Introduction
Lung cancer is the leading cause of cancer
mortalities worldwide (1). Among
all cases, ~80% are classified as non-small cell lung cancer
(NSCLC) and the remaining 20% are identified as SCLC. In addition
to genetic lesions, including gene mutation, genomic
insertion/deletion and translocation, erroneous epigenetic
modifications are often involved in the development and progression
of cancer (2). Silencing of tumor
suppressor genes owing to aberrant promoter DNA methylation
(3) and faulty activation of
oncogenes caused by genomic DNA hypomethylation (4) are common in cancer cells.
Additionally, overexpression of histone deacetylases (HDACs), which
induce transcriptional silencing by catalyzing the removal of
acetyl moieties from histones, represents another modality of
epigenetic defect that contributes to cancer development (5,6). The
use of small-molecule chemical agents to reactivate the expression
of tumor suppressor genes or to repress oncogenes epigenetically
has emerged as a promising approach to eradicate cancer.
Accordingly, inhibitors of DNA methyltransferases (DNMTi) and HDACs
(HDACi) represent the two major classes of epigenetic antitumor
agents.
In addition to protein coding genes, the expression
of non-coding RNA transcripts, including microRNAs (miRNAs), is
often dysregulated at the epigenetic level in cancer cells
(7,8). miRNAs are small RNAs (~22 nucleotides)
that regulate gene expression by binding to the 3′-untranslated
regions of target gene transcripts to induce translational
repression or transcript degradation. Depending on the biological
function of the target gene products, miRNAs are involved in
diverse biological processes, including cell proliferation and
differentiation. With regard to cancer development, miRNAs were
shown to exhibit oncogenic (9–11) and
tumor suppressive (12–14) properties, respectively. Treatment of
cancer cells with HDACi and DNMTi separately or in combination was
shown to modulate miRNA expression (15–21),
indicating the possibility of suppressing cancer cell growth and
spread by targeting miRNA expression.
In addition to DNA methylation and histone
acetylation, histone lysine methylation is involved in the
epigenetic regulation of gene expression and represents another
target of dysregulation. Depending on the position of the lysine
residues to be methylated, histone methylation is involved in
transcriptional activation and repression. Notably, the mono- and
di-methylation of histone H3 at lysine 9 (H3K9me1 and H3K9me2) are
associated with transcriptional repression in euchromatin (22). The enzyme responsible for H3K9me1
and H3K9me2 formation is G9a histone methyltransferase (23). G9a expression is upregulated in
various types of human cancer (24,25),
which indicates that the enzymatic activity is oncogenic.
Consistent with this, the promoter regions of the aberrantly
silenced tumor suppressor genes are marked by an increased level of
H3K9me2 in cancer cells (26), and
H3K9me1 and H3K9me2 are erased from the promoters of reactivated
tumor suppressor genes (27).
Additionally, the silencing of G9a expression by RNA interference
reduces the invasiveness and metastatic potential of human lung
cancer cells (28) and inhibits the
growth of prostate cancer cells (29). These observations indicate a
functional association between G9a activity and cancer development.
Treatment of cells with BIX01294, a chemical inhibitor specific to
G9a, results in a decline of the cellular H3K9me2 content (30). The reduction of proliferation,
motility and invasiveness of human neuroblastoma cells following
BIX01294 treatment (31) further
indicates the use of this chemical as an antitumor agent. To
examine whether specific miRNAs are involved in the tumor
suppressive effect of G9a inhibition, a microarray analysis was
performed in the current study to probe the global change in miRNA
expression levels in human NSCLC H1299 cells following BIX01294
treatment.
Materials and methods
Cell culture
The human NSCLC cells, H1299 (CRL-5803) were
obtained from the American Type Culture Collection (ATCC; Manassas,
VA, USA) and cultured in RPMI-1640 medium (Life Technologies,
Carlsbad, CA, USA) supplemented with 10% non heat-inactivated fetal
bovine serum (ATCC) and 1% antibiotic-antimycotic solution (Corning
Inc., Acton, MA, USA). Four hours prior to drug treatment,
5×104 proliferating H1299 cells were seeded into each
well of a 12-well culture plate. BIX01294 (Stemgent, Cambridge, MA,
USA) was reconstituted in dimethyl sulfoxide (DMSO), and diluted 10
times in 1× phosphate-buffered saline (PBS) immediately prior to
use. The working BIX01294 solution was added directly to the
culture medium to a final concentration of 4 μM. For the cells that
were receiving the mock treatment, an equal volume of PBS-diluted
DMSO was added. The cells were incubated at 37°C in a 5%
CO2 atmosphere for 48 h prior to sample collection.
Total RNA extraction
H1299 cells were lysed in TRIzol reagent (Life
Technologies). The total RNA fraction was harvested following a
chloroform extraction and further purified using the Direct-Zol
purification kit (Zymo Research Corporation, Irvine, CA, USA). RNA
quantity and quality were analyzed using a a NanoVue
spectrophotometer (GE Healthcare, Pittsburg, PA, USA) and
Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA),
respectively.
miRNA microarray analysis
A genome-wide miRNA expression profiling experiment
was performed by LC Sciences (Houston, TX, USA). The probes for a
total of 2,019 unique mature human miRNAs (Sanger miRBase Release
19.0; Wellcome Trust Sanger Institute, Hinxton, UK) were printed on
the microarray in quadruplicate. Equal quantities of total RNA from
three independent preparations of each sample group (mock versus
BIX01294 treatment) were pooled for the miRNA microarray
experiment. Fluorescent signals were background subtracted and
normalized using the locally weighted scatterplot smoothing method.
A two-tailed t-test (P<0.01 was identified to indicate a
statistically significant difference) was performed to identify the
differentially expressed miRNAs.
Quantitative real-time polymerase chain
reaction (qPCR) analysis of miRNAs
qPCR analysis of the expression level of individual
miRNAs was performed according to instructions from Life
Technologies (User bulletin no. 4465407, Jan 2013 version C) with
minor modifications. Briefly, reverse transcription (RT) was
conducted using the Taqman microRNA Reverse Transcription kit. In
each reaction, 50 ng total RNA was reverse transcribed in the
presence of 6 μl 100-fold diluted RT primer stock solution, 2 mM
deoxyribonucleotide triphosphate, 3.8 units of RNase inhibitor and
150 units of MultiScribe reverse transcriptase. The RT product was
diluted five times in nuclease-free water. In each subsequent PCR
reaction, 8 μl of the diluted RT product was used with 1× Taqman
microRNA assay and 1× Taqman Universal Master Mix (Life
Technologies). Triplicate measurements for each miRNA were
performed, and the analysis was performed with the three
independent preparations of total RNA samples harvested from each
sample group. The expression level of individual miRNAs was
normalized to that of small nucleolar RNA, RNU24, and the change in
expression level was calculated using the 2−ΔΔCt method.
A two-tailed t-test (P<0.05 was identified to indicate a
statistically significant difference) was performed to identify the
differentially expressed miRNAs. All Taqman miRNA assays for mature
human miRNAs and RNU24 were purchased from Life Technologies.
miRNA target prediction
Potential target genes of miRNAs were predicted
using the miRNA Target Prediction and Functional Study Database
(www.mirdb.org) (32,33).
Enrichment analysis for disease-associated genes was performed
using the WEB-based Gene Set Analysis Toolkit (http://bioinfo.vanderbilt.edu/webgestalt/) (34). The functional annotation of the
genes was further inquired from the Gene References Into Functions
(GeneRIFs) on the National Center for Biotechnology Information
website (http://www.ncbi.nlm.nih.gov/gene/).
Statistical analysis
All statistical analyses were performed using a
two-tailed Student’s t-test. P<0.01 and P<0.05 were
considered to indicate a statistically significant difference in
miRNA microarray and qPCR analysis, respectively.
Results
G9a regulates the expression of miRNAs in
human lung cancer cells
To examine whether G9a regulates the expression of
miRNAs in human lung cancer cells, a microarray analysis was
performed to study the change in the global miRNA expression
pattern in H1299 cells in the presence and absence of BIX01294.
Among the 2,019 mature human miRNAs scrutinized, only 51 of them
were found to be differentially expressed (Table I). To identify the miRNAs that
showed a robust change in expression level, the focus was on the
miRNAs that exhibited a signal intensity of 500 units in at least
one of the sample groups (35). Of
the 51 differentially expressed miRNAs, 10 passed this selection
criterion. Among the 10 miRNAs, six showed downregulation and the
remaining four showed upregulation, following BIX01294
treatment.
 | Table IHuman miRNAs that showed a
differential expression in H1299 cells following BIX01294 treatment
through microarray analysis. |
Table I
Human miRNAs that showed a
differential expression in H1299 cells following BIX01294 treatment
through microarray analysis.
| Reporter term | Mock | Treatment | Fold change
(Treatment/Mock) | P-value | miRNA sequence (5′ to
3′) |
|---|
|
|
|---|
| Mean | SD | Mean | SD |
|---|
| Transcripts with high
microarray signal levels (signal >500) |
| hsa-miR-15b-3p | 604 | 24 | 336 | 20 | 0.56 |
1.72×10−5 |
CGAAUCAUUAUUUGCUGCUCUA |
| hsa-miR-5096 | 3,782 | 492 | 1,289 | 318 | 0.34 |
1.40×10−3 |
GUUUCACCAUGUUGGUCAGGC |
| hsa-miR-106b-3p | 545 | 19 | 311 | 33 | 0.57 |
2.39×10−3 |
CCGCACUGUGGGUACUUGCUGC |
| hsa-miR-1229-5p | 260 | 29 | 576 | 137 | 2.22 |
4.12×10−3 |
GUGGGUAGGGUUUGGGGGAGAGCG |
| hsa-miR-301b | 982 | 50 | 433 | 94 | 0.44 |
5.11×10−3 |
CAGUGCAAUGAUAUUGUCAAAGC |
|
hsa-miR-188-5p | 675 | 93 | 1,128 | 192 | 1.67 |
5.33×10−3 |
CAUCCCUUGCAUGGUGGAGGG |
|
hsa-miR-151a-3p | 665 | 61 | 364 | 73 | 0.55 |
5.96×10−3 |
CUAGACUGAAGCUCCUUGAGG |
|
hsa-miR-374c-5p | 1,257 | 48 | 747 | 112 | 0.59 |
7.68×10−3 |
AUAAUACAACCUGCUAAGUGCU |
|
hsa-miR-3613-3p | 2,960 | 195 | 9,363 | 3,142 | 3.16 |
8.55×10−3 |
ACAAAAAAAAAAGCCCAACCCUUC |
| hsa-miR-1290 | 1,205 | 73 | 2,211 | 390 | 1.83 |
9.56×10−3 |
UGGAUUUUUGGAUCAGGGA |
| Transcripts with
low microarray signal levels (signal <500) |
|
hsa-miR-335-5p | 49 | 13 | 0 | 0 | 0.00 |
1.08×10−5 |
UCAAGAGCAAUAACGAAAAAUGU |
|
hsa-miR-1207-5p | 231 | 12 | 343 | 24 | 1.49 |
2.86×10−4 |
UGGCAGGGAGGCUGGGAGGGG |
|
hsa-miR-550a-3-5p | 100 | 6 | 157 | 13 | 1.57 |
3.07×10−4 |
AGUGCCUGAGGGAGUAAGAG |
| hsa-miR-577 | 44 | 27 | 0 | 0 | 0.00 |
5.14×10−4 |
UAGAUAAAAUAUUGGUACCUG |
| hsa-miR-4440 | 200 | 13 | 147 | 9 | 0.74 |
8.02×10−4 |
UGUCGUGGGGCUUGCUGGCUUG |
| hsa-miR-1303 | 108 | 19 | 50 | 7 | 0.46 |
1.16×10−3 |
UUUAGAGACGGGGUCUUGCUCU |
| hsa-miR-5707 | 253 | 36 | 447 | 43 | 1.76 |
1.16×10−3 |
ACGUUUGAAUGCUGUACAAGGC |
|
hsa-miR-501-5p | 157 | 21 | 89 | 12 | 0.57 |
1.48×10−3 |
AAUCCUUUGUCCCUGGGUGAGA |
|
hsa-miR-16-2-3p | 393 | 23 | 239 | 28 | 0.61 |
1.69×10−3 |
CCAAUAUUACUGUGCUGCUUUA |
| hsa-miR-657 | 64 | 9 | 34 | 5 | 0.53 |
1.71×10−3 |
GGCAGGUUCUCACCCUCUCUAGG |
| hsa-miR-4669 | 254 | 25 | 394 | 42 | 1.55 |
1.90×10−3 |
UGUGUCCGGGAAGUGGAGGAGG |
|
hsa-miR-224-5p | 368 | 28 | 203 | 33 | 0.55 |
2.14×10−3 |
CAAGUCACUAGUGGUUCCGUU |
|
hsa-miR-548au-5p | 25 | 19 | 0 | 0 | 0.00 |
2.15×10−3 |
AAAAGUAAUUGCGGUUUUUGC |
| hsa-let-7a-3p | 120 | 27 | 51 | 11 | 0.43 |
2.22×10−3 |
CUAUACAAUCUACUGUCUUUC |
|
hsa-miR-4749-3p | 166 | 12 | 113 | 13 | 0.68 |
2.34×10−3 |
CGCCCCUCCUGCCCCCACAG |
|
hsa-miR-6511b-3p | 241 | 48 | 117 | 9 | 0.49 |
2.45×10−3 |
CCUCACCACCCCUUCUGCCUGCA |
|
hsa-miR-339-5p | 233 | 25 | 154 | 7 | 0.66 |
2.46×10−3 |
UCCCUGUCCUCCAGGAGCUCACG |
| hsa-miR-596 | 154 | 10 | 112 | 11 | 0.73 |
2.56×10−3 |
AAGCCUGCCCGGCUCCUCGGG |
|
hsa-miR-101-3p | 339 | 13 | 185 | 23 | 0.55 |
2.76×10−3 |
UACAGUACUGUGAUAACUGAA |
| hsa-miR-4258 | 201 | 17 | 134 | 16 | 0.67 |
2.99×10−3 |
CCCCGCCACCGCCUUGG |
|
hsa-miR-339-3p | 115 | 15 | 71 | 10 | 0.61 |
3.48×10−3 |
UGAGCGCCUCGACGACAGAGCCG |
|
hsa-miR-3156-5p | 168 | 12 | 273 | 41 | 1.62 |
3.80×10−3 |
AAAGAUCUGGAAGUGGGAGACA |
|
hsa-miR-130b-5p | 228 | 11 | 137 | 16 | 0.60 |
3.83×10−3 |
ACUCUUUCCCUGUUGCACUAC |
| hsa-miR-4417 | 98 | 9 | 132 | 10 | 1.36 |
4.41×10−3 |
GGUGGGCUUCCCGGAGGG |
| hsa-miR-6073 | 112 | 30 | 318 | 94 | 2.84 |
4.64×10−3 |
GGUAGUGAGUUAUCAGCUAC |
| hsa-miR-3943 | 94 | 10 | 33 | 9 | 0.35 |
4.78×10−3 |
UAGCCCCCAGGCUUCACUUGGCG |
|
hsa-miR-371a-5p | 98 | 9 | 143 | 18 | 1.45 |
5.20×10−3 |
ACUCAAACUGUGGGGGCACU |
|
hsa-miR-30d-3p | 177 | 12 | 70 | 18 | 0.40 |
5.32×10−3 |
CUUUCAGUCAGAUGUUUGCUGC |
|
hsa-miR-493-3p | 71 | 13 | 116 | 14 | 1.63 |
5.85×10−3 |
UGAAGGUCUACUGUGUGCCAGG |
|
hsa-miR-140-3p | 283 | 9 | 200 | 19 | 0.71 |
5.96×10−3 |
UACCACAGGGUAGAACCACGG |
| hsa-miR-4278 | 124 | 30 | 307 | 88 | 2.47 |
6.25×10−3 |
CUAGGGGGUUUGCCCUUG |
|
hsa-miR-539-3p | 60 | 23 | 171 | 45 | 2.86 |
6.39×10−3 |
AUCAUACAAGGACAAUUUCUUU |
|
hsa-miR-654-5p | 57 | 20 | 19 | 7 | 0.34 |
7.03×10−3 |
UGGUGGGCCGCAGAACAUGUGC |
|
hsa-miR-24-2-5p | 291 | 24 | 192 | 28 | 0.66 |
7.33×10−3 |
UGCCUACUGAGCUGAAACACAG |
|
hsa-miR-140-5p | 86 | 31 | 27 | 8 | 0.32 |
7.62×10−3 |
CAGUGGUUUUACCCUAUGGUAG |
|
hsa-miR-3162-3p | 127 | 6 | 108 | 6 | 0.85 |
7.76×10−3 |
UCCCUACCCCUCCACUCCCCA |
|
hsa-miR-296-5p | 222 | 28 | 159 | 13 | 0.72 |
8.19×10−3 |
AGGGCCCCCCCUCAAUCCUGU |
|
hsa-miR-222-5p | 237 | 29 | 91 | 27 | 0.38 |
8.24×10−3 |
CUCAGUAGCCAGUGUAGAUCCU |
| hsa-miR-564 | 285 | 19 | 193 | 30 | 0.68 |
8.65×10−3 |
AGGCACGGUGUCAGCAGGC |
|
hsa-miR-642b-3p | 241 | 24 | 333 | 40 | 1.38 |
8.93×10−3 |
AGACACAUUUGGAGAGGGACCC |
|
hsa-miR-576-5p | 86 | 29 | 25 | 13 | 0.29 |
8.96×10−3 |
AUUCUAAUUUCUCCACGUCUUU |
Subsequently, a qPCR experiment was performed to
validate the differential expression patterns of the 10 miRNAs. At
the time of the experiment, the Taqman miRNA assay for one of the
miRNAs (hsa-miR-1229-5p) was not available; therefore, it was
excluded from the analysis. In addition, hsa-miR-3613-3p was found
to be undetectable in the sample groups in qPCR analysis. The
normalized and averaged expression levels of the remaining eight
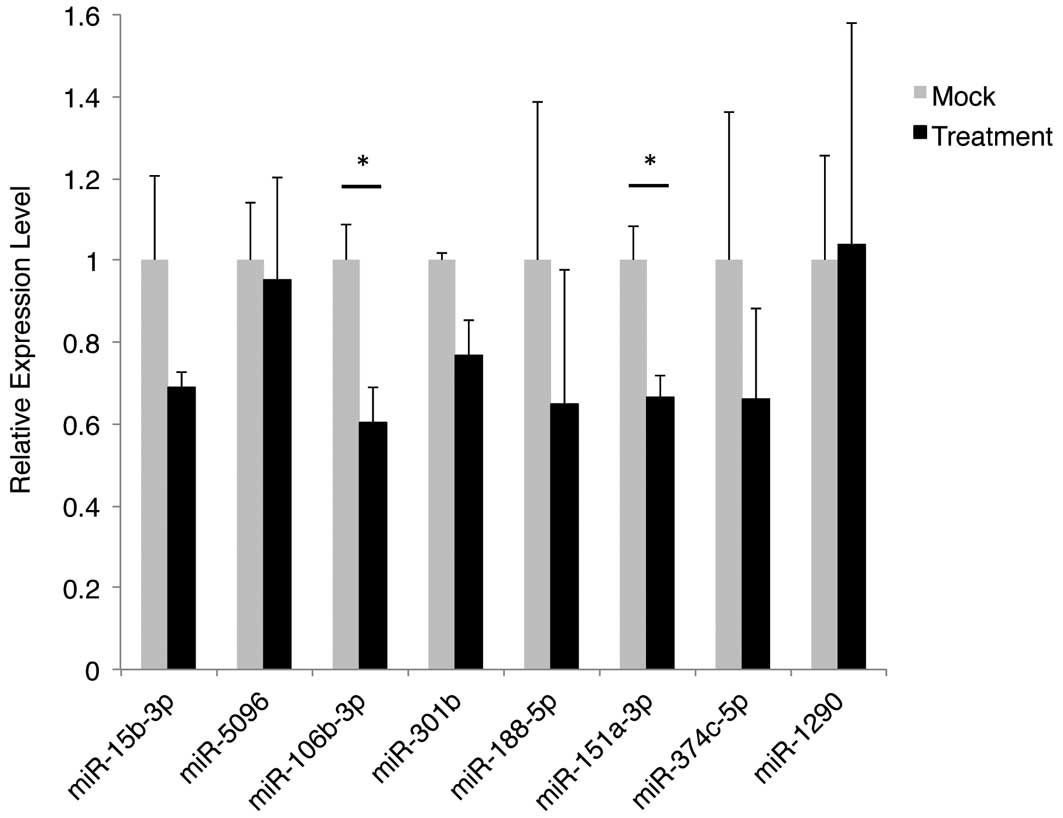
miRNAs are shown in Fig. 1. Two of
the miRNAs, hsa-miR-106b-3p and hsa-miR-151a-3p, exhibited a
significant reduction (40 and 33%, respectively) in expression
level that is consistent with the result that was obtained from the
microarray analysis.
Certain target genes of hsa-miR-151a-3p
are associated with cancerous diseases
A search was conducted for the genes whose
expression may be regulated by these two miRNAs. A total of 14 and
182 genes were predicted to be the targets of hsa-miR-106b-3p and
hsa-miR-151a-3p, respectively (Table
II). The small number of genes identified for hsa-miR-106b-3p
precluded the performance of a robust prediction of the associated
biological functions or disorders. A gene ontology analysis of the
182 genes that were potentially regulated by hsa-miR-151a-3p
revealed that the most significantly associated biological process
was the negative regulation of branching that is involved in
ureteric bud morphogenesis. To examine if there is an association
of human diseases with the predicted target genes, an enrichment
analysis for disease-associated genes was performed. A total of 10
classes of disease (neoplasm metastasis, neoplastic processes,
syndrome, adhesion, carcinoma, fasciculation, eye abnormalities,
brain injuries, schizophrenia, optic nerve diseases) were found to
be associated with the 182 genes. Among these diseases neoplasm
metastasis, neoplastic processes, adhesion and carcinoma are
relevant to cancer development and propagation. The genes
associated with these four diseases are listed in Table III.
 | Table IIPutative target genes of
hsa-miR-106b-3p and hsa-miR-151a-3p. |
Table II
Putative target genes of
hsa-miR-106b-3p and hsa-miR-151a-3p.
| miRNA | Putative target
gene |
|---|
|
hsa-miR-106b-3p | LOC347411, SOCS7,
TFAP2C, C15orf26, PCDHB13, C4orf39, RNGTT, ARHGAP17, IRAK2, TNRC6A,
BICD2, KCTD2, SLC35A1, MNT |
|
hsa-miR-151a-3p | UPP2, FXR1, PKN2,
ZFAND5, GABRA6, ZMAT1, KCNH8, ME1, CLASP2, ZNF326, PGM3, RPS6KA5,
EIF2C2, ATP2A2, CLK1, PITPNA, CHL1, GFM2, DCTN4, ITK, HIF1A, CASD1,
FAM104A, LIG4, SIX1, FAM76B, ADAM7, PURB, RGS6, CRK, KLHL4, AQP4,
SLC8A1, ANKRD44, PTGER3, HMGN2, OXR1, TRA2B, API5, FAM5C, STXBP4,
ZFPM2, RYBP, YSK4, ZNF24, ACTR2, PTPRZ1, ARMC8, NEURL1B, PANK2,
PANX3, CAPZA2, ARHGAP23, ZEB1, DISC1, SNX18, MFAP5, FAM59A, TRDN,
RERG, FBXL3, QKI, GHR, CCNDBP1, ZNF415, C5orf28, DBT, TSC1, CREBZF,
ZNF254, IL26, SPIRE2, PFN2, RBM27, CEP95, RNF20, SOS1, RBM5, DSCC1,
CAB39, NKAIN3, PCBP2, CYTIP, LPIN2, C1orf9, PTPN12, MANEA, SOCS5,
DTX3L, YTHDF3, SLCO3A1, TWIST1, PRKACB, CUX1, MRPS25, PYROXD1,
IKZF3, HSDL1, BTLA, PLEKHF2, CAST, YIPF6, SETD6, C9orf117, GPD2,
C3orf43, CGA, CASC4, HMGA2, SPIRE1, DTHD1, OSBPL3, ACAP2, GLS,
LOC100506156, SERPINA1, PHC1, FAM199X, SUSD5, RAB3GAP1, KIAA1217,
PCDHB7, C15orf41, ECT2L, SIAH3, OPA3, STMN2, FAM211A, LGSN, SLA,
NIPBL, SEC22C, BMPR1B, HCCS, ZSCAN29, TAF5L, LOC100653121, PPFIA1,
DUSP19, HPDL, UCHL1, HTR1F, SLC35E2B, ZNF345, GREM1, TET2, NIPAL2,
LOC100652774, C3orf17, FAM170A, MYLK4, CEACAM5, ARL1, NETO2,
SLC35E2, THBS1, C8orf84, SMARCAD1, PGR, TMEM99, KBTBD2, CALD1,
DSG3, GRM3, PGM2L1, LAPTM5, UHMK1, TACSTD2, MAGEA2B, FAM120AOS,
PAPOLG, C10orf10, ZNF264, MAGEA2, PRR23C, CALCR, PTPRS, RPRD2,
TIAM1, NAMPT, MCTP2, RPL27A |
 | Table IIIPutative target genes of
hsa-miR-151a-3p that are associated with diseases relevant to
cancer. |
Table III
Putative target genes of
hsa-miR-151a-3p that are associated with diseases relevant to
cancer.
| Human disease | Putative target
gene |
|---|
| Neoplasm
metastasis | SIX1, HIF1A, ZEB1,
TACSTD2, TWIST1, CEACAM5, TIAM1, PGR, THBS1 |
| Neoplastic
processes | HMGA2, SIX1, CRK,
HIF1A, ZEB1, TWIST1, CEACAM5, TIAM1, PGR, THBS1 |
| Adhesion | CYTIP, PCDHB7, CRK,
PTPN12, TACSTD2, CEACAM5, CHL1, PPFIA1, TIAM1, DSG3, THBS1 |
| Carcinoma | RBM5, HMGA2, SIX1,
HIF1A, ZEB1, TWIST1, CEACAM5, TIAM1, PGR, THBS1 |
Discussion
The involvement in cell growth and susceptibility to
epigenetic dysregulation highlights the role of miRNAs in cancer
development. For this reason, these small RNA species may serve as
potential therapeutic targets against cancer. The role of H3K9
methylation in the regulation of miRNA expression in human lung
cancer cells has not been established. In the present study, it was
found that the blockade of G9a activity, and thus histone H3K9
methylation, modulated the expression of miRNAs in the invasive
H1299 lung cancer cell line. By interrogating the change in miRNA
expression pattern with microarray analysis, it was found that only
a particularly small portion of the human miRNA collection (51 out
of 2,019; 2.5%) exhibited differential expression following
BIX01294 treatment. This observation indicates that the regulatory
activity of G9a may be specific towards a subset of miRNAs in these
cells. Coupled with qPCR analysis, the two miRNAs that were
identified, hsa-miR-106b-3p and hsa-miR-151a-3p, were downregulated
in H1299 cells following BIX01294 treatment.
The biological function of hsa-miR-106b-3p and
hsa-miR-151a-3p in lung cancer development has not been
characterized. Based on their genomic location, hsa-miR-106b-3p and
hsa-miR-151a-3p are known to reside in chromosome 7q22.1 and
8q24.3, respectively. The two genomic loci are frequently amplified
in various cancers and the overexpression of the embedded genes has
been shown to promote malignancy (36–40).
Furthermore, the amplified and overexpressed functional non-coding
RNA species participate in cancer development (41). Therefore, the downregulation of
hsa-miR-106b-3p and hsa-miR-151a-3p expression by BIX01294
treatment may exert a tumor suppressive effect, presumably through
the derepression of their target gene expression at the
post-transcriptional level. The silencing of G9a expression
inhibits the migration and invasion potential of lung cancer cells
by enhancing the transactivation of expression of the cell adhesion
molecule, epithelial cell adhesion molecule (28). It is likely that specific target
genes of these miRNAs may encode adhesion molecules, which promote
cell-cell adhesion and limit cell motility.
The target genes of hsa-miR-106b-3p and
hsa-miR-151a-3p were searched for, and their biological activities
and associated human diseases were identified. The small number of
genes identified for hsa-miR-106b-3p precluded the performance of a
robust prediction of their associated biological functions or human
diseases. For hsa-miR-151a-3p, it was found that a subset of its
target genes is involved in the cell adhesion process. Notably,
certain genes (PCDHB7, PTPN12, CHL1, PPF1A1 and THBS1) encode cell
adhesion or cell junction molecules that have been demonstrated or
indicated to inhibit cell invasion (42–45).
In addition, the genes exhibiting a similar function (TFAP2C,
PCDHB13 and MNT) (46,47) are also target genes of
hsa-miR-106b-3p. As a result, the inhibition of G9a activity by
BIX01294 treatment may suppress metastasis by downregulating the
expression level of miRNAs that block the translation of genes
encoding the cell adhesion molecules. By contrast, another subset
of the target genes of hsa-miR-151a-3p were identified, which are
involved in neoplasm formation and metastasis; however, their
involvement in the action of BIX01294 on H1299 cells remains
unclear. The extent of derepression of the individual target genes
may determine the overall cellular response to the downregulation
of hsa-miR-106b-3p and hsa-miR-151a-3p. Alternatively, the
biological effect of BIX01294 on H1299 cells may involve the
interplay among these gene products.
The change in expression level of specific miRNAs
upon inhibition of G9a activity strongly indicates a role for
miRNAs in the mediation of the malignancy-promoting effect of G9a.
Since mono- and di-methylation of H3K9 are involved in
transcriptional silencing (22), a
blockade of G9a activity, and thus H3K9me1 and H3K9me2 formation,
is expected to reactivate the transcription of genes, including
miRNAs. By contrast, the mechanism of downregulation of miRNA
expression by G9a suppression remains unclear and requires further
investigation.
In conclusion, the findings of the present study
indicate that the suppression of G9a activity by BIX01294 treatment
modulates the expression of specific miRNAs in H1299 cells. Further
studies are required to establish the functional role, and
prognostic and diagnostic potential of hsa-miR-106b-3p and
hsa-miR-151a-3p in lung cancer development.
Acknowledgements
The authors are supported by the Intramural Research
Program of Eunice Kennedy Shriver National Institute of Child
Health and Human Development, National Institutes of Health,
USA.
References
|
1
|
American Cancer Society. Lung Cancer
(Non-Small Cell). http://www.cancer.org/cancer/lungcancer-non-smallcell/detaile-dguide/non-small-cell-lung-cancer-key-statistics.
Accessed August 16, 2013
|
|
2
|
Geutjes EJ, Bajpe PK and Bernards R:
Targeting the epigenome for treatment of cancer. Oncogene.
31:3827–3844. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zöchbauer-Müller S, Fong KM, Virmani AK,
Geradts J, Gazdar AF and Minna JD: Aberrant promoter methylation of
multiple genes in non-small cell lung cancers. Cancer Res.
61:249–255. 2001.PubMed/NCBI
|
|
4
|
Hur K, Cejas P, Feliu J, et al:
Hypomethylation of long interspersed nuclear element-1 (LINE-1)
leads to activation of proto-oncogenes in human colorectal cancer
metastasis. Gut. 63:635–646. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jung KH, Noh JH, Kim JK, et al: HDAC2
overexpression confers oncogenic potential to human lung cancer
cells by deregulating expression of apoptosis and cell cycle
proteins. J Cell Biochem. 113:2167–2177. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hayashi A, Horiuchi A, Kikuchi N, et al:
Type-specific roles of histone deacetylase (HDAC) overexpression in
ovarian carcinoma: HDAC1 enhances cell proliferation and HDAC3
stimulates cell migration with downregulation of E-cadherin. Int J
Cancer. 127:1332–1346. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lujambio A and Esteller M: How epigenetics
can explain human metastasis: a new role for microRNAs. Cell Cycle.
8:377–382. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Baer C, Claus R and Plass C: Genome-wide
epigenetic regulation of miRNAs in cancer. Cancer Res. 73:473–477.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He L, Thomson JM, Hemann MT, et al: A
microRNA polycistron as a potential human oncogene. Nature.
435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song SJ, Ito K, Ala U, et al: The
oncogenic microRNA miR-22 targets the TET2 tumor suppressor to
promote hematopoietic stem cell self-renewal and transformation.
Cell Stem Cell. 13:87–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen P, Price C, Li Z, et al: miR-9 is an
essential oncogenic microRNA specifically overexpressed in mixed
lineage leukemia-rearranged leukemia. Proc Natl Acad Sci USA.
110:11511–11516. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Takamizawa J, Konishi H, Yanagisawa K, et
al: Reduced expression of the let-7 microRNAs in human lung cancers
in association with shortened postoperative survival. Cancer Res.
64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cimmino A, Calin GA, Fabbri M, et al:
miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu YY, Wu HJ, Ma HD, Xu LP, Huo Y and Yin
LR: MicroRNA-503 suppresses proliferation and cell-cycle
progression of endometrioid endometrial cancer by negatively
regulating cyclin D1. FEBS J. 280:3768–3779. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rhodes LV, Nitschke AM, Segar HC, et al:
The histone deacetylase inhibitor trichostatin A alters microRNA
expression profiles in apoptosis-resistant breast cancer cells.
Oncol Rep. 27:10–16. 2012.
|
|
16
|
Scott GK, Mattie MD, Berger CE, Benz SC
and Benz CC: Rapid alteration of microRNA levels by histone
deacetylase inhibition. Cancer Res. 66:1277–1281. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chatterjee N, Wang WL, Conklin T, Chittur
S and Tenniswood M: Histone deacetylase inhibitors modulate miRNA
and mRNA expression, block metaphase and induce apoptosis in
inflammatory breast cancer cells. Cancer Biol Ther. 14:658–671.
2013. View Article : Google Scholar
|
|
18
|
Bandres E, Agirre X, Bitarte N, et al:
Epigenetic regulation of microRNA expression in colorectal cancer.
Int J Cancer. 125:2737–2743. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Saito Y, Liang G, Egger G, et al: Specific
activation of microRNA-127 with downregulation of the
proto-oncogene BCL6 by chromatin-modifying drugs in human cancer
cells. Cancer Cell. 9:435–443. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee EM, Shin S, Cha HJ, et al:
Suberoylanilide hydroxamic acid (SAHA) changes microRNA expression
profiles in A549 human non-small cell lung cancer cells. Int J Mol
Med. 24:45–50. 2009.PubMed/NCBI
|
|
21
|
Heller G, Weinzierl M, Noll C, et al:
Genome-wide miRNA expression profiling identifies miR-9–3 and
miR-193a as targets for DNA methylation in non-small cell lung
cancers. Clin Cancer Res. 18:1619–1629. 2012.
|
|
22
|
Rice JC, Briggs SD, Ueberheide B, et al:
Histone methyltransferases direct different degrees of methylation
to define distinct chromatin domains. Mol Cell. 12:1591–1598. 2003.
View Article : Google Scholar
|
|
23
|
Tachibana M, Sugimoto K, Nozaki M, et al:
G9a histone methyltransferase plays a dominant role in euchromatic
histone H3 lysine 9 methylation and is essential for early
embryogenesis. Genes Dev. 16:1779–1791. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang J, Dorsey J, Chuikov S, et al: G9a
and Glp methylate lysine 373 in the tumor suppressor p53. J Biol
Chem. 285:9636–9641. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kondo Y, Shen L, Suzuki S, et al:
Alterations of DNA methylation and histone modifications contribute
to gene silencing in hepatocellular carcinomas. Hepatol Res.
37:974–983. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nguyen CT, Weisenberger DJ, Velicescu M,
et al: Histone H3-lysine 9 methylation is associated with aberrant
gene silencing in cancer cells and is rapidly reversed by
5-aza-2′-deoxycytidine. Cancer Res. 62:6456–6461. 2002.PubMed/NCBI
|
|
27
|
McGarvey KM, Fahrner JA, Greene E, Martens
J, Jenuwein T and Baylin SB: Silenced tumor suppressor genes
reactivated by DNA demethylation do not return to a fully
euchromatic chromatin state. Cancer Res. 66:3541–3549. 2006.
View Article : Google Scholar
|
|
28
|
Chen MW, Hua KT, Kao HJ, et al: H3K9
histone methyltransferase G9a promotes lung cancer invasion and
metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer
Res. 70:7830–7840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kondo Y, Shen L, Ahmed S, et al:
Downregulation of histone H3 lysine 9 methyltransferase G9a induces
centrosome disruption and chromosome instability in cancer cells.
PLoS One. 3:e20372008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kubicek S, O’Sullivan RJ, August EM, et
al: Reversal of H3K9me2 by a small-molecule inhibitor for the G9a
histone methyltransferase. Mol Cell. 25:473–481. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu Z, Tian Y, Salwen HR, et al:
Histone-lysine methyltransferase EHMT2 is involved in
proliferation, apoptosis, cell invasion, and DNA methylation of
human neuroblastoma cells. Anticancer Drugs. 24:484–493. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang X: miRDB: a microRNA target
prediction and functional annotation database with a wiki
interface. RNA. 14:1012–1017. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang X and El Naqa IM: Prediction of both
conserved and nonconserved microRNA targets in animals.
Bioinformatics. 24:325–332. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang J, Duncan D, Shi Z and Zhang B:
WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013.
Nucleic Acids Res. 41:W77–W83. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Validation of miRNA Microarray Results
with Real-Time qPCR. Technical Bulletin. LC Sciences; Houston, TX,
USA:
|
|
36
|
Law FB, Chen YW, Wong KY, et al:
Identification of a novel tumor transforming gene GAEC1 at 7q22
which encodes a nuclear protein and is frequently amplified and
overexpressed in esophageal squamous cell carcinoma. Oncogene.
26:5877–5888. 2007. View Article : Google Scholar
|
|
37
|
Nagel S, Leich E, Quentmeier H, et al:
Amplification at 7q22 targets cyclin-dependent kinase 6 in T-cell
lymphoma. Leukemia. 22:387–392. 2008. View Article : Google Scholar
|
|
38
|
Kwei KA, Shain AH, Bair R, et al: SMURF1
amplification promotes invasiveness in pancreatic cancer. PLoS One.
6:e239242011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mu D, Chen L, Zhang X, et al: Genomic
amplification and oncogenic properties of the KCNK9 potassium
channel gene. Cancer Cell. 3:297–302. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ho JC, Cheung ST, Patil M, Chen X and Fan
ST: Increased expression of glycosyl-phosphatidylinositol anchor
attachment protein 1 (GPAA1) is associated with gene amplification
in hepatocellular carcinoma. Int J Cancer. 119:1330–1337. 2006.
View Article : Google Scholar
|
|
41
|
Kan T, Sato F, Ito T, et al: The
miR-106b-25 polycistron, activated by genomic amplification,
functions as an oncogene by suppressing p21 and Bim.
Gastroenterology. 136:1689–1700. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Long MJ, Wu FX, Li P, Liu M, Li X and Tang
H: MicroRNA-10a targets CHL1 and promotes cell growth, migration
and invasion in human cervical cancer cells. Cancer Lett.
324:186–196. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
John AS, Hu X, Rothman VL and Tuszynski
GP: Thrombospondin-1 (TSP-1) up-regulates tissue inhibitor of
metalloproteinase-1 (TIMP-1) production in human tumor cells:
exploring the functional significance in tumor cell invasion. Exp
Mol Pathol. 87:184–188. 2009. View Article : Google Scholar
|
|
44
|
Espejo R, Rengifo-Cam W, Schaller MD,
Evers BM and Sastry SK: PTP-PEST controls motility, adherens
junction assembly, and Rho GTPase activity in colon cancer cells.
Am J Physiol Cell Physiol. 299:C454–C463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tan KD, Zhu Y, Tan HK, et al:
Amplification and overexpression of PPFIA1, a putative 11q13
invasion suppressor gene, in head and neck squamous cell carcinoma.
Genes Chromosomes Cancer. 47:353–362. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhou Q, Huang T, Wang YF, Zhang KS, Chen D
and Peng BG: Identification of Max binding protein as a novel
binding protein of Nck1 and characterization of its role in
inhibiting human liver cancer SK-HEP-1 cells. Chin Med J (Engl).
125:3336–3339. 2012.PubMed/NCBI
|
|
47
|
Penna E, Orso F, Cimino D, et al:
microRNA-214 contributes to melanoma tumour progression through
suppression of TFAP2C. EMBO J. 30:1990–2007. 2011. View Article : Google Scholar : PubMed/NCBI
|