Introduction
Colorectal cancer (CRC) is a major health concern
worldwide as one of the leading causes of cancer-associated
mortality and the third most common cancer diagnosed in males and
females (1,2). The global incidence rate of CRC is
estimated to be 1.2 million new cases per year with a 50% mortality
rate (3), and >90% of CRCs are
adenocarcinomas that originate from benign adenomas of the lining
layer of the bowel. Sporadic cases with no previous relevant
history account for 70–75% of CRCs (4). CRCs of familial and sporadic origins
are caused by a specific, but not necessarily similar, genetic
instability (5,6). Increasing evidence is now available
with regard to the significance of mutations that inactivate tumor
suppressor genes, including adenomatous polyposis coli (APC), p53,
PTEN and KRAS, in the tumorigenesis of colon cancer (7). A mutation in the APC gene is an early
step in the tumorigenesis of colon cancer and is considered to be a
hallmark of sporadic non-hereditary polyposis (6). The inactivation of tumor suppressor
genes in colon cancer tumorigenesis is believed to be associated
with invasiveness (8). The
progression of genetic instability in colon cancer may follow
various pathways, including chromosomal instability (CIN), which is
detected in up to 80% of CRCs and may be accompanied by a loss of
heterozygosity (LOH) and chromosomal rearrangement (9). Another pathway is marked by the
occurrence of microsatellite instability (MSI), which represents a
mutator phenotype that is observed in a number of CRC cases. MSI is
believed to be caused by accumulated mutations in the CPG islands
of hMLH1 and/or the hypermethylation of its promoter, which impairs
the function of the key mismatch repair (MMR) protein (5,10). In
brief, MSI is a change in the length of a repetitive DNA sequence
in tumor tissues compared with the original length in the normal
counterpart of the same patient (11). MSI occurs in the genome through a
slippage process during DNA replication and causes frequent
deletions/insertions of repeat units (12). Such replication errors may occur due
to a decreased fidelity of the replication apparatus, which worsens
if it is added to an impaired MMR system (12). Although germline and somatic
mutations have been detected in various MMR genes, hMLH1 and hMSH2
have shown the strongest correlations with MSI in colon cancer
(13). hMLH1 has been prevalently
connected with sporadic and familial colon cancer pathogeneses
(14). The main chemotherapy regime
for advanced colon cancer cases is 5-fluorouracil, to which MSI
colon cancers have shown resistance (15). Furthermore, MSI has been noted as an
earlier step in the CRC progression pathway in >90% of
hereditary non-polyposis colorectal cancer (HNPCC) patients and in
15–20% of sporadic cases (2,13).
Similar forms of instability have been diagnosed in numerous other
tumors (16,17). These findings highlight the higher
tendency of MMR-deficient tissues to accumulate mutations, develop
MSI and eventually progress to a cancerous state. Hence, it has
been recommended that assessing the MSI status in CRC tumors is
important in obtaining an improved diagnosis, interpretation and
prediction of the disease outcome. In 1997, the National Cancer
Institute (NCI) workshop developed the Bethesda guidelines
(18), which recommend the use of a
five-marker panel to assess the presence and extent of MSI in colon
cancers. The NCI panel includes two mononucleotide repeat sequences
(Bat-25 and Bat-26) and three dinucleotide repeat sequences
(D2S123, D5S346 and D17S250). An MSI status determination is
performed by comparing DNA from tumor and normal tissues. The
results are classified according to the number of altered
microsatellite sequences. If alterations are present in two or more
of the five microsatellite sequences, the cancer is classified as
high MSI (MSI-H). If only one marker is mutated, the cancer is
classified as low MSI (MSI-L). If no changes are present among the
five microsatellites, the tumor is considered to be microsatellite
stable (MSS). Since the investigations of MSI and other
instabilities in CRCs may not be sufficient to fully comprehend the
instabilities and their causes, combining immunohistochemical
techniques to assess MMR protein expression has also been
recommended (13). The Evaluation
of Genomic Applications in Practice and Prevention Working Group
demonstrated that MSI detection is useful for diagnosing suspected
HNPCC (19). In addition, several
studies have suggested a prognostic value for MSI and LOH
assessment (20,21). A number of regions around the globe,
including the United Arab Emirates (UAE), lack adequate
epidemiological data or active long-term cancer registries.
Furthermore, the number of clinicopathological studies is scant. In
2008, a study was conducted at Howard University on archived colon
cancers from the Sultanate of Oman to estimate the MSI status and
MMR defects in this population (22). The study of the Omani subjects
revealed an MSI incidence that was comparable with that of the
United States. The present study analyzed the existence and extent
of MSI, LOH and MMR deficiencies in archived CRCs from the
Department of Pathology at Tawam Hospital (Abu Dhabi, UAE) between
2005 and 2008. This study is the second in the region and the first
to investigate the frequency of MSI, LOH and MMR deficiencies in
colon cancer patients in the UAE.
Materials and methods
Study design and tumor collection
Colorectal carcinomas specimens were archived
between 2005 and 2008 by the Department of Pathology at Tawam
Hospital. Approval for this study was obtained from the Al Ain
Medical District Human Research Ethics Committee and the Faculty of
Medicine and Health Sciences at UAE University (Al Muwaiji, Al Ain,
UAE). Written informed consent was obtained from the patients.
Specimens were collected in paraffin-embedded blocks for the
genetic analyses of MSI and LOH and an immunohistochemical analysis
of MMR protein expression. A total of 114 specimens from 38 CRC
patients were analyzed. All specimens were screened against five
microsatellite markers (NCI panel) and immunohistochemistry was
performed for hMLH1 and hMSH2 expression, as they are the most
frequently reported genes to harbor MMR defects in CRC patients.
The sample of patients consisted of 21 (55.3%) males and 17 (44.7%)
females, resulting in a male-to-female ratio of 12:10. The age and
gender of the patients and the TNM classification of malignant
tumors (TNM) are displayed in Table
I.
 | Table ICRC patients; basic information, TNM
staging and results of MSI, LOH and MMR protein expression. |
Table I
CRC patients; basic information, TNM
staging and results of MSI, LOH and MMR protein expression.
| Patient no. | Age, years | Gender | Tumor stage, TMN | Stage | MSI detected in | LOH detected in | hMLH1 | hMSH2 |
|---|
| 1 | 68 | F | T3, N2, M1 | 4 | MSS | APC | − | + |
| 2 | 61 | M | T2, N2, M0 | 3 | MSS | No LOH | + | + |
| 3 | 49 | M | T3, N1, M0 | 3 | MSS | No LOH | + | + |
| 4 | 64 | F | T3, N2, M0 | 3 | Bat25 | No LOH | − | + |
| 5 | 67 | M | T3, N1, M0 | 3 | MSS | No LOH | + | + |
| 6 | 70 | F | T1, N0, M0 | 1 | MSS | No LOH | + | + |
| 7 | 48 | M | T3, N2, M0 | 3 | Bat25, Bat26, APC,
Mfd15 | No LOH | − | + |
| 8 | 73 | M | T3, N0, Mx | 2 | MSS | No LOH | + | + |
| 9 | 65 | F | T4, N2, M1 | 4 | Bat25, APC, Mfd15,
AFM093xh3 | No LOH | − | − |
| 10 | 69 | M | T3, N1, M0 | 3 | Bat26 | No LOH | + | − |
| 11 | 66 | F | T3, N2, M1 | 4 | MSS | No LOH | + | + |
| 12 | 58 | M | T3, N0, Mx | 2 | MSS | No LOH | + | + |
| 13 | 72 | M | T3, N0, M0 | 2 | MSS | No LOH | + | + |
| 14 | 59 | F | T3, N2, M0 | 3 | MSS | No LOH | + | + |
| 15 | 38 | M | T2, N1, M0 | 3 | Bat25 | No LOH | − | + |
| 16 | 53 | F | T3, N2, M1 | 4 | MSS | No LOH | + | + |
| 17 | 67 | M | T2, N0, M0 | 1 | MSS | No LOH | + | + |
| 18 | 62 | M | T3, N2, M1 | 4 | MSS | Bat26 | + | + |
| 19 | 55 | F | T2, N1, M0 | 3 | MSS | No LOH | + | + |
| 20 | 49 | M | T3, N2, M0 | 3 | Bat25 | No LOH | + | + |
| 21 | 71 | F | T4, N2, M1 | 4 | Bat25, Bat26, APC,
Mfd15, AFM093xh3 | No LOH | − | + |
| 22 | 60 | M | T2, N0, M0 | 1 | MSS | No LOH | + | + |
| 23 | 59 | M | T3, N0, M0 | 2 | APC | No LOH | + | + |
| 24 | 68 | F | T3, N0, Mx | 2 | MSS | No LOH | + | − |
| 25 | 58 | M | T3, N0, M0 | 2 | MSS | No LOH | + | + |
| 26 | 64 | F | T3, N2, M0 | 3 | APC | No LOH | − | + |
| 27 | 67 | M | T2, N1, M0 | 3 | MSS | No LOH | + | + |
| 28 | 60 | M | T3, N1, M0 | 3 | MSS | No LOH | + | + |
| 29 | 59 | F | T2, N0, M0 | 1 | MSS | No LOH | + | + |
| 30 | 52 | M | T3, N2, M1 | 4 | MSS | APC | + | + |
| 31 | 67 | F | T3, N1, M0 | 3 | Bat25 | No LOH | + | + |
| 32 | 69 | F | T2, N1, M0 | 3 | MSS | No LOH | + | + |
| 33 | 68 | M | T2, N0, M0 | 1 | N/A | N/A | + | + |
| 34 | 59 | F | T3, N0, M0 | 2 | N/A | N/A | + | + |
| 35 | 61 | F | T3, N2, M1 | 4 | N/A | N/A | + | + |
| 36 | 57 | M | T3, N1, M0 | 3 | N/A | N/A | + | + |
| 37 | 63 | M | T3, N0, M0 | 2 | N/A | N/A | + | + |
| 38 | 58 | F | T3, N1, M1 | 4 | N/A | N/A | + | + |
Genetic analysis
DNA extraction from the formalin-fixed and
paraffin-embedded tissues was conducted using a QIAamp DNA FFPE
Tissue kit from Qiagen Inc. (Valencia, CA, USA), which included a
specific column to purify PCR-grade DNA from prefixed and
paraffin-embedded tissues. However, in six patients (Table I, Patient nos. 33–38), no DNA was
extracted or the quality of the extracted DNA was not suitable for
the required PCR reactions. Hence, no microsatellite analysis was
assessed for those patients.
Microsatellite analyses were performed using
fluorescently labeled primers from Applied Biosystems (Carlsbad,
CA, USA). These sequences have been described previously (23). Specific information with regard to
the microsatellite marker panel, including the sequences that were
used, is provided in Table II.
 | Table IICharacteristics of microsatellite
markers analyzed. |
Table II
Characteristics of microsatellite
markers analyzed.
| Name (locus) | Primer sequence,
5′→3′ | Unit of
repeats |
PCR-Tmc | Dye | Size, bp |
|---|
| Bat-25 |
| F | TCG CCT CCA AGA ATG
TAA GT | | | | |
| R | TCT GCA TTT TAA CTA
TGG CTC | 1 | 55°C | NED | 119–124 |
| Bat-26 |
| F | TGA CTA CTT TTG ACT
TCA GCC | | | | |
| R | AAC CAT TCA ACA TTT
TTA ACC C | 1 | 55°C | FAM | 112–127 |
| Mfd15
(D17S250) |
| F | GGA AGA ATC AAA TAG
ACA AT | | | | |
| R | GCT GGC CAT ATA TAT
ATT TAA ACC | 2 | 50°C | VIC | 147–163 |
| APC (D5S346) |
| F | ACT CAC TCT AGT GAT
AAA TCG | | | | |
| R | AGC AGA TAA GAC AGT
ATT ACT AGT T | 2 | 55°C | FAM | 107–131 |
| AFM093xh3
(D2S123) |
| F | AAA CAG GAT GCC TGC
CTT TA | | | | |
| R | GGA CTT TCC ACC TAT
GGG AC | 2 | 58°C | PET | 209–232 |
PCR and fragment analysis
Single and multiplex PCR reactions were conducted
using a Biometra T3000 Thermocycler (Biometra, Göttingen, Germany)
to amplify the five markers recommended by NCI (Bat25, Bat26,
D2S123, D5S346 and D17S250). The amplification reactions were
carried out in a 20 μl reaction volume consisting of 1X Gold
Amplitaq Master Mix (Applied Biosystems), with the addition of 100
ng purified genomic DNA and adjusted to a final primer
concentration of 0.2 μM. The cycling conditions consisted of an
initial denaturation cycle at 95°C for 5 min, followed by 29 cycles
at 94°C for 1 min, 50, 55 or 58°C for 45 sec and 72°C for 50 sec,
with a final 12-min extension at 70°C. A fragment analysis was
conducted by loading the PCR products onto an ABI 3130 genetic
analyzer (Applied Biosystems). The fragments were sized and
compared using the Gene Mapper software (Version 4) from the same
manufacturer. All runs included the use of an internal molecular
weight control (LIZ 500 Genescan; Applied Biosystems).
Using capillary array electrophoresis, MSI may be
demonstrated using two main features: de novo alleles that
appear as new peaks (i.e., peaks that did not exist in the normal
tissue genotype) and slipped pre-existing alleles for the few base
pairs (23,24). However, a partial (>35%) to
complete signal loss of one heterozygote allele is an indicator of
LOH (25,26).
MMR protein expression
Tissues were emeddded in paraffin and microdissected
at a thickness of 4 μm for the MMR protein expression analysis. The
TP-125 HLX Ultra Vision Plus Anti-Polyvalent HRP detection system
(Lab Vision, Fremont, CA, USA) was applied using specific
monoclonal antibodies for hMLH1 and hMSH2 (Cell Marque, Rocklin,
CA, USA). Healthy tissue from each patient was used as an internal
control, in addition to the standard controls that were provided by
the manufacturer. The results were designated as positive when a
≥10% proportion of the cell nuclei were stained positively.
Statistical analysis
Statistical analyses of categorical variables were
performed using Fisher’s exact test, the Mann-Whitney U test and
the Kruskal-Wallis test. Hypothesis testing for associations
between MMR and MSI and for LOH was conducted using Fisher’s exact
test. The correlations between gender and MMR, MSI and LOH,
respectively, were also analyzed using Fisher’s exact test. When
MSI was subdivided into MSS and MSI (MSI-L+MSI-H) categories, a
Mann-Whitney U test was conducted to evaluate their associations
with cancer stage and age, respectively. When the three original
MSI categories (MSS, MSI-L and MSI-H) were kept, a Kruskal-Wallis
test was conducted to evaluate their associations with cancer stage
and age, respectively. A Mann-Whitney U test was used to evaluate
the associations between cancer stage and MMR and LOH,
respectively, and the associations between age and MMR and LOH,
respectively. The exact two-sided P-values were calculated and
P<0.05 was considered to indicate a statistically significant
difference (27). All the results
were calculated using SAS software (release 9.2; SAS Institute
Inc., Cary, NC, USA).
Results
MSI and LOH
MSI-H and -L-positive events were observed in 10 of
32 patients (31.3%), including three MSI-H patients (9.4%) and
seven MSI-L patients (21.9%). LOH was detected in another three
patients (9.4%), but the remaining 25 patients (78.1%) showed no
instability and were classified as MSS. The frequencies of MSI and
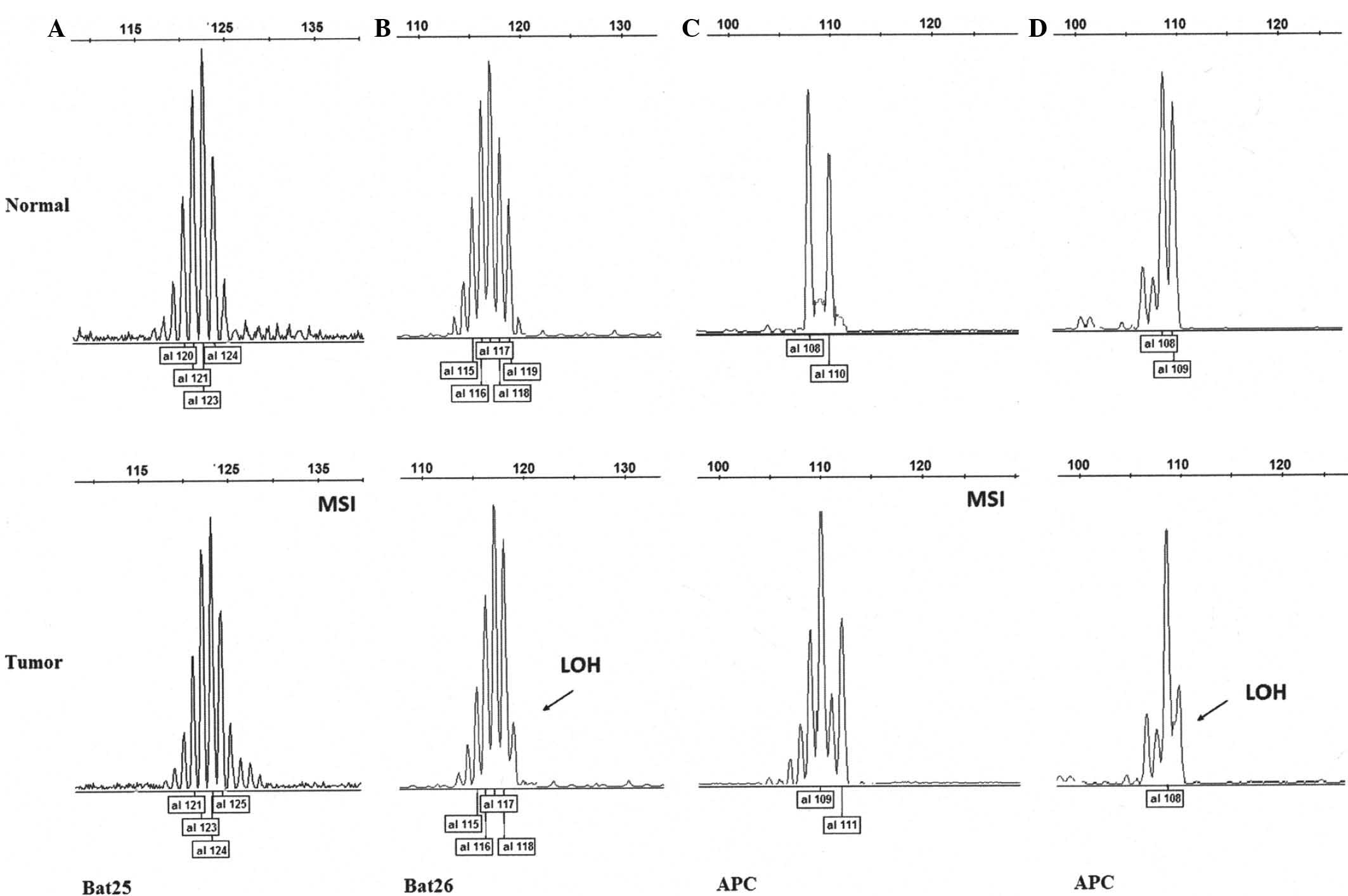
LOH at the individual marker levels are shown in Table I. Bat25 presented with MSI in seven
patients (21.9 %) with no evidence of LOH. Bat26 showed MSI in
three patients (9.4%) and LOH in one patient (3.1%). MSI at APC
(D5S346) was evident in five patients (15.6%) and LOH was
identified in two patients (6.3%; Fig.
1). Whereas AFM093xh3 (D2S123) and Mfd15 (D17S250) harbored MSI
in two (6.3%) and three (9.4%) patients, respectively, there was no
evidence of LOH events in the two loci.
Age and gender
The same number of MSIs were detected in males and
females (five of each gender), with a relatively higher incidence
in females (29.4%) versus males (23.8%). Of the three MSI-H tumors,
two were identified in females. Despite the relatively small study
sample, four of the five male MSI patients were younger than their
female counterparts (38, 48, 49, 59 and 69 years versus 64, 64, 65,
67 and 71 years, respectively; Table
I).
Tumor stage (TNM) and lymph node
involvement
The tumor stages and MSI and LOH statuses are
summarized in Table I. A large
number of the sampled tumors were stage III (44.8%), while stage II
and IV tumors were represented equally with 21% each. Stage I
tumors accounted for 13.1% of the samples and six of the seven
MSI-L tumors were at stage III and one was at stage II. Of the
three MSI-H tumors, two were classed as stage IV and one as stage
III. In total, two of the LOH tumors were at stage IV and one was
at stage III. A pathological assessment of lymph node involvement
estimated that tumor cells were evident in nine of the 10 MSI
tumors (three MSI-H and six MSI-L).
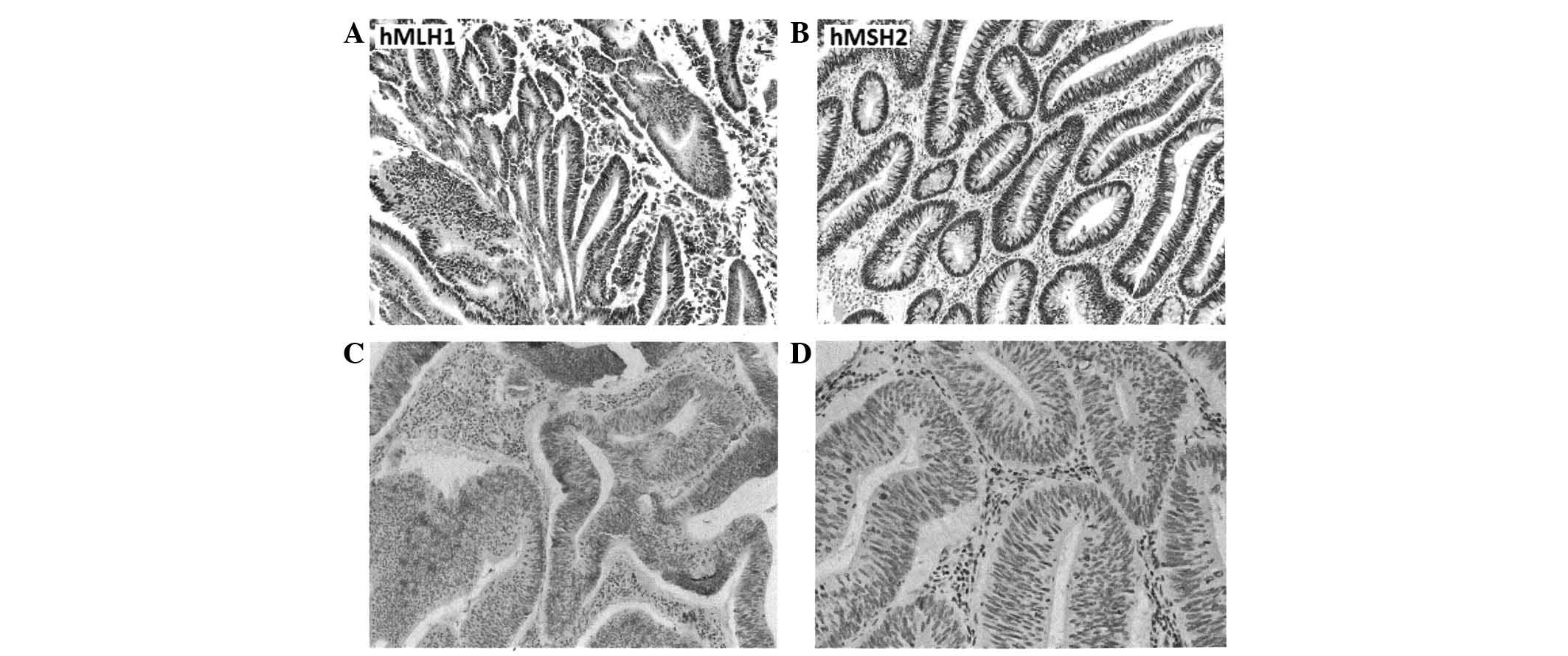
Expression of MMR proteins
An immunohistochemical analysis showed that the
expression of the two studied MMR proteins, hMLH1 and hMSH2, was
deficient in nine patients (23.7%), six of which were deficient in
hMLH1, two in hMSH2 and one in both proteins (Table I). MSI-H events were evident in two
of the six patients who showed hMLH1 deficiencies alone and in the
single patient that was deficient in both MMR proteins. Fig. 2 shows examples of MMR protein
expression in the patients.
Statistical findings
Of the 38 patients in the studied population, 21
(55.3%) were male and 17 (44.7%) were female. The mean age of the
population was 61.4 years and when subdivided by gender, the mean
ages for the females and males were 63.3 and 59.9 years,
respectively. The demographical and clinical characteristics are
displayed in Table III.
 | Table IIIDemographics and clinical
characteristics of the studied population. |
Table III
Demographics and clinical
characteristics of the studied population.
| Characteristic | Subset where MSI
and LOH were analyzed | All |
|---|
| Mean age ± SD,
years | 61.5±8.2 | 61.4±7.6 |
| Gender, n |
| Male | 18 | 21 |
| Female | 14 | 17 |
| Cancer stage,
n |
| I | 4 | 5 |
| II | 6 | 8 |
| III | 15 | 16 |
| IV | 7 | 9 |
| MMR, n |
| hMLH1 |
| (+) | 25 | 31 |
| (−) | 7 | 7 |
| hMSH2 |
| (+) | 29 | 35 |
| (−) | 3 | 3 |
Incidence of MSI, LOH, and MMR
MSI was detected in 10 of 32 patients (31.3%; 95%
CI, 16.1–50.0%). A total of seven of these 10 patients were
classified as MSI-L and three were classified as MSI-H. LOH was
detected in three of the 32 patients (9.4%; 95% CI, 2.0–25.0). MMR
deficiencies, with regard to hMLH1, were detected in seven of 38
patients (18.4%; 95% CI, 7.7–34.3), and a loss of hMSH2 expression
was detected in three of the 38 patients (7.9%; 95% CI,
1.7–21.4).
Association tests
A lack of hMLH1 expression was identified in six of
the patients accompanied with MSI, whereas only four of the 25
patients expressing hMLH1 showed MSI. A statistical analysis using
Fisher’s exact test showed significant associations between hMLH1
and MSI when classified as MSS, MSI-L or MSI-H (P=0.0003), and
between hMLH1 and MSI when subcategorized into MSS or MSI
(MSI-L+MSI-H; P=0.0014). No association was observed between MSI
and hMSH2, age, gender or cancer stage.
The tumors from the three patients showing LOH were
classified as stage IV, and a statistical analysis using a
Mann-Whitney U test revealed a significant association between LOH
and the cancer stage (P=0.0079). No such association was observed
for abnormal MMR protein expression, age, cancer stage or gender.
The association tests are displayed in Table IV.
 | Table IVCorrelations between MSI or LOH and
hMLH1and hMSH2 and between these endpoints and age, gender and
cancer stage, using different statistical methods. |
Table IV
Correlations between MSI or LOH and
hMLH1and hMSH2 and between these endpoints and age, gender and
cancer stage, using different statistical methods.
| Variables | Test of
Association | P-value |
|---|
| MSIa, hMLH1 | Fisher’s exact
test | 0.0003 |
| MSIa, hMSH2 | Fisher’s exact
test | 0.2238 |
| MSIa, age | Kruskal-Wallis
test | 0.7907 |
| MSIa, gender | Fisher’s exact
test | 0.8560 |
| MSIa, cancer stage | Kruskal-Wallis
test | 0.1865 |
| MSIb, hMLH1 | Fisher’s exact
test | 0.0014 |
| MSIb, hMSH2 | Fisher’s exact
test | 0.2238 |
| MSIb, age | Mann-Whitney U
test | 0.5954 |
| MSIb, gender | Fisher’s exact
test | 0.7120 |
| MSIb, cancer stage | Mann-Whitney U
test | 0.2576 |
| LOH, hMLH1 | Fisher’s exact
test | 0.5363 |
| LOH, hMSH2 | Fisher’s exact
test | 1.0000 |
| LOH, age | Mann-Whitney U
test | 0.8615 |
| LOH, gender | Fisher’s exact
test | 1.0000 |
| LOH, cancer
stage | Mann-Whitney U
test | 0.0079 |
| hMLH1, age | Mann-Whitney U
test | 0.9342 |
| hMLH1, gender | Fisher’s exact
test | 0.2074 |
| hMLH1, cancer
stage | Mann-Whitney U
test | 0.0537 |
| hMSH2, age | Mann-Whitney U
test | 0.1127 |
| hMSH2, gender | Fisher’s exact
test | 0.5768 |
| hMSH2, cancer
stage | Mann-Whitney U
test | 0.7046 |
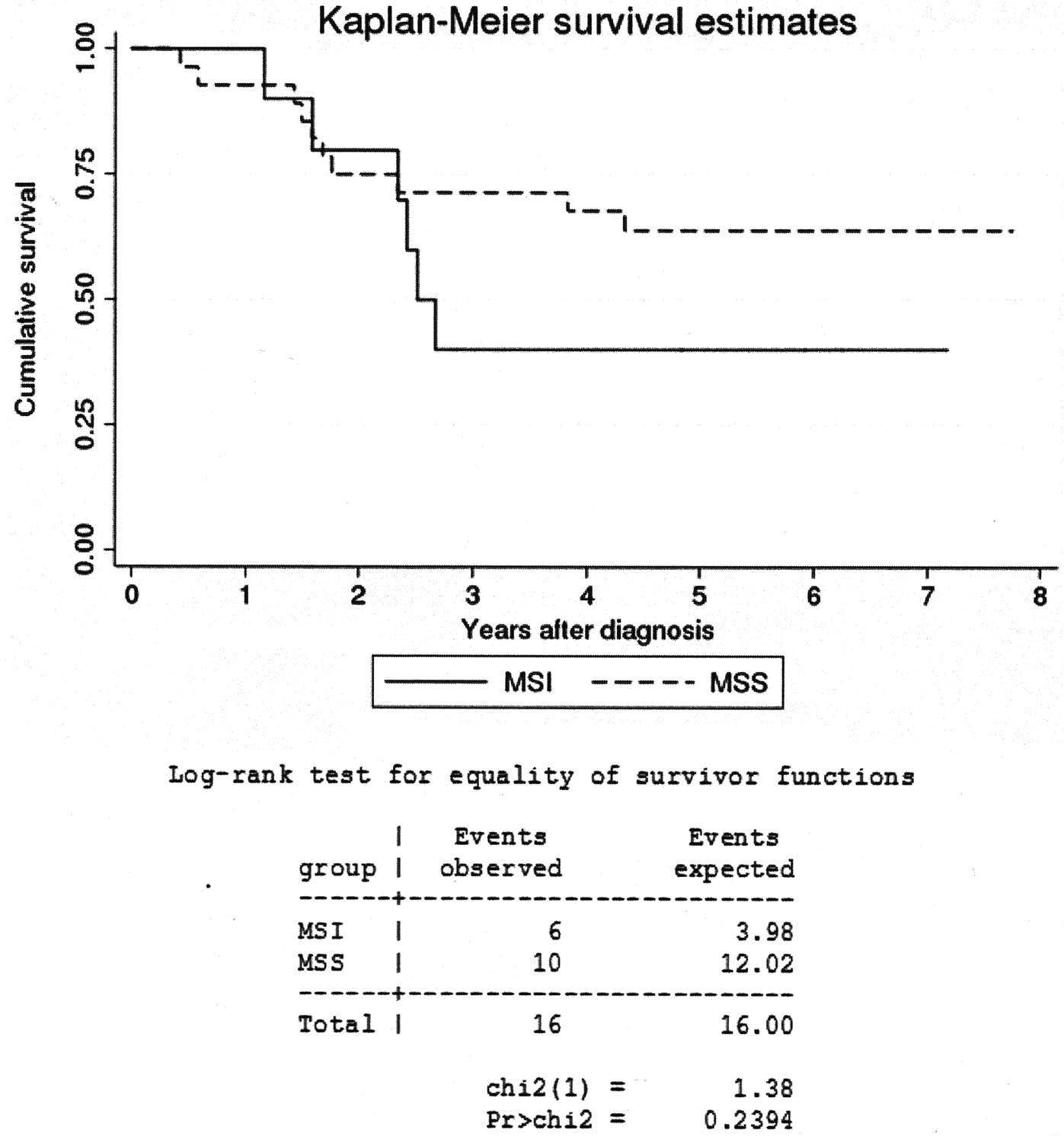
The length of patient survival following the
diagnosis was averaged at 4.1 years. A Kaplan-Meier survival curve
(Fig. 3) and a log-rank test
revealed no significant difference in the survival times between
the patients with MSI-positive tumors and patients with MSS
tumors.
Discussion
Genetic instability is a hallmark of most, if not
all, known cancers. Genetic instability pathways have been shown to
underlie the tumorigenesis of CRC. Of particular interest is MSI,
which shows a marked variation in its incidence between hereditary
and sporadic CRCs (85 and 15–20%, respectively) (2,13).
Studies have elucidated the mechanisms that are believed to play a
pivotal role in MSI occurrence to a certain extent, and a
malfunctioning or inactivated MMR repair apparatus is considered to
be a main cause (9,12). The resultant sequential accumulation
of mutations and mismatches that accidently occurs during DNA
replication is normally corrected by an intact MMR apparatus
(12).
The present study is the first to be designed to
reveal the incidence and clinical significance of MSI and LOH in
patients with CRC in the UAE. An MSI incidence rate of 26.3% (7.9%
MSI-H and 18.4% MSI-L) and an LOH rate of 7.9% were observed. An
MSI-H incidence (MSI in two or more loci) was evident in 7.9% of
CRC patients (three of 38). This rate is lower than the 12.2% that
was detected in a previous study of another population (Omani
patients) in this region (22). The
difference was not statistically significant (Fisher’s exact test)
and was most likely to be due to the relatively small size of the
sample population. In line with the Omani study, MSI was detected
in younger patients compared with studies of other populations
(28,29), particularly among males (Table I). According to the available
medical history, none of the studied patients had a family history
of cancer predisposition. At least one female patient under our
observation developed an ovarian tumor, which may indicate that she
carried a germline mutation that may have predisposed her to HNPCC.
MMR pathway inactivation is widely accepted as a mechanism for
mediating MSI in colorectal carcinoma, particularly hMLH1 and
hMSH2, which are the main two genes involved (6,30). In
hereditary forms of CRC, it is well established that a mutation or
epigenetic alteration, including hypermethylation of an hMLH1
promoter, are typical of this syndrome (9). Although hMLH1 and hMSH2 are implicated
in sporadic CRC mutations, hMLH1 has been observed to be affected
more frequently (31). In the
present results, hMLH1 protein expression was deficient in seven
patients (18.4%) and the hMSH2 protein expression was deficient in
three patients (7.9%). The hMLH1 deficiency correlated with
incidences of MSI-H and MSI-L. These results must be confirmed with
a larger cohort of patients. LOH was detected in three patients
(7.9%), including two at APC (5.3%) and one at Bat26 (2.6%). MMR
protein expression was evidently intact in the three patients with
LOH (Table I). The highest rate of
MSI was detected in Bat25, in concordance with the reported data
that mononucleotide markers are more sensitive to MSI than
dinucleotide repeats (32–34). Of particular interest are the MSI
and LOH events at APC, which were screened using the D5S346 marker.
A total of five patients showed MSI and another two suffered LOH at
this locus. Younger males, aged 48, 59 and 52, respectively, were
identified to display two MSI and one LOH event at APC. The
significance of these findings is emphasized by the frequently
reported role of the inactivation of this tumor suppressor gene in
CRC (35). In fact, it has been
identified that APC is inactivated by LOH or epigenetic alteration
(methylation) in several tumors, including CRC (36,37).
In addition, in familial adenomatous polyposis (FAP), patients who
inherit a germline mutation in the APC gene, the lifetime risk of
developing CRC approaches 100% (38). It is worth noting that the same
types of instability have been detected in hereditary and sporadic
colon cancers, with varied rates and clinical significance
(2). MSI and LOH statuses have been
considered to be valuable and independent prognostic markers in CRC
patients (20). Furthermore, the
type of genetic instability inherited in patients with CRC may play
a role in their survival time, as patients with HNPCC have a
survival time that is estimated to be 5 years longer than that of
patients with inherited FAP (39).
Another significant link was identified between MSI and the
response to certain chemotherapy agents. Colon cancers with MSI
were noticeably resistant to 5-fluoruracil, which remains the
treatment of choice in advanced CRC cases (15).
In terms of the correlation between tumor stage and
lymph node involvement, of the 38 sampled tumors, 25 were in stages
III or IV (16 in stage III and 9 in stage IV) and 13 were observed
in stages I or II (5 in stage I and 8 in stage II). Stage IV tumors
were detected in two of the three MSI-H cases (66.7%). Lymph node
involvement was confirmed in nine of the 10 MSI tumors (90%), which
may indicate why these tumors behaved more aggressively (Table I).
In conclusion, the incidence of MSI in CRC patients
from the UAE is within the rates that have been reported in other
studies of various ethnic and geographical backgrounds. The present
patient group produced noteworthy molecular and clinical findings
in terms of earlier ages of detection, heterozygosity losses and
relations between MSI, lymph node involvement and tumor stage.
Thus, these incidence rates and clinical findings must be verified
through a larger study cohort or population-based study.
Acknowledgements
The authors would like to thank Dr Jan-Olov Persson
and Dr Elin Alvehag from the Department of Mathematics at Stockholm
University for their valuable work on the statistical analysis and
Khaled Al-qawasmeh from the Department of Oncology at Tawam
Hospital, UAE, for collecting the patient data. This study was
supported by a grant from the Sven and Lilly Lawski Foundation.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
2
|
Mishra J, Drummond J, Quazi SH, et al:
Prospective of colon cancer treatments and scope for combinatorial
approach to enhanced cancer cell apoptosis. Crit Rev Oncol Hematol.
2013.PubMed/NCBI
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
4
|
Jasperson KW, Blazer KR, Lowstuter K and
Weitzel JN: Working through a diagnostic challenge: colonic
polyposis, Amsterdam criteria, and a mismatch repair mutation. Fam
Cancer. 7:281–285. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sanchez JA, Krumroy L, Plummer S, et al:
Genetic and epigenetic classifications define clinical phenotypes
and determine patient outcomes in colorectal cancer. Br J Surg.
96:1196–1204. 2009. View
Article : Google Scholar
|
|
6
|
Saif MW and Chu E: Biology of colorectal
cancer. Cancer J. 16:196–201. 2010. View Article : Google Scholar
|
|
7
|
Fearon ER: Molecular genetics of
colorectal cancer. Annu Rev Pathol. 6:479–507. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fearnhead NS, Wilding JL and Bodmer WF:
Genetics of colorectal cancer: hereditary aspects and overview of
colorectal tumorigenesis. Br Med Bull. 64:27–43. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jass JR: Classification of colorectal
cancer based on correlation of clinical, morphological and
molecular features. Histopathology. 50:113–130. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dahlin AM, Palmqvist R, Henriksson ML, et
al: The role of the CpG island methylator phenotype in colorectal
cancer prognosis depends on microsatellite instability screening
status. Clin Cancer Res. 16:1845–1855. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bellizzi AM and Frankel WL: Colorectal
cancer due to deficiency in DNA mismatch repair function: a review.
Adv Anat Pathol. 16:405–417. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sinicrope FA and Sargent DJ: Molecular
pathways: microsatellite instability in colorectal cancer:
prognostic, predictive, and therapeutic implications. Clin Cancer
Res. 18:1506–1512. 2012. View Article : Google Scholar
|
|
13
|
Cicek MS, Lindor NM, Gallinger S, et al:
Quality assessment and correlation of microsatellite instability
and immunohistochemical markers among population- and clinic-based
colorectal tumors results from the Colon Cancer Family Registry. J
Mol Diagn. 13:271–281. 2011. View Article : Google Scholar
|
|
14
|
Kaur G, Masoud A, Raihan N, Radzi M,
Khamizar W and Kam LS: Mismatch repair genes expression defects
& association with clinicopathological characteristics in
colorectal carcinoma. Indian J Med Res. 134:186–192. 2011.
|
|
15
|
Xavier CP, Lima CF, Rohde M and
Pereira-Wilson C: Quercetin enhances 5-fluorouracil-induced
apoptosis in MSI colorectal cancer cells through p53 modulation.
Cancer Chemother Pharmacol. 68:1449–1457. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fehringer G, Boyd NF, Knight JA, et al:
Family-based genetic association study of insulin-like growth
factor I microsatellite markers and premenopausal breast cancer
risk. Breast Cancer Res Treat. 118:415–424. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Leite M, Corso G, Sousa S, et al: MSI
phenotype and MMR alterations in familial and sporadic gastric
cancer. Int J Cancer. 128:1606–1613. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Boland CR, Thibodeau SN, Hamilton SR, et
al: A National Cancer Institute Workshop on Microsatellite
Instability for cancer detection and familial predisposition:
development of international criteria for the determination of
microsatellite instability in colorectal cancer. Cancer Res.
58:5248–5257. 1998.
|
|
19
|
Evaluations of Genomic Applications in
Practice and Prevention (EGAPP) Working Group. Recommendations from
the EGAPP Working Group: genetic testing strategies in newly
diagnosed individuals with colorectal cancer aimed at reducing
morbidity and mortality from Lynch syndrome in relatives. Genet
Med. 11:35–41. 2009. View Article : Google Scholar
|
|
20
|
Chang SC, Lin JK, Lin TC and Liang WY:
Loss of heterozygosity: an independent prognostic factor of
colorectal cancer. World J Gastroenterol. 11:778–784. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Migliore L, Migheli F, Spisni R and
Coppede F: Genetics, cytogenetics, and epigenetics of colorectal
cancer. J Biomed Biotechnol. 2011:7923622011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ashktorab H, Brim H, Al-Riyami M, et al:
Sporadic colon cancer: mismatch repair immunohistochemistry and
microsatellite instability in Omani subjects. Dig Dis Sci.
53:2723–2731. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dietmaier W, Wallinger S, Bocker T,
Kullmann F, Fishel R and Rüschoff J: Diagnostic microsatellite
instability: definition and correlation with mismatch repair
protein expression. Cancer Res. 57:4749–4756. 1997.PubMed/NCBI
|
|
24
|
Castagnaro A, Marangio E, Verduri A, et
al: Microsatellite analysis of induced sputum DNA in patients with
lung cancer in heavy smokers and in healthy subjects. Exp Lung Res.
33:289–301. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Green MR, Jardine P, Wood P, et al: A new
method to detect loss of heterozygosity using cohort heterozygosity
comparisons. BMC Cancer. 10:1952010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Powierska-Czarny J, Miścicka-Sliwka D,
Czarny J, et al: Analysis of microsatellite instability and loss of
heterozygosity in breast cancer with the use of a well
characterized multiplex system. Acta Biochim Pol. 50:1195–1203.
2003.
|
|
27
|
Agresti A: Categorical Data Analysis. 2nd
edition. Wiley; Hoboken, NJ: 2002, View Article : Google Scholar
|
|
28
|
Vilar E and Gruber SB: Microsatellite
instability in colorectal cancer-the stable evidence. Nat Rev Clin
Oncol. 7:153–162. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Poynter JN, Haile RW, Siegmund KD, et al:
Colon Cancer Family Registry: Associations between smoking, alcohol
consumption, and colorectal cancer, overall and by tumor
microsatellite instability status. Cancer Epidemiol Biomarkers
Prev. 18:2745–2750. 2009. View Article : Google Scholar
|
|
30
|
Samowitz WS: Genetic and epigenetic
changes in colon cancer. Exp Mol Pathol. 85:64–67. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Samowitz WS, Albertsen H, Herrick J, et
al: Evaluation of a large, population-based sample supports a CpG
island methylator phenotype in colon cancer. Gastroenterology.
129:837–845. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Imai K and Yamamoto H: Carcinogenesis and
microsatellite instability: the interrelationship between genetics
and epigenetics. Carcinogenesis. 29:673–680. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hile SE, Wang X, Lee MY and Eckert KA:
Beyond translesion synthesis: polymerase κ fidelity as a potential
determinant of microsatellite stability. Nucleic Acids Res.
40:1636–1647. 2012.
|
|
34
|
Deschoolmeester V, Baay M, Wuyts W, et al:
Detection of microsatellite instability in colorectal cancer using
an alternative multiplex assay of quasi-monomorphic mononucleotide
markers. J Mol Diagn. 10:154–159. 2008. View Article : Google Scholar
|
|
35
|
Sasikumar R, Rejitha JR, Binumon PK and
Manoj M: Role of heterozygous APC mutation in niche succession and
initiation of colorectal cancer - a computational study. PLoS One.
6:e227202011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Peng DF, Kanai Y, Sawada M, et al: DNA
methylation of multiple tumor-related genes in association with
overexpression of DNA methyltransferase 1 (DNMT1) during multistage
carcinogenesis of the pancreas. Carcinogenesis. 27:1160–1168. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Feng Q, Hawes SE, Stern JE, et al: DNA
methylation in tumor and matched normal tissues from non-small cell
lung cancer patients. Cancer Epidemiol Biomarkers Prev. 17:645–654.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Burn J, Mathers J and Bishop DT: Genetics,
inheritance and strategies for prevention in populations at high
risk of colorectal cancer (CRC). Recent Results Cancer Res.
191:157–183. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Emery J, Lucassen A and Murphy M: Common
hereditary cancers and implications for primary care. Lancet.
358:56–63. 2001. View Article : Google Scholar : PubMed/NCBI
|