotic (e.g. BaX) and
proapoptotic (e.g. Bcl-2 and Bcl-XL) proteins.
Antiapoptotic Bax and proapoptotic Bcl-2 appear to form
heterodimers, and the existence of Bcl-2/Bax heterodimers are
responsible for the suppression and acceleration of apoptosis
(20). It is known that members of
the Bcl-2 family contain two conserved regions, Bcl-2 homology 1
and 2 (BH1 and BH2). Previous studies have shown that the deletion
of BH1 and BH2 is important in order to form heterodimers with BAX.
Overexpression of the Bcl-2 protein leads to Bcl-2/Bax
heterodimers, which inhibit apoptosis (21,22).
Thus, the ratio of Bcl-2 and Bax determines apoptosis sensitivity.
Further investigations have shown (23) that NSAIDs may inhibit the expression
of antiapoptotic protein Bcl-XL, resulting in an altered
ratio of BAX to Bcl-XL and subsequently, to
mitochondria-mediated cell death. Chung et al(14) hypothesized that the overexpression
of Bcl-2 or Bcl-x(L) prevents ROS production and the subsequent
loss of mitochondrial membrane potential (Δψm), thereby, inhibiting
apoptotic cell death. In the current study, IN induced the
decreased expression of BfL-1, which may indicate the induced
apoptosis of the HCT116 cells. Additionally, BfL-1 has two highly
conserved regions known as BH1 and BH2 domains, which are necessary
for the interaction between Bcl-2 or Bcl-XL and BAX
(22). It may be noteworthy to
determine the additional BfL-1 heterodimers with BAX.
Mitogen-activated protein kinases (MAPK) are
important signal transduction systems from extracellular signaling
to intracellular reactions in eukaryotic cells. MAPK pathways are
involved in a diverse set of responses affecting cell fate,
including cell proliferation and differentiation, adaptation to
environmental stress and apoptosis (24). MAPK cascades are key signaling
pathways involved in the regulation of normal cell proliferation,
survival and differentiation. The aberrant regulation of MAPK
cascades contributes to cancer and other human diseases. The MAPKs
are conserved proteins that regulate cell growth, division and
death. To date, four types of MAPK pathways have been defined in
mammalian cells. These include the extracellular signal regulated
protein kinase pathway, the c-Jun NH2-terminal
kinase/stress activated protein kinase pathway, the big MAPK/ERK5
pathway and the MAPKp38 pathway (25,26).
The MAPK module includes three kinases that establish a sequential
activation pathway comprising of MAPK kinase kinase (MKKK), MAPK
kinase and MAPK. At present, it has been hypothesized that
activation of the MAPK pathway contributes to cell hyperplasia and
that the aberrant MAPK pathway contributes to apoptosis inhibition.
The MAPK signal transduction pathway has been shown to be
significant for the development of cell malignant transformation
and tumor invasion and metastasis (27,28).
The current study identified that the 3rd
and 20th differential protein spots exhibited decreased
expression in IN-treated cells. These spots were identified as the
Ras-related protein, Rab-39A, and MAPK3, p44 MAPK. The results show
that the Ras-associated protein, Rab-39A, and the p44 MAPK protein
are action targets of IN. Ligation of numerous receptors leads to
the activation of MAPKerk1/2 through the activation of
Ras. This was inferred from the observation that Ras activates
MKKKraf1 and that activated MKKKraf1 is
sufficient to stimulate the MAPKerk1/2 signaling pathway
(24). It has been hypothesized
that the activated p38 and p44 MAPK may direct ERKs into the
cellular nucleus, which induces transcription factor
phosphorylation, cell proliferation and differentiation. p44 MAPK
is an isomer of ERK, named ERK1. Berger et al(29) showed that seven cell lines were
noted to have mutations of K-ras, while seven cell lines did not.
No difference in the expression of Raf-1 was identified between the
K-ras mutant and non-mutant cell lines. However, there was a
significant increase in MAPK activity in the non-mutant cell lines
compared with the cell lines with Ras mutations (P=0.026). This may
be associated with the active expression of Raf-1 in these cell
lines. Schwenger et al(30)
hypothesized that IN fails to induce a significant activation of
p38 MAPK, but that salicylates induce p38 MAPK activation. In the
current study, the p44 MAPK protein was downregulated in the
IN-treated cells. The results showed that IN inhibited the
proliferation of the HCT116 cells through the inhibited expression
of the p44 MAPK protein in the MAPK signaling pathway. These
results are consistent with the inhibited proliferation of
pancreatic tumor cells with NSAID therapy via the MEK-ERK signaling
pathway (31).
Overall, the use of functional proteomics in the
present study showed that IN may induce apoptosis and inhibit the
proliferation of HCT116 cells by independent-COX. This confirms a
new method to study the antitumor mechanisms of IN.
Acknowledgements
The present study was supported by a grant from the
National Natural Science Foundation of China (no. 30271516)
Abbreviations:
|
NSAIDs
|
non-steroidal anti-inflammatory
drugs
|
|
COX
|
cyclooxygenase
|
|
2-DE
|
two-dimensional gel
electrophoresis
|
|
PMF
|
peptide mass fingerprint
|
|
IN
|
indomethacin
|
|
MALDI-TOF-MS
|
matrix-assisted laser
desorption/ionization time of flight mass spectrometry
|
References
|
1
|
Cole BF, Logan RF, Halabi S, Benamouzig R,
Sandler RS, Grainge MJ, Chaussade S and Baron JA: Aspirin for the
chemoprevention of colorectal adenomas: meta-analysis of the
randomized trials. J Natl Cancer Inst. 101:256–266. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Martin C, Connelly A, Keku TO, Mountcastle
SB, Galanko J, Woosley JT, Schliebe B, Lund PK and Sandler RS:
Nonsteroidal anti-inflammatory drugs, apoptosis, and colorectal
adenomas. Gastroenterology. 123:1770–1777. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cheng J, Imanishi H, Amuro Y and Hada T:
NS-398, a selective cyclooxygenase 2 inhibitor, inhibited cell
growth and induced cell cycle arrest in human hepatocellular
carcinoma cell lines. Int J Cancer. 99:755–761. 2002. View Article : Google Scholar
|
|
4
|
Shureiqi I, Xu X, Chen D, Lotan R, Morris
JS, Fischer SM and Lippman SM: Nonsteroidal anti-inflammatory drugs
induce apoptosis in esophageal cancer cells by restoring
15-lipoxygenase-1 expression. Cancer Res. 61:4879–4884.
2001.PubMed/NCBI
|
|
5
|
Tang C, Wang C and Tang L: Effects of
combined octreotide and aspirin on the growth of gastric cancer.
Chin Med J (Engl). 116:373–377. 2003.PubMed/NCBI
|
|
6
|
Molina MA, Sitja-Arnau M, Lemoine MG,
Frazier ML and Sinicrope FA: Increased cyclooxygenase-2 expression
in human pancreatic carcinomas and cell lines: growth inhibition by
nonsteroidal anti-inflammatory drugs. Cancer Res. 59:4356–4362.
1999.
|
|
7
|
Baron JA, Cole BF, Sandler RS, Haile RW,
Ahnen D, Bresalier R, McKeown-Eyssen G, Summers RW, Rothstein R,
Burke CA, et al: A randomized trial of aspirin to prevent
colorectal adenomas. N Engl J Med. 348:891–899. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Y, Eu W, Seow-Choen F and Cheah Y:
Differential cytostatic effect of sodium salicylate in human
colorectal cancers using an individualized histoculture system.
Cancer Chemother Pharmacol. 49:473–478. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Boudreau MD, Sohn KH, Rhee SH, Lee SW,
Hunt JD and Hwang DH: Suppression of tumor cell growth both in nude
mice and in culture by n-3 polyunsaturated fatty acids: mediation
through cyclooxygenase-independent pathways. Cancer Res.
61:1386–1391. 2001.
|
|
10
|
Elder DJ, Halton DE, Crew TE and Paraskeva
C: Apoptosis induction and cyclooxygenase-2 regulation in human
colorectal adenoma and carcinoma cell lines by the
cyclooxygenase-2-selective non-steroidal anti-inflammatory drug
NS-398. Int J Cancer. 86:553–560. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lönnroth C, Andersson M and Lundholm K:
Indomethacin and telomerase activity in tumor growth retardation.
Int J Oncol. 18:929–937. 2001.PubMed/NCBI
|
|
12
|
Yamamoto Y, Yin MJ, Lin KM and Gaynor RB:
Sulindac inhibits activation of the NF-kappaB pathway. J Biol Chem.
274:27307–27314. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rice PL, Washington M, Schleman S, Beard
KS, Driggers LJ and Ahnen DJ: Sulindac sulfide inhibits epidermal
growth factor-induced phosphorylation of extracellular-regulated
kinase 1/2 and Bad in human colon cancer cells. Cancer Res.
63:616–620. 2003.
|
|
14
|
Chung YM, Bae YS and Lee SY: Molecular
ordering of ROS production, mitochondrial changes, and caspase
activation during sodium salicylate-induced apoptosis. Free Radic
Biol Med. 34:434–442. 2003. View Article : Google Scholar
|
|
15
|
Cheng YL, Zhang GY, Xiao ZQ and Tang FQ:
Two-dimensional polyacrylamide gel electrophoresis analysis of
indomethacin-treated human colon cancer cells. World J
Gastroenterol. 11:2420–2425. 2005. View Article : Google Scholar
|
|
16
|
Corbett JM, Dunn MJ, Posch A and Görg A:
Positional reproducibility of protein spots in two-dimensional
polyacrylamide gel electrophoresis using immobilised pH gradient
isoelectric focusing in the first dimension: an interlaboratory
comparison. Electrophoresis. 15:1205–1211. 1994. View Article : Google Scholar
|
|
17
|
Haberkorn U, Altmann A and Eisenhut M:
Functional genomics and proteomics - the role of nuclear medicine.
Eur J Nucl Med Mol Imaging. 29:115–132. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pennica D, Swanson TA, Welsh JW, Roy MA,
Lawrence DA, Lee J, Brush J, Taneyhill LA, Deuel B, Lew M, et al:
WISP genes are members of the connective tissue growth factor
family that are up-regulated in wnt-1-transformed cells and
aberrantly expressed in human colon tumors. Proc Natl Acad Sci USA.
95:14717–14722. 1998. View Article : Google Scholar
|
|
19
|
Su F, Overholtzer M, Besser D and Levine
AJ: WISP-1 attenuates p53-mediated apoptosis in response to DNA
damage through activation of the Akt kinase. Genes Dev. 16:46–57.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Reed JC: Bcl-2 and the regulation of
programmed cell death. J Cell Biol. 124:1–6. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zha H, Aimé-Sempé C, Sato T and Reed JC:
Proapoptotic protein Bax heterodimerzies with Bcl-2 and with Bax
via a novel domain (BH3) distinct from BH1 and BH2. J Biol chem.
271:7440–7444. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yin XM, Oltvai ZN and Korsmeyer SJ: BH1
and BH2 domains of Bcl-2 are required for inhibition of apoptosis
and heterodimerization with Bax. Nature. 369:321–323. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang L, Yu J, Park BH, Kinzler KW and
Vogelstein B: Role of BAX in the apoptotic response to anticancer
agents. Science. 290:989–992. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Widmann C, Gibson S, Jarpe MB and Johnson
GL: Mitogen-activated protein kinase: conservation of a
three-kinase module from yeast to human. Physiol Rev. 79:143–180.
1999.PubMed/NCBI
|
|
25
|
Kyriakis JM and Avruch J: Mammalian
mitogen-activated protein kinase signal transduction pathways
activated by stress and inflammation. Physiol Rev. 81:807–869.
2001.PubMed/NCBI
|
|
26
|
Hayashi M, Fearns C, Eliceiri B, Yang Y
and Lee JD: Big mitogen-activated protein kinase 1/extracellular
signal-regulated kinase 5 signaling pathway is essential for
tumor-associated angiogenesis. Cancer Res. 65:7699–7706. 2005.
|
|
27
|
Hilger RA, Scheulen ME and Strumberg D:
The Ras-Raf-MEK-ERK pathway in the treatment of cancer. Onkologie.
25:511–518. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Denkert C, Koch I, Berger S, Köbel M,
Siegert A and Hauptmann S: Cytokine-suppressive anti-inflammatory
drugs (CSAIDs) inhibit invasion and MMP-1 production of ovarian
carcinoma cells. Cancer Lett. 195:101–109. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Berger DH, Jardines LA, Chang H and
Ruggeri B: Activation of Raf-1 in human pancreatic adenocarcinoma.
J Surg Res. 69:199–204. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schwenger P, Alpert D, Skolnik EY and
Vilcek J: Cell-type-specific activation of c-Jun N-terminal kinase
by salicylates. J Cell Physiol. 179:109–114. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yip-Schneider MT and Schmidt CM: MEK
inhibition of pancreatic carcinoma cells by U0126 and its effect in
combination with sulindac. Pancreas. 27:337–344. 2003. View Article : Google Scholar : PubMed/NCBI
|