Introduction
Lung cancer is a leading cause of cancer-associated
mortality in males and females, with a five-year survival rate of
<15% (1). Increasing
experimental evidence indicates that the abnormal expression of
different types of receptor may influence tumor behavior and
patient survival. Numerous types of tumor express the epidermal
growth factor receptor (EGFR), which regulates cell proliferation,
migration and differentiation (2).
c-Kit is the receptor for stem cell factor and has been implicated
in the pathogenesis of various types of solid tumor (3). More than 70% of cases of small cell
lung cancer (SCLC) express the c-Kit receptor (4). However, little is known regarding the
presence of these receptors in non-small cell lung cancer (NSCLC)
and how the receptors predict patient survival.
The present study aimed to investigate whether the
expression of EGFR and/or c-Kit, in tumor tissue samples from
patients who had undergone surgery for NSCLC, was capable of
predicting patient survival. A patient cohort of 146 subjects with
NSCLC was investigated and specific immunostaining for c-Kit as
well as fluorescent in situ hybridization (FISH) for EGFR
was performed using the tumor samples. Patient survival was
compared retrospectively between groups of patients expressing
c-Kit and/or EGFR.
Patients and methods
Patients and clinical samples
The present study was approved by the Ethics Review
Board of Shanghai First People’s Hospital affiliated to Shanghai
Jiaotong University (Shanghai, China). Written informed consent was
obtained from all patients prior to enrollment in the present
study. Tumor samples from 146 patients with NSCLC who had undergone
surgical resection at Shanghai First People’s Hospital between
January 2009 and August 2012 were obtained. All records were
anonymized in order to protect individual confidentiality. The
tumor tissues were retrieved from paraffin-embedded blocks. Slides
from the lung resection specimens were analyzed by two
pathologists. In all cases, the diagnosis of NSCLC was established
according to the 2011 World Health Organization’s classification of
tumors. The clinical staging was determined according to the
recommendations of the 7th International Association for the Study
of Lung Cancer (5).
Immunohistochemistry (IHC) and FISH
IHC and FISH were performed using formalin-fixed,
paraffin-embedded tissue samples obtained during surgery. Sections
from the tissue blocks containing >80% representative tumor
tissue were used for all analyses.
For the IHC, endogenous peroxidase activity was
blocked using 3% hydrogen peroxide for 20 min and the sections were
de-waxed and stained with rabbit polyclonal primary antibodies
against human c-Kit (phospho Y703; DakoCytomation, Glostrup,
Denmark) overnight at 4°C using automatic immunostaining. The
sections were incubated with reagents from a high-sensitivity
detection kit (Envision detection kit; Dako, Carpinteria, CA, USA).
according to the manufacturer’s instructions. Tumors were
considered to be negative for c-Kit if the staining was either
completely absent or observed in <5% of the neoplastic cells.
Sections that were not incubated with the primary antibody served
as the controls. All lung cancer cases were histologically analyzed
by two pathologists and the most representative areas of the viable
tumor cells were selected.
For FISH, gene copy numbers (GCNs) of EGFR per
nucleus were determined using an LSI EGFR/centromeric probe for
chromosome 7 (CEP 7) probe mix (Abbott Laboratories S.A., Shanghai,
China). The EGFR gene was visualized as an orange signal, the CEP7
as green and the nucleus was shown as a blue signal using a
4,6-diamidini-2-phenylindone (DAPI) filter (Carl Zeiss, Oberkochen,
Germany).
For FISH, 4-μm thick sections of lung cancer tissue
were composed and subsequently deparaffinized, dehydrated, immersed
in 0.2 N HCl and boiled in a microwave in citrate buffer (pH 6.0).
The probe mixtures were added to the slides, which were incubated
in a humidified atmosphere at 73°C for 5 min in order to denature
the probes and target the DNA. The slides were cooled and incubated
at 37°C for 19 h to allow hybridization. The slides were washed
with 0.4X saline-sodium citrate (SSC)/0.3% NP-40 for 2 min at room
temperature, followed by nuclear counterstaining with 2X SSC/0.1%
NP-40 for 5 min at 73°C. DAPI and the anti-face compound,
p-phenylenediamine were subsequently added. The signals for each
probe were analyzed under a microscope equipped with a triple pass
filter (DAPI/green/orange; Carl Zeiss). At least 100 tumor cell
nuclei were counted per case. FISH analysis was performed
independently by two pathologists, who were blinded to the
patients’ clinical characteristics and to any additionally
molecular variables (2). All FISH
staining was performed by CSPC Ouyi Pharmaceutical Co., Ltd.
(Shijiazhuang, China).
In the present study, an average value of ≥2.4 EGFR
GCN signals per nucleus was determined as the cut-off for EGFR
gain, although an EGFR/CEP7 ratio of ≥2.0 is the true amplification
cut-off, by counting 120 cells. A Sartore-Bianchi cut-off of 2.4
GCN per nucleus was used, as it was the lowest value that was
reported in previous studies. This value preserved the sensitivity
and specificity of the assessments in the specific clinical setting
of the present study, where only carcinomas that were identified
using screening procedures and analyzed at the point of diagnosis
were included. Gene amplification of EGFR as a result of polysomy
of chromosome 7 was considered to be indicated by an increase in
EGFR signals (≥2 signals per nucleus) as well as an increase in
chromosome 7, as measured by the number of CEP7 green signals per
nucleus.
Statistical analysis
Statistical analyses were performed using SPSS 17.0
software (SPSS, Inc, Chicago, IL, USA). All of the patients were
included in the statistical calculations and were followed-up until
November 1, 2012. The χ2 test and Fisher’s exact test
were used to assess the association between molecular marker
expression and various clinicopathological parameters. Unadjusted
survival estimates based on the c-Kit expression status were
calculated using the Kaplan-Meier method and compared using the
log-rank test. All tests of statistical significance were two-sided
and P<0.05 was considered to indicate a statistically
significant difference.
Results
Patient baseline characteristics
Demographic and clinical variables are shown in
Table I. The median age of the
patients was 63.3 years (range, 37–79 years) and the majority were
male (57.5%). The NSCLC tumors comprised 68 squamous cell
carcinomas and 78 adenocarcinomas. A total of 60 patients were
found to be lymph node-negative, while 66 patients exhibited lymph
node metastases. Due to nodal metastasis or non-radical surgical
margins, 47 (32.2%) patients received postoperative radiotherapy
and/or treatment with targeted agents; however, there were no
evident differences in receptor expression between the patients
with and without lymph node metastases.
 | Table IClinicopathological characteristics of
patients with non-small cell lung cancer who underwent
immunohistochemical analysis to detect c-Kit expression and FISH
analysis to detect EGFR expression. |
Table I
Clinicopathological characteristics of
patients with non-small cell lung cancer who underwent
immunohistochemical analysis to detect c-Kit expression and FISH
analysis to detect EGFR expression.
| c-Kit | EGFR |
|---|
|
|
|
|---|
| Characteristic | − | + | − | + |
|---|
| Total patients
(n=146) | 78 | 68 | 88 | 58 |
| Gender |
| Male (n=84) | 45 | 39 | 52 | 32 |
| Female (n=62) | 33 | 29 | 36 | 26 |
| P-value | 0.319 | - | 0.416 | - |
| Age (years) |
| ≤65 (n=60) | 27 | 33 | 25 | 35 |
| >65 (n=86) | 51 | 35 | 63 | 23 |
| P-value | 0.806 | - | 0.223 | - |
| Tumor size (cm) |
| ≤3 (n=70) | 40 | 30 | 40 | 30 |
| >3 (n=76) | 38 | 38 | 48 | 28 |
| P-value | 0.501 | - | 0.187 | - |
| Differentiation |
| Well (n=40) | 20 | 20 | 17 | 23 |
| Moderate (n=61) | 33 | 28 | 40 | 21 |
| Poor (n=45) | 25 | 20 | 31 | 14 |
| P-value | 0.043 | - | 0.023 | - |
| Histology |
| SCC (n=68) | 37 | 31 | 42 | 26 |
| Adenocarcinoma
(n=78) | 41 | 37 | 46 | 32 |
| P-value | 0.280 | - | 0.632 | - |
| Pathological
stage |
| I (n=50) | 27 | 23 | 32 | 18 |
| II (n=49) | 23 | 26 | 26 | 23 |
| III (n=36) | 25 | 11 | 24 | 12 |
| IV (n=11) | 3 | 8 | 6 | 5 |
| P-value | 0.923 | | 0.223 | |
| Nodal status |
| No (n=80) | 40 | 40 | 52 | 28 |
| Yes (n=66) | 38 | 28 | 36 | 30 |
| P-value | 0.418 | - | 0.069 | - |
| Vascular
infiltration |
| No (n=90) | 51 | 39 | 54 | 36 |
| Yes (n=56) | 27 | 29 | 34 | 22 |
| P-value | 0.516 | - | 0.176 | - |
| Nerve
infiltration |
| No (n=93) | 45 | 48 | 58 | 35 |
| Yes (n=53) | 33 | 20 | 30 | 23 |
| P-value | 0.658 | - | 0.138 | - |
| Smoking history |
| Never (n=41) | 19 | 22 | 29 | 12 |
| Current (n=32) | 19 | 13 | 16 | 16 |
| Former (n=73) | 50 | 23 | 43 | 30 |
| P-value | 0.032 | - | 0.044 | - |
| Involving the
pleura |
| No (n=100) | 53 | 47 | 54 | 46 |
| Yes (n=46) | 25 | 21 | 34 | 12 |
| P-value | 0.007 | - | 0.150 | - |
| TNM |
| T1–T2 (n=124) | 74 | 50 | 76 | 48 |
| T3–T4 (n=22) | 4 | 18 | 12 | 10 |
| P-value | 0.416 | - | 0.480 | - |
Expression of c-Kit and EGFR
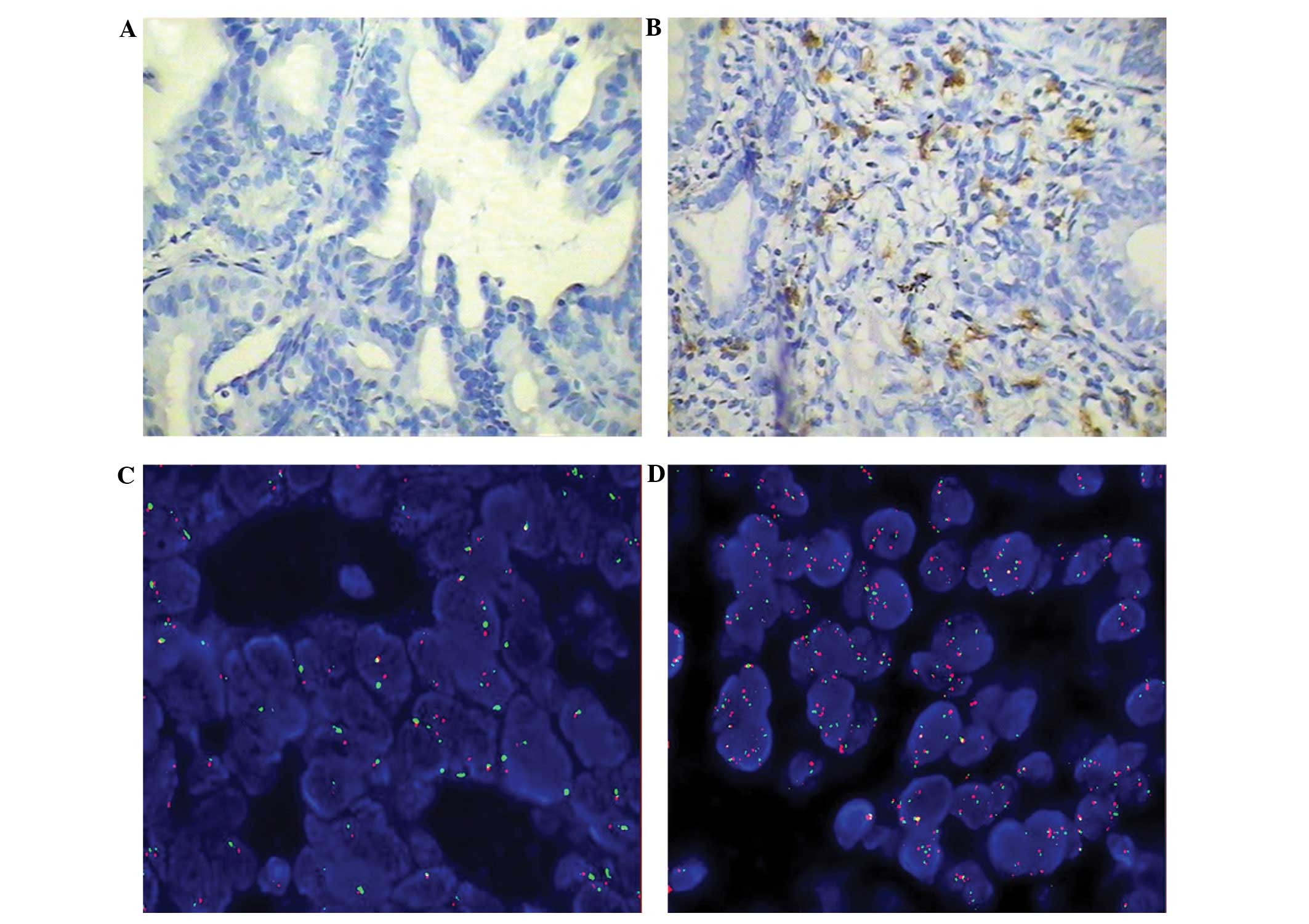
The immunohistochemical expression of c-Kit is shown
in Fig. 1, comparing representative
samples of a c-Kit-negative tumor (Fig.
1A) and a c-Kit-positive tumor (Fig. 1B). In the c-Kit-positive slide, a
number of cells that were positive for c-Kit were observed. The
number of cells that were positive for c-Kit varied marginally
among the sections, however, was calculated to be ~20% of the total
cells that were visualized. The expression of c-Kit was correlated
with poor tumor differentiation, pleura involvement and smoking
history (P=0.043, 0.007 and 0.032, respectively; data not
shown).
The GCNs of EGFR that were revealed using FISH are
shown in Fig. 1C and D, which show
negative and positive expression of EGFR (red) expression in tumors
from different patients. In the EGFR-positive tumor sample, FISH
revealed that the majority of the cells were expressing EGFR.
Furthermore, EGFR gene amplification as a result of polysomy of
chromosome 7 was found to be negatively correlated with poor tumor
differentiation (52.9 vs. 23.5%; P=0.023; data not shown). EGFR
gene amplification was not identified to differ significantly in
patients with regard to gender, age, tumor location,
tumor-node-metastasis stage, tumor size, lymph node metastases or
involvement of the pleura. However, the absence of a history of
smoking was identified to be significantly correlated with reduced
EGFR gene amplification (odds ratio: 2.5) compared with a history
of smoking (P=0.044).
Table II shows the
distribution of c-Kit and EGFR expression in patients with NSCLC
using IHC and FISH, respectively.
 | Table IIc-Kit and EGFR distribution in
patients with NSCLC. |
Table II
c-Kit and EGFR distribution in
patients with NSCLC.
| EGFR, n (%) | |
|---|
|
| |
|---|
| c-Kit | − | + | Total, n |
|---|
| − | 42 (28.76) | 36 (24.65) | 78 |
| + | 46 (31.51) | 22 (15.07) | 68 |
| Total | 88 | 58 | 146 |
Mortality
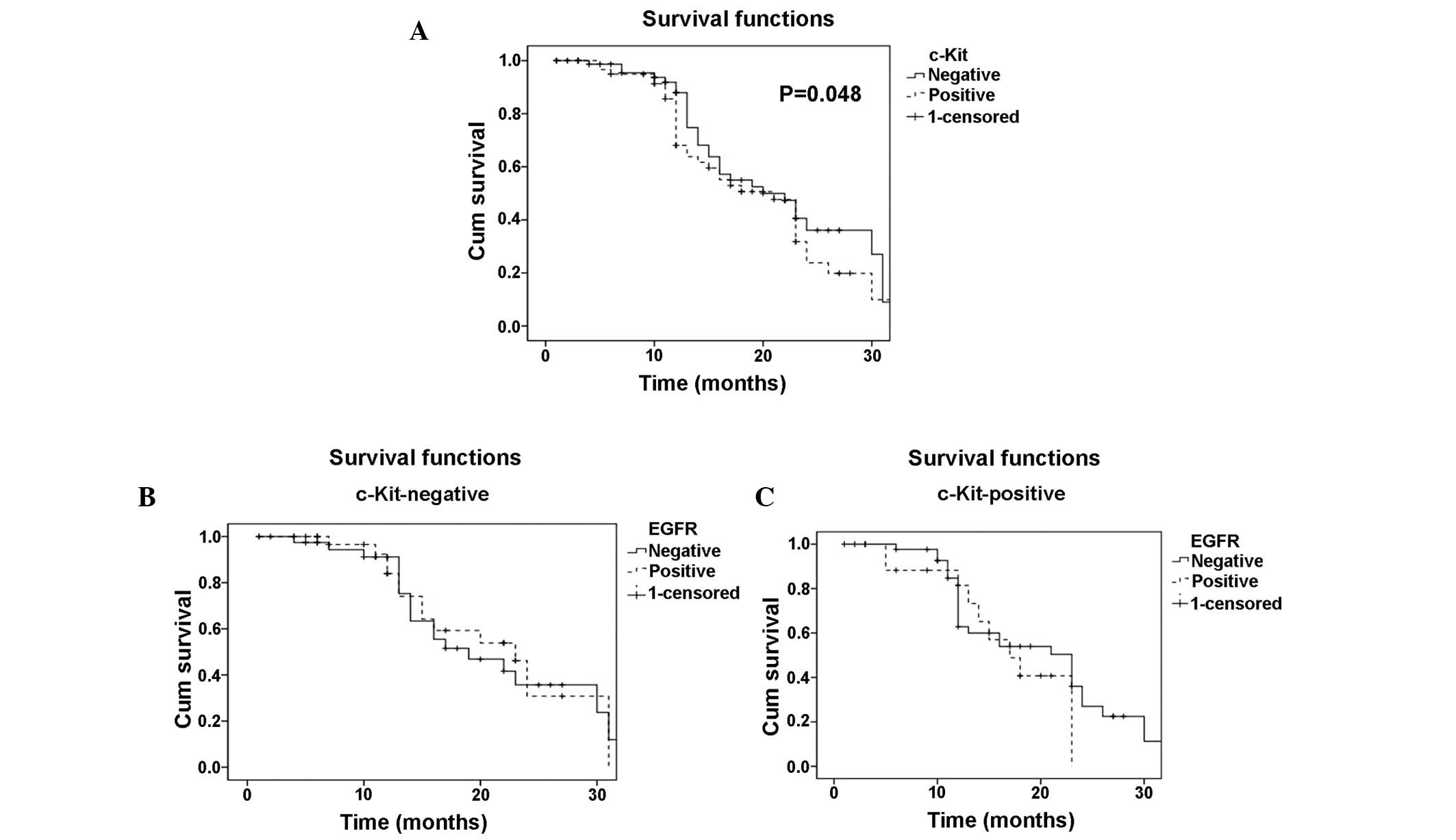
Fig. 2A demonstrates
patient mortality between the time of surgery and 30 months
post-surgery in patients with c-Kit-positive and -negative tumors.
Mortality was found to be significantly higher in patients with
c-Kit-positive tumors compared with those with c-Kit-negative
tumors (P=0.048). In the patients with c-Kit-positive tumors,
positive EGFR expression, which was detected using FISH (Fig. 2C), was observed to be associated
with an increase in mortality compared with the patients with
negative EGFR expression, as no patients with c-Kit- and
EGFR-positive expression were found to have survived 30 months
post-surgery. However, this difference was not statistically
significant (P=0.06). In the patients with c-Kit-negative tumors, a
similar pattern (although not significant) of increased mortality
in patients with EGFR-positive tumors compared with those with
EGFR-negative tumors was observed (Fig.
2B).
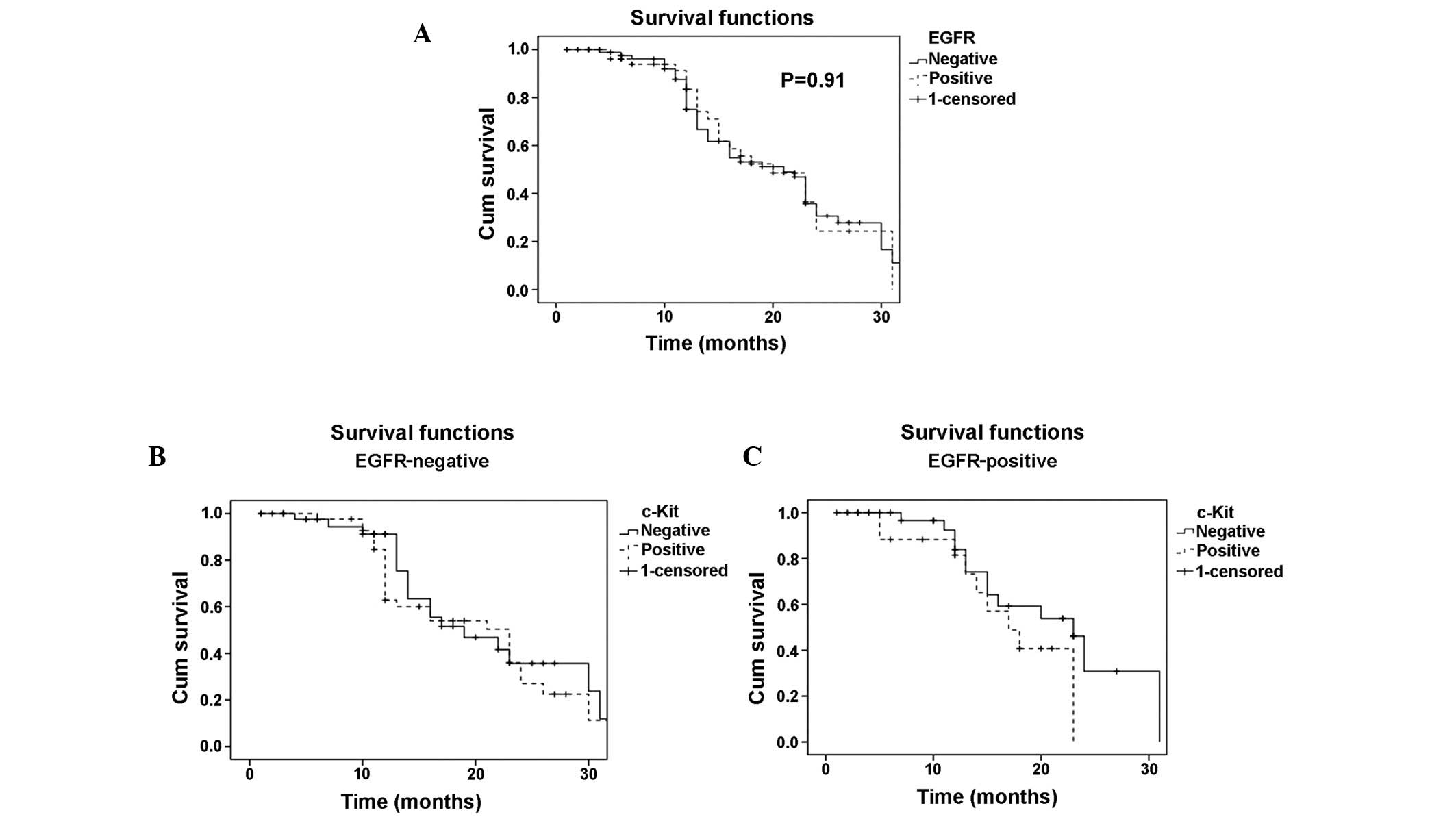
Fig. 3A shows the
rate of mortality between the time of surgery and 30 months
post-surgery in patients with EGFR-positive and -negative tumors.
EGFR-negative tumors were observed to be associated with reduced
mortality compared with EGFR-positive tumors, although this
difference was not statistically significant (Fig. 3A; P=0.91). Furthermore, the
expression of c-Kit was not found to affect mortality in patients
with EGFR-negative tumors (Fig.
2B). However, in the patients with EGFR- and c-Kit-positive
tumors, the rate of mortality was found to be 100% after two years,
with an increased mortality rate detected in those patients with
EGFR-positive tumors who were also expressing c-Kit. However, the
effect of c-Kit expression in patients with varying EGFR expression
was not identified to be significant (P=0.616).
After 30 months, the patients in all of the groups
deteriorated and by 48 months all of the patients had succumbed
(data not shown). Post-surgery therapy, where reported, did not
significantly influence the effect of c-Kit and/or EGFR expression
on mortality (data not shown).
Discussion
The present study identified that certain patients
with NSCLC have tumors, which express c-Kit, also known as cluster
of differentiation 117, and among those patients, the GCN of EGFR
varies. Mortality was found to be influenced by c-Kit and/or EGFR
expression in the tumor, with patients with c-Kit-positive tumors
found to have an increased mortality rate compared with those with
c-Kit-negative tumors, 30 months post-surgery. Furthermore, the
co-expression of c-Kit and EGFR was found to increase the rate of
mortality in patients with NSCLC.
IHC for c-Kit in surgically resected NSCLC tissues
revealed that patients could be divided into two subgroups based on
positive or negative c-Kit expression. In addition, the GCNs of
EGFR in the tumor cells was found to differ markedly between
patients. These findings emphasize that NSCLC is a complex disease,
which may be divided into multiple subsets, which express different
proteins/genes, even though the light-microscopic histopathology
may appear similar between patients. Potentially, other surface
markers and gene expression profiles may divide tumors and,
therefore, patients into additional subgroups, beyond grouping
determined by the expression of c-Kit and the gene amplification of
EGFR. However, this hypothesis is outside the scope of the present
study. Other molecules which may further subdivide lung cancers
include Ki67 (6), p53 (7), carcinoembryonic antigen (8), vascular endothelial growth factor
(9–11), epithelial membrane antigen (11) and cytokeratin 5/6 (12), all of which have been suggested to
influence disease progression and patient survival. A major problem
for individualizing therapy is understanding the tumor
characteristics of individual patients. The present study indicates
that c-Kit and EGFR should be involved in such profiling.
One of the key findings of the present study was
that the expression of c-Kit in NSCLC tumors was associated with
increased mortality up to 30 months, whereas long-term survival up
to 48 months was not affected. This finding is similar to reports
regarding various other types of tumors, including breast cancer
(13) and large cell neuroendocrine
carcinoma of the lung (14),
although not SCLC (15,16). Yoo et al (17) proposed that c-Kit expression in
NSCLC did not influence survival; therefore, this requires further
investigation. However, there are certain important differences
between the present study, which indicates that c-Kit expression
may affect patient survival, and the study by Yoo et al
(17). The study by Yoo et
al (17) used fewer patient
samples compared with the present study and there were certain
differences in the staining protocols that were used. Although such
parameters may have influenced the results, further investigations
are required to assess the effect of c-Kit expression on mortality
in patients with NSCLC.
In the present study, the EGFR GCN in single NSLC
cells was found to significantly differ between patients,
furthermore, it significantly influenced mortality, albeit not as
strongly as c-Kit. This finding is consistent with a study by Tsao
et al (18), who found that
EGFR gene amplification as a result of polysomy of chromosome 7,
which was analyzed using FISH, was assocaited with mortality. A
previous study proposed that EGFR expression influences survival in
NSCLC (19), however, the study was
significantly smaller than the present study and survival favored
EGFR-positive patients who received concurrent therapy. Thus, the
present study significantly strengthens the conclusion that EGFR
GCNs do not influence disease progression and mortality. Survival
may be significantly influenced by c-Kit and EGFR expression in
patients with NSCLC.
In the present study, the expression of c-Kit was
correlated with smoking. A total of 25 patients quit smoking within
six months of commencing treatment for NSCLC and 33 continued to
smoke. In those who stopped smoking at the time of diagnosis, the
relative risk of NSCLC was 9 [confidence interval (CI), 4.2–21],
whereas in those who continued to smoke, it was 23 (CI, 12–59).
Thus, smoking may cause KIT and EGFR mutations; however, the
underlying mechanism requires further functional
investigations.
A limitation of the present study is that it was
retrospective, and the cancer treatment was not controlled on an
individual basis. However, an advantage is that the present study
was relatively large compared with previous studies (20). In the present study, the overall
survival beyond two years was higher than in previous studies in
other institutions and countries, which is difficult to explain.
Patient diagnostic criteria and/or the disease stage may be
different in the previous studies. Another limitation of the
present study is that it does not address other potential
predictive EGFR pathway biomarkers, including EGFR mutations or
functional EGFR protein expression. Of note, c-Kit positivity in
NSCLC has been reported to have the potential to predict
responsiveness to its inhibitor, imatinib, as this drug has been
shown to be ineffective in SCLC (21,22).
In conclusion, the present study showed that c-Kit
is a strong and independent prognostic factor in NSCLC, and the
prognostic impact is highly associated with poor differentiation,
pleura involvement and smoking history. Furthermore, mortality was
observed to be significantly higher in patients with c-Kit-positive
tumors. In addition, the present study demonstrated that the
expression of EGFR was an independent negative prognostic factor,
which is associated with differentiation and smoking history;
however, patient survival was not identified to be influenced by
EGFR GCN. Although this study was limited by its retrospective
nature, we predict that this study will aid the establishment of
novel molecular targeted therapies in the field of lung cancer.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81072448),
the Key Projects of Shanghai Science and Technology Foundation
Office (grant no. 11JC1410300) and the Shanghai Jiaotong University
training fund. Dr Xiao’s study period at Krefting Research Centre,
Gothenburg, Sweden was funded by Shanghai Jiaotong University.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012.
|
|
2
|
American Thoracic Society; Infectious
Diseases Society of America. Guidelines for the management of
adults with hospital-acquired, ventilator-associated, and
healthcare-associated pneumonia. Am J Respir Crit Care Med.
171:388–416. 2005.
|
|
3
|
Hirota S, Isozaki K, Moriyama Y, et al:
Gain-of-function mutations of c-kit in human gastrointestinal
stromal tumors. Science. 279:577–580. 1998.
|
|
4
|
Thiel G, Ekici M and Rössler OG:
Regulation of cellular proliferation, differentiation and cell
death by activated Raf. Cell Commun Signal. 7:82009.
|
|
5
|
Mirsadraee M, Oswal D, Alizade Y, et al:
The 7th lung cancer TNM classification and staging system: Review
of the changes and implications. World J Radiol. 4:128–134.
2012.
|
|
6
|
Lei B, Liu S, Qi W, et al: PBK/TOPK
expression in non-small-cell lung cancer: its correlation and
prognostic significance with Ki67 and p53 expression.
Histopathology. 63:696–703. 2013.
|
|
7
|
Erler BS, Presby MM, Finch M, et al:
CD117, Ki-67, and p53 predict survival in neuroendocrine
carcinomas, but not within the subgroup of small cell lung
carcinoma. Tumour Biol. 32:107–111. 2011.
|
|
8
|
Mohammad T, Garratt J, Torlakovic E, Gilks
B and Churg A: Utility of a CEA, CD15, calretinin, and CK5/6 panel
for distinguishing between mesotheliomas and pulmonary
adenocarcinomas in clinical practice. Am J Surg Pathol.
36:1503–1508. 2012.
|
|
9
|
Cheng D, Kong H and Li Y: Prognostic
values of VEGF and IL-8 in malignant pleural effusion in patients
with lung cancer. Biomarkers. 18:386–390. 2013.
|
|
10
|
Chatterjee S, Heukamp LC, Siobal M, et al:
Tumor VEGF:VEGFR2 autocrine feed-forward loop triggers angiogenesis
in lung cancer. J Clin Invest. 123:1732–1740. 2013.
|
|
11
|
Yang Y, Li J, Mao S and Zhu H: Comparison
of immunohistology using pan-CK and EMA in the diagnosis of lymph
node metastasis of gastric cancer, particularly micrometastasis and
isolated tumor cells. Oncol Lett. 5:768–772. 2013.
|
|
12
|
Kanapathy Pillai SK, Tay A, Nair S and
Leong CO: Triple-negative breast cancer is associated with EGFR,
CK5/6 and c-KIT expression in Malaysian women. BMC Clin Pathol.
12:182012.
|
|
13
|
Al-Dubai SA, Qureshi AM, Saif-Ali R,
Ganasegeran K, Alwan MR and Hadi JI: Awareness and knowledge of
breast cancer and mammography among a group of Malaysian women in
Shah Alam. Asian Pac J Cancer Prev. 12:2531–2538. 2011.
|
|
14
|
Casali C, Stefani A, Rossi G, et al: The
prognostic role of c-kit protein expression in resected large cell
neuroendocrine carcinoma of the lung. Ann Thorac Surg. 77:247–253.
2004.
|
|
15
|
Boldrini L, Ursino S, Gisfredi S, et al:
Expression and mutational status of c-kit in small-cell lung
cancer: prognostic relevance. Clin Cancer Res. 10:4101–4108.
2004.
|
|
16
|
López-Martin A, Ballestín C,
Garcia-Carbonero R, et al: Prognostic value of KIT expression in
small cell lung cancer. Lung Cancer. 56:405–413. 2007.
|
|
17
|
Yoo J, Kim CH, Song SH, et al: Expression
of c-kit and p53 in non-small cell lung cancers. Cancer Res Treat.
36:167–172. 2004.
|
|
18
|
Tsao MS, Sakurada A, Cutz JC, et al:
Erlotinib in lung cancer - molecular and clinical predictors of
outcome. N Engl J Med. 353:133–144. 2005.
|
|
19
|
Hirsch FR, Herbst RS, Olsen C, et al:
Increased EGFR gene copy number detected by fluorescent in situ
hybridization predicts outcome in non-small-cell lung cancer
patients treated with cetuximab and chemotherapy. J Clin Oncol.
26:3351–3357. 2008.
|
|
20
|
Park EA, Lee HJ, Kim YT, et al: EGFR gene
copy number in adenocarcinoma of the lung by FISH analysis:
investigation of significantly related factors on CT, FDG-PET, and
histopathology. Lung Cancer. 64:179–186. 2009.
|
|
21
|
Dy GK, Miller AA, Mandrekar SJ, et al: A
phase II trial of imatinib (ST1571) in patients with c-kit
expressing relapsed small-cell lung cancer: a CALGB and NCCTG
study. Ann Oncol. 16:1811–1816. 2005.
|
|
22
|
Schneider BJ, Kalemkerian GP, Ramnath N,
et al: Phase II trial of imatinib maintenance therapy after
irinotecan and cisplatin in patients with c-Kit-positive,
extensive-stage small-cell lung cancer. Clin Lung Cancer.
11:223–227. 2010.
|