Introduction
Gastric cancer is the fourth most common type of
cancer and the second leading cause of cancer-related mortality
worldwide (1), with approximately
one million new cases diagnosed each year. One of the major factors
that controls tumor cell death is the tumor suppressor, p53
(2). The importance of cell death
to tumor suppression is exemplified by p53 (3). In response to various forms of
cellular stress, including DNA damage, hypoxia and oncogene
activation, p53 levels are elevated (2). p53 has also been linked to another
cell process that controls cell death known as autophagy (4,5).
Autophagy is a vesicular trafficking process that mediates the
degradation of long-lived proteins and is the only pathway within
the cell for the degradation of organelles (6). In tumor development, autophagy is
considered to act in either an oncogenic or tumor suppressive
capacity and p53 has been reported to be an inducer of autophagy
(4,5). Moreover, the discovery that
damage-regulated autophagy regulator (DRAM), a p53 target gene
which is required for p53-induced autophagy, is frequently
downregulated in squamous cancers underscores the theory that
autophagy is a component of tumor suppression downstream of p53
(5).
DRAM has been identified as an effector molecule
that is critical for p53-mediated apoptosis, thus further
supporting the tumor-suppressive role of autophagy (5,7,8). The
discovery of DRAM revealed a novel role for autophagy in
p53-induced apoptotic cell death (5), and DRAM is considered to be a crucial
modulator in apoptosis and autophagy. The present study aimed to
investigate the effects of AdMax-pDC315-DRAM-EGFP on growth,
apoptosis and autophagy of gastric cancer cells in vitro,
and to compare the infection efficiency, biological and molecular
mechanisms of AdMax-pDC315-DRAM-EGFP.
Materials and methods
Reagents
The SGC7901 gastric cancer cell line was purchased
from the Shanghai Institute of Cell Biology, Chinese Academy of
Sciences (Shanghai, China). The RPMI-1640 medium was purchased from
Gibco-BRL (Rockville, MD, USA). Fetal bovine serum (FBS) was
obtained from Hangzhou Sijiqing Biological Engineering Material
Co., Ltd. (Hangzhou, China), and L-glutamine and MTT were provided
by Sigma (St. Louis, MO, USA). Antibodies against p53 (1:500;
Rabbit monoclonal anti-human), B cell lymphoma 2 (Bcl 2; 1:500;
Rabbit monoclonal anti-human), Beclin1 (1:700; Rabbit monoclonal
anti-human) and p21 (1;500; Rabbit monoclonal anti-human) were
supplied by Cell Signaling Technology, Inc. (Beverly, MA, USA).
Adenoviral vectors and infections
The adenoviral vectors and NC-RNAi-GFP-AD were
purchased from Shanghai Jikai Biological Technology Co., Ltd.
(Shanghai, China). Stocks of replication-defective adenoviral
vectors expressing green fluorescent protein (GFP)
(AdMax-pDC315-DRAM-EGFP) were stored at −80°C. NC-RNAi-GFP-AD was
used as a control which was also stored at −80°C. Infections were
performed at 70–75% confluence in Dulbecco’s modified Eagle’s
medium supplemented with 2% fetal calf serum (FCS). The cells were
subsequently incubated at 37°C for at least 4 h, followed by the
addition of fresh medium. Cells were then subjected to functional
analyses at fixed time points following infection as described for
individual experimental conditions (9).
Determination of optimal multiplicity of
infection (MOI)
The SGC7901 cells (1×104 cells/well) were
seeded in 96-well plates and reached 60–70% confluence. Different
MOI (MOI = 10, 20, 30, 50 and 100) values of the NC-RNAi-GFP-AD
100-μl diluted infected cells were added to the plates and, after 8
h, RPMI-1640 medium containing 10% FBS was added. After 48 h of
culture, the cells were counted under a fluorescence microscope
(Leica DMI4000B; Leica Microsystems Wetzlar GmbH, Wetzlar, Germany)
to calculate the number of cells expressing GFP.
Cell culture and viability assay
The SGC7901 cells were maintained in RPMI-1640
medium containing 10% heat-inactivated FBS and 0.03% L-glutamine,
and incubated in an atmosphere of 5% CO2 at 37°C. The
cells in a mid-log phase were used in the experiments. Cell
viability was assessed by the MTT assay. To determine the effects
of AdMax-pDC315-DRAM-EGFP, the SGC7901 cells were plated into
96-well microplates (7×104 cells/well) and
AdMax-pDC315-DRAM-EGFP was added to the culture medium. Cell
viability was assessed by the MTT assay 24 h after
AdMax-pDC315-DRAM-EGFP treatment. MTT (Sigma) solution was added to
the culture medium (500 μg/ml final concentration) for 4 h prior to
the end of treatment and the reaction was inhibited by the addition
of 10% acid sodium dodecyl sulfate (100 μl; Beijing Biosea
Biotechnology Co., Ltd., Beijing, China). The absorbance value (A)
at 570 nm was measured using an automatic multi-well
spectrophotometer (Bio-Rad, Richmond, CA, USA). The percentage of
cell proliferation was calculated as follows: Cell proliferation
(%)= (1−A of experiment well/A of positive control well) × 100.
Visualization of MDC-labeled
vacuoles
Exponentially growing cells were plated on
24-chamber culture slides, cultured for 24 h and then incubated
with the drug in 10% FCS/RPMI-1640 medium for 12 and 24 h.
Autophagic vacuoles were labeled with MDC (Sigma) (10) by incubating cells with 0.001 mmol/l
MDC in RPMI-1640 at 37°C for 10 min. Following incubation, cells
were washed three times with phosphate-buffered saline (PBS) and
immediately analyzed with a fluorescence Nikon Eclipse TE300
microscope (Nikon, Tokyo, Japan) equipped with a filter system
(V-2A excitation filter, 380–420 nm; barrier filter, 450 nm).
Images were captured with a charged couple device camera (CoolSNAP
ES, Roper Scientific; Trenton, NJ, USA) and imported into
Photoshop.
Immunofluorescent staining
The SGC7901 cells were seeded onto 24-chamber
culture slides and treated with AdMax-pDC315-DRAM-EGFP. Following
fixation in methanol for 10 min, cells were blocked with a buffer
containing 1% bovine serum albumin (BSA; Hangzhou Sijiqing
Biological Engineering Material Co., Ltd.) and 0.1% Triton X-100
(Nanjing KeyGen Biotech., Co., Ltd., Nanjing, China) for 1 h. The
cells were then incubated with the primary antibody against LC3
(diluted 1:200; Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) and PBS containing 1% BSA at 4°C overnight, and then incubated
for 1 h with secondary ghost against rabbit cy3 fluorescence
conjugated antibodies (1:500; Sigma) to visualize the binding sites
of the primary antibody with laser confocal microscopy (Leica
Microsystems Wetzlar GmbH).
Total cell protein extraction and western
blot analysis
For extraction of total cell proteins, cells were
washed with pre-cooled PBS and subsequently lysed in pre-cooled
radioimmunoprecipitation assay lysis buffer [50 mM Tris-HCl (pH
7.4), 150 mM NaCl, 1 mM dithiothreitol, 0.25% sodium deoxycholate
and 0.1% NP-40] containing 1 mM phenylmethysulfonyl fluoride, 50 mM
sodium pyrophosphate, 1 mM Na3VO4, 1 mM NaF,
5 mM EDTA, 5 mM EGTA and protease inhibitors cocktail (Nantong
Biyuntian Biological Technology Co., Ltd., Nantong, China). Cell
lysis was performed on ice for 30 min. Clear protein extracts were
obtained by centrifugation 12,000 × g for 30 min at 4°C. Protein
extraction from the SGC7901 gastric cancer cells was performed as
previously described (11). Protein
concentration was determined with a Bradford protein assay kit
(Nanjing KeyGen Biotech., Co., Ltd.). Proteins were resolved on
8.5% polyacrylamide gels (Nantong Biyuntian Biological Technology
Co., Ltd.) and subsequently transferred onto nitrocellulose
membranes (Nanjing KeyGen Biotech., Co., Ltd.). For immunoblotting,
nitrocellulose membranes were incubated with specific antibodies
recognizing target proteins overnight at 4°C. The membranes were
washed as previously described and then incubated with horseradish
peroxidase-conjugated goat anti-rabbit IgG monoclonal secondary
antibody (1:20,000; Amersham Pharmacia Biotech, Arlington Heights,
IL, USA) for 1 h at room temperature and and visualized by
autoradiograpy. β-actin protein (1:5,000; Sigma) was used as the
loading control. The membrane was washed three times with
Tris-buffered saline and Tween 20 [10 mM Tris-HCl (pH 8.0), 150 mM
NaCl and 0.5% Tween-20] and developed using the enhanced
chemiluminescence detection system (Amersham Pharmacia Biotech).
The intensity of the immunoreactive bands was quantified using a
densitometer (SI, Molecular Dynamics, Sunnyvale, CA, USA).
Statistical analysis
All data are presented as the mean ± standard
deviation. Statistical analysis was performed by analysis of
variance followed by Dunnett’s test. P<0.05 was considered to
indicate a statistically significant difference.
Results
AdMax-pDC315-DRAM-EGFP treatment
increases cell viability
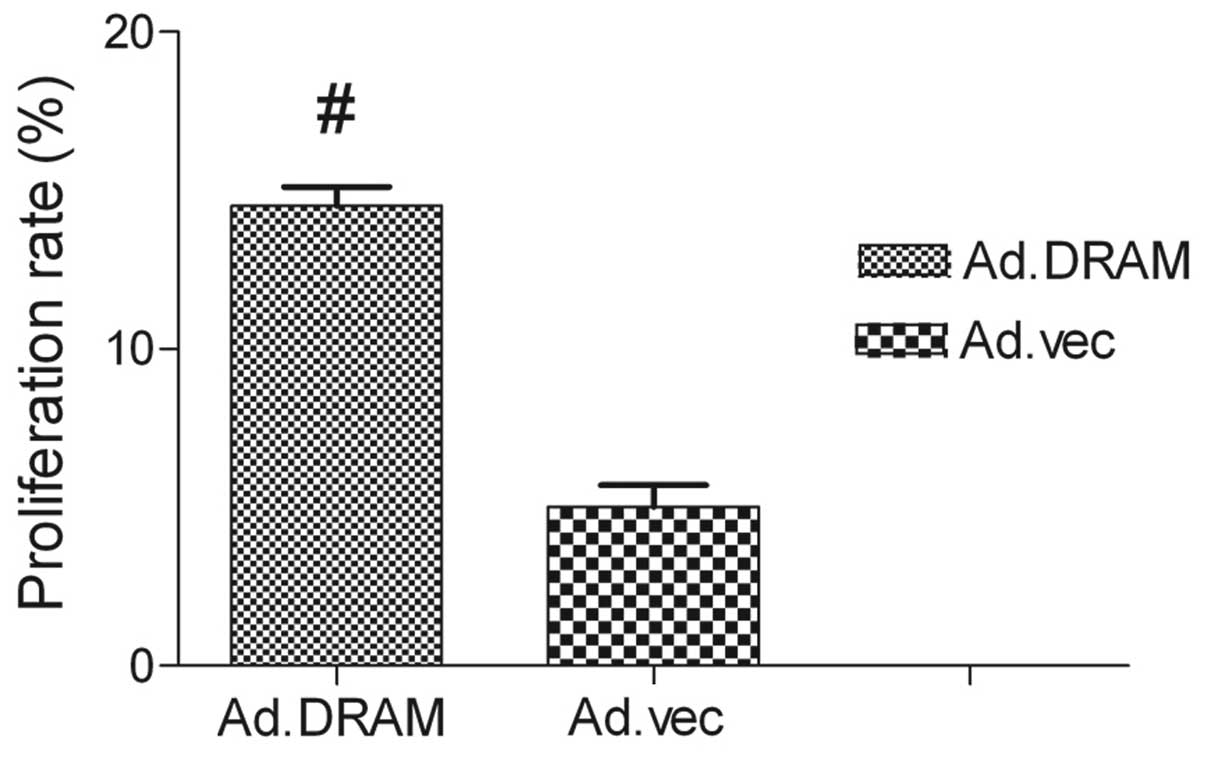
The MTT assay showed that the proliferation capacity
of gastric cancer cells infected with AdMax-pDC315-DRAM-EGFP was
significant1y higher than AdMax-pDC315-EGFP (MOI, 60) (P<0.05).
The DRAM gene promoted the proliferation of cell viability. After
24 h of treatment, the rate of proliferation had reached
14.71±4.13% (Fig. 1).
Infection efficiency and cell
morphology
Following AdMax-pDC315-DRAM-EGFP (MOI, 60) infection
(12 h) of SGC7901 cells, the cell body appeared swollen, rounded
and the cells revealed deformation, with the cells showing further
deformation after 24 h. After infection for 24 h, the SGC7901 cells
were counted under a fluorescence microscope to determine the
percentage of the of infected cells (Fig. 2). Infection efficiency did not
increase with increasing MOI and the time of infection. The
infection efficiency and cell viability are dependent on the
correct MOI (60) and the time of infection (24 h). It was
determined that at an MOI of 60, the infection efficiency was
93±5.4%.
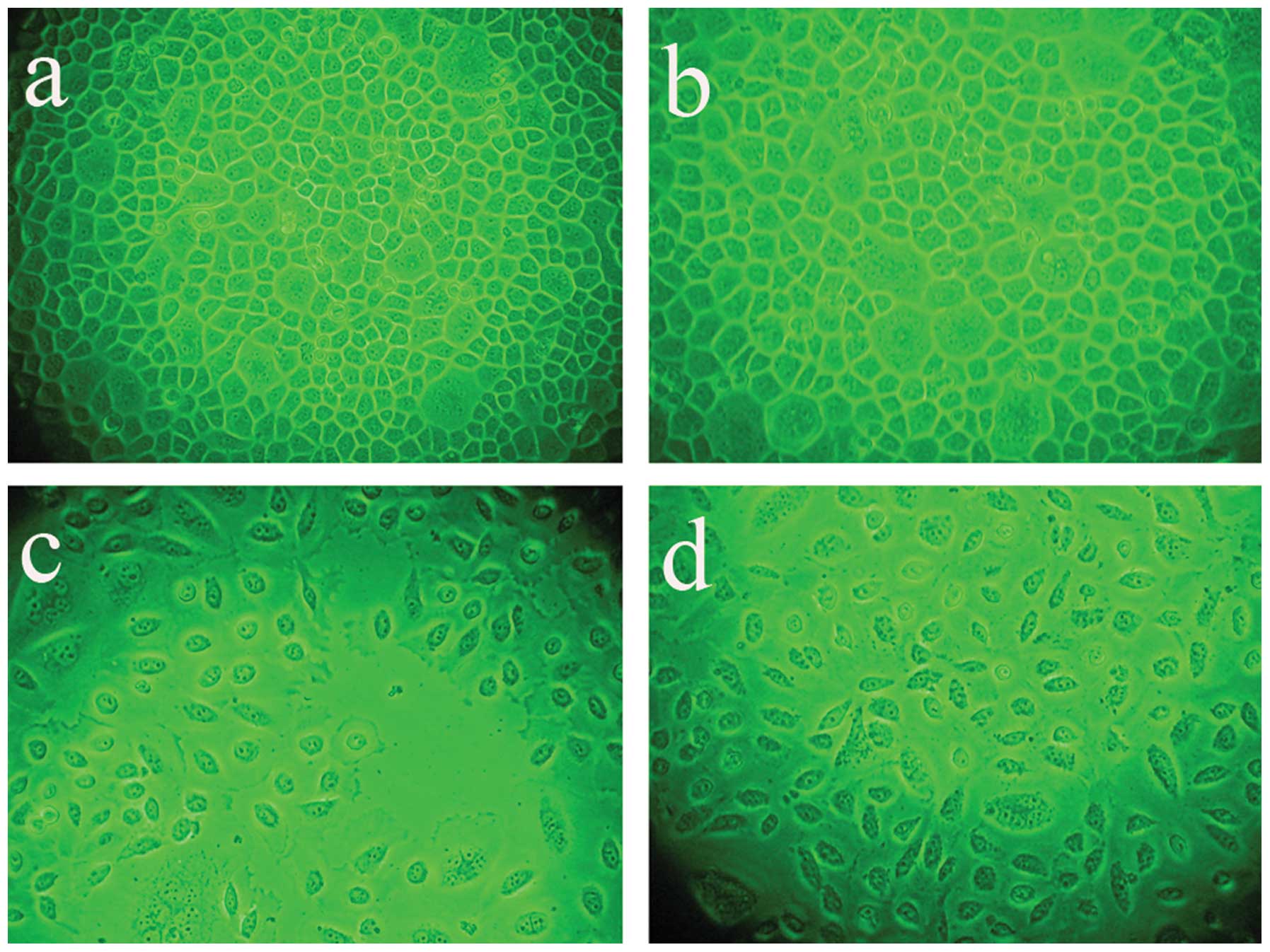 | Figure 2Infection efficiency and cell
morphology were analyzed under a fluorescence microscope following
AdMax-pDC315-DRAM-EGFP (MOI, 60) and AdMax-pDC315-EGFP (MOI, 60)
treatment. The SGC7901 cells were incubated with
AdMax-pDC315-DRAM-EGFP (MOI, 60) for the indicated time. (A)
Control, (B) AdMax-pDC315-EGFP, (C) 12 h after
AdMax-pDC315-DRAM-EGFP (MOI, 60) treatment and (D) 24 h after
AdMax-pDC315-DRAM-EGFP (MOI, 60) treatment. Magnification, ×200.
DRAM, damage-regulated autophagy regulator; MOI, multiplicity of
infection. |
AdMax-pDC315-DRAM-EGFP infection
increases autophagic vacuoles
The autofluorescent substance, MDC, is a marker for
late autophagic vacuoles (L-AVs), but not endosomes (12). The dye is trapped in acidic,
membrane-rich organelles and exhibits an increased fluorescence
quantum yield in response to the compacted lipid bilayers present
in L-AVS (10). When cells are
analyzed under a fluorescent microscope, AVs stained by MDC appear
as distinct dot-like structures distributed within the cytoplasm or
localizing in the perinuclear regions. In this study, an increase
in the number of MDC-labeled vesicles following infection with
AdMax-pDC315-DRAM-EGFP (MOI, 60) from 12 to 24 h was observed
(Fig. 3).
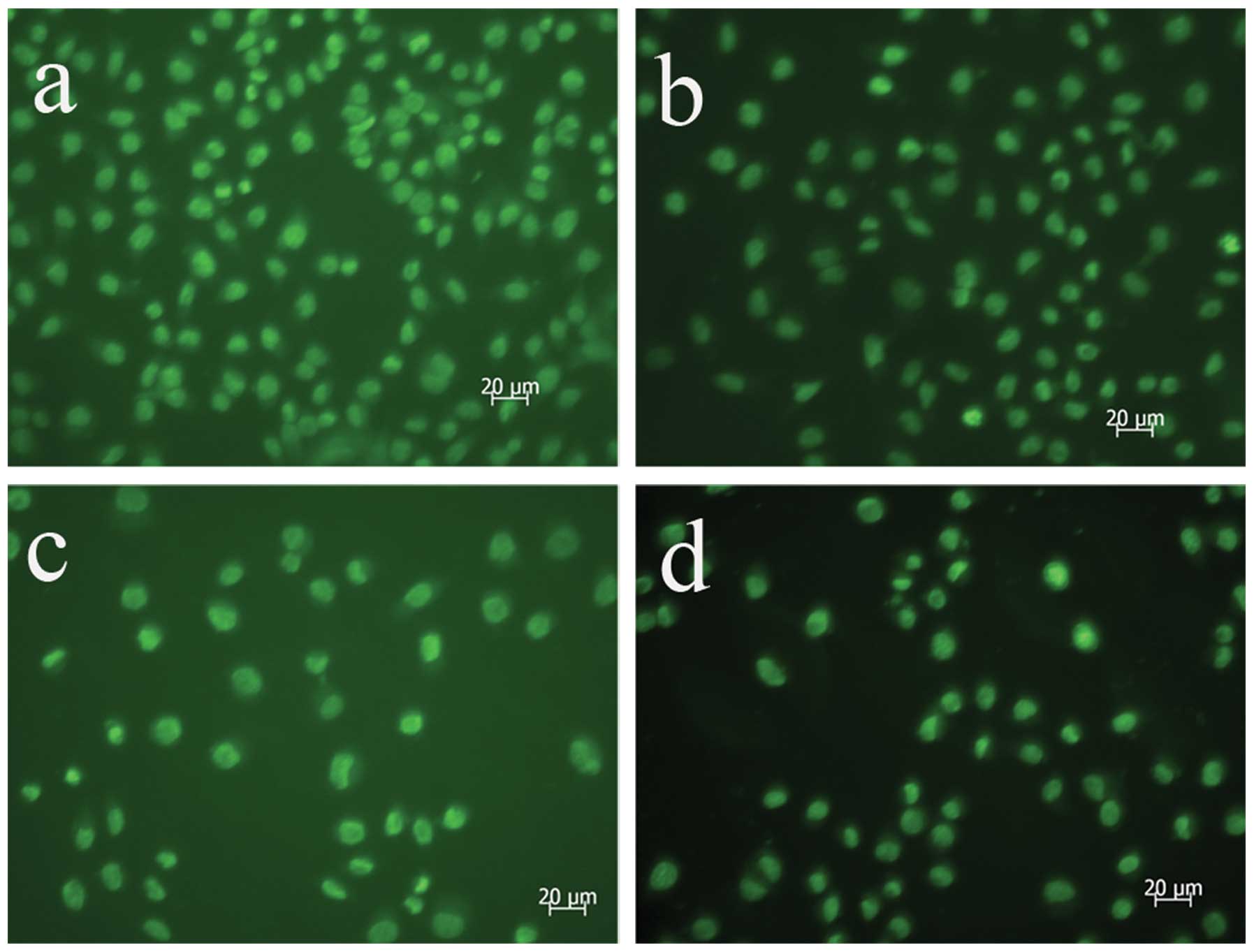 | Figure 3MDC staining revealed autophagy was
activated following AdMax-pDC315-DRAM-EGFP (MOI, 60) and
AdMax-pDC315-EGFP (MOI, 60) treatment. The SGC7901 cells were
incubated with AdMax-pDC315-DRAM-EGFP (MOI, 60) for an indicated
time and stained with MDC (100 μmol/l). Fluorescent particles
revealed late autophagic vacuoles. (A) Control, (B)
AdMax-pDC315-EGFP, (C) 12 h after AdMax-pDC315-DRAM-EGFP (MOI, 60)
treatment and (D) 24 h after AdMax-pDC315-DRAM-EGFP (MOI, 60)
treatment. Magnification, ×1,000. MDC, monodansylcadaverin; MOI,
multiplicity of infection; DRAM, damage-regulated autophagy
regulator. |
AdMax-pDC315-DRAM-EGFP infection
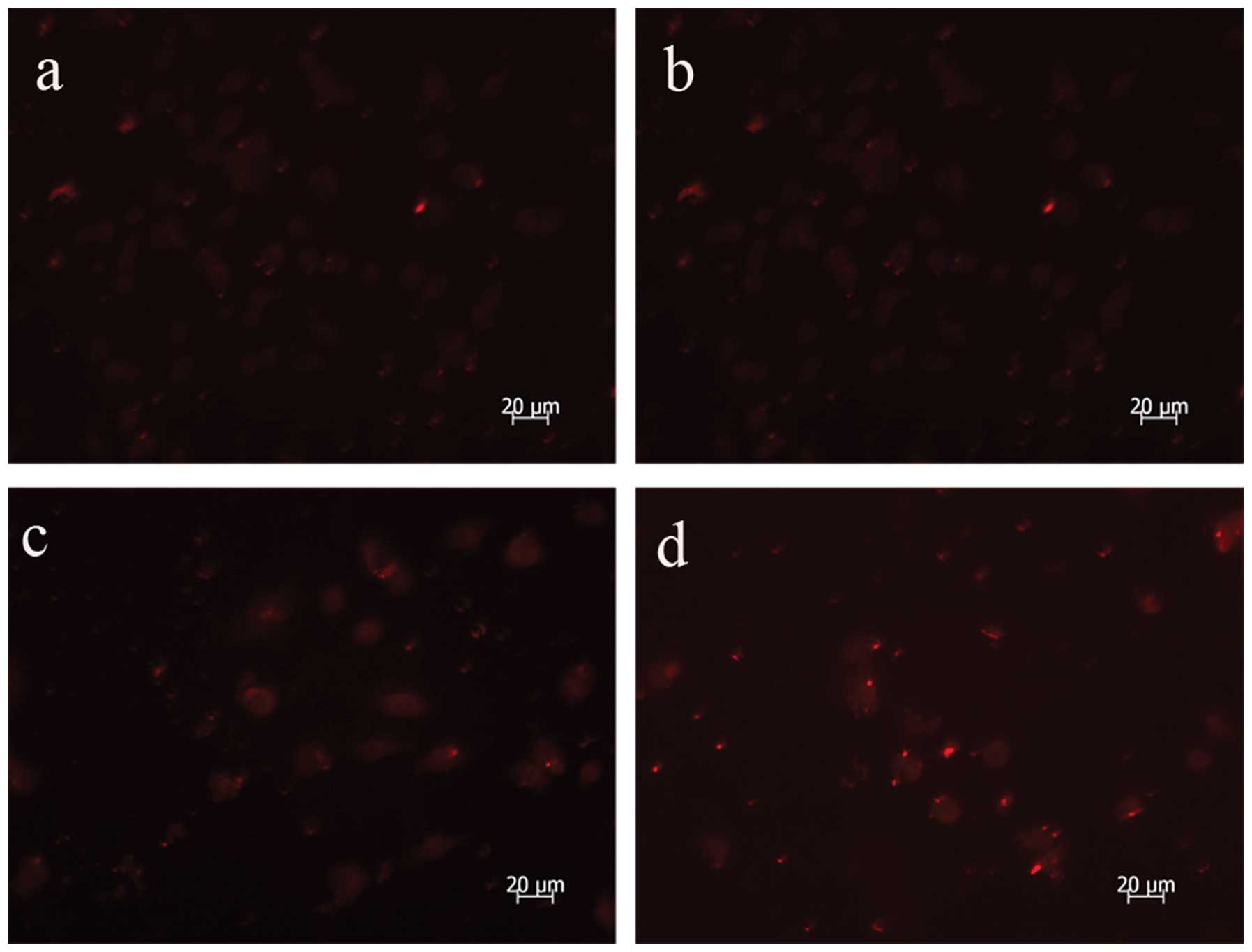
upregulates the expression of LC3
Microtubule-associated protein 1 LD3, the mammalian
ontology of Atg8, targets to the autophagosomal membranes in an
Atg5-dependent manner and remains there even after Atg12-Atg5
dissociates. LC3 is considered to be the only credible marker of
the autophagosome in mammalian cells (13). The present study used
immunofluorescence to analyze the expression and location of LC3
and identified an increased formation of autophagosomes following
AdMax-pDC315-DRAM-EGFP (MOI, 60) infection (Fig. 4).
AdMax-pDC315-DRAM-EGFP infection
upregulates the expression of Beclin1 and p53
To investigate the effects of AdMax-pDC315-DRAM-EGFP
(MOI, 60) infection on the expression of autophagic-related
proteins, western blot analysis was used to detect the expression
of p53 and Beclin1. The findings revealed that the basal level of
Beclin1 and p53 in the SGC7901 cells was low. Following incubation
with AdMax-pDC315-DRAM-EGFP (MOI, 60), the Beclin1 and p53 protein
expression levels significantly increased from 12 to 24 h (Fig. 5).
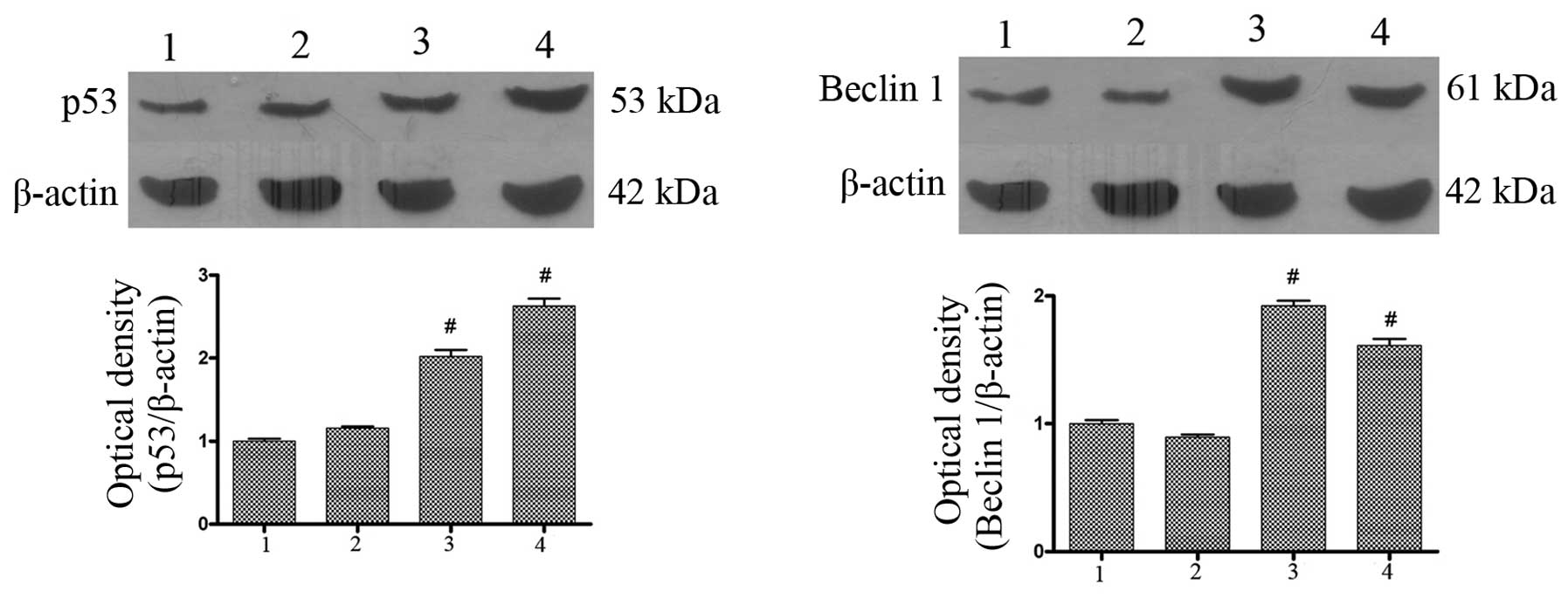 | Figure 5Effect of AdMax-pDC315-DRAM-EGFP (MOI,
60) infection on p53 and Beclin1 protein expression. The SGC7901
cells were treated with AdMax-pDC315-DRAM-EGFP (MOI, 60) and
AdMax-pDC315-EGFP (MOI, 60) for 12 and 24 h, then harvested for
extraction of total proteins. AdMax-pDC315-DRAM-EGFP upregulated
the expression of p53 and Beclin1 protein. 1, normal group; 2,
AdMax-pDC315-EGFP group; 3, AdMax-pDC315-DRAM-EGFP treatment for 12
h; 4, AdMax-pDC315-DRAM-EGFP treatment for 24 h. Statistical
comparisons were performed using Dunnett’s test (n=3). Values are
expressed as the mean ± standard deviation. #P<0.01,
compared with the control group. Cont, control; DRAM,
damage-regulated autophagy regulator; MOI, multiplicity of
infection. |
AdMax-pDC315-DRAM-EGFP infection
increases the expression of p21 and decreases the expression of
Bcl-2
To determine whether AdMax-pDC315-DRAM-EGFP (MOI,
60) infection affects the expression of apoptotic-related proteins,
western blot analysis was used to detect the expression of Bcl-2
and p21 (Fig. 6). The findings
revealed that the basal level of p21 protein in SGC7901 cells was
low; however, following incubation with DRAM, the p21 protein
expression levels were significantly increased from 12 to 24 h. By
contrast, the Bcl-2 protein expression levels were downregulated
with the addition of AdMax-pDC315-DRAM-EGFP (MOI, 60) (Fig. 6).
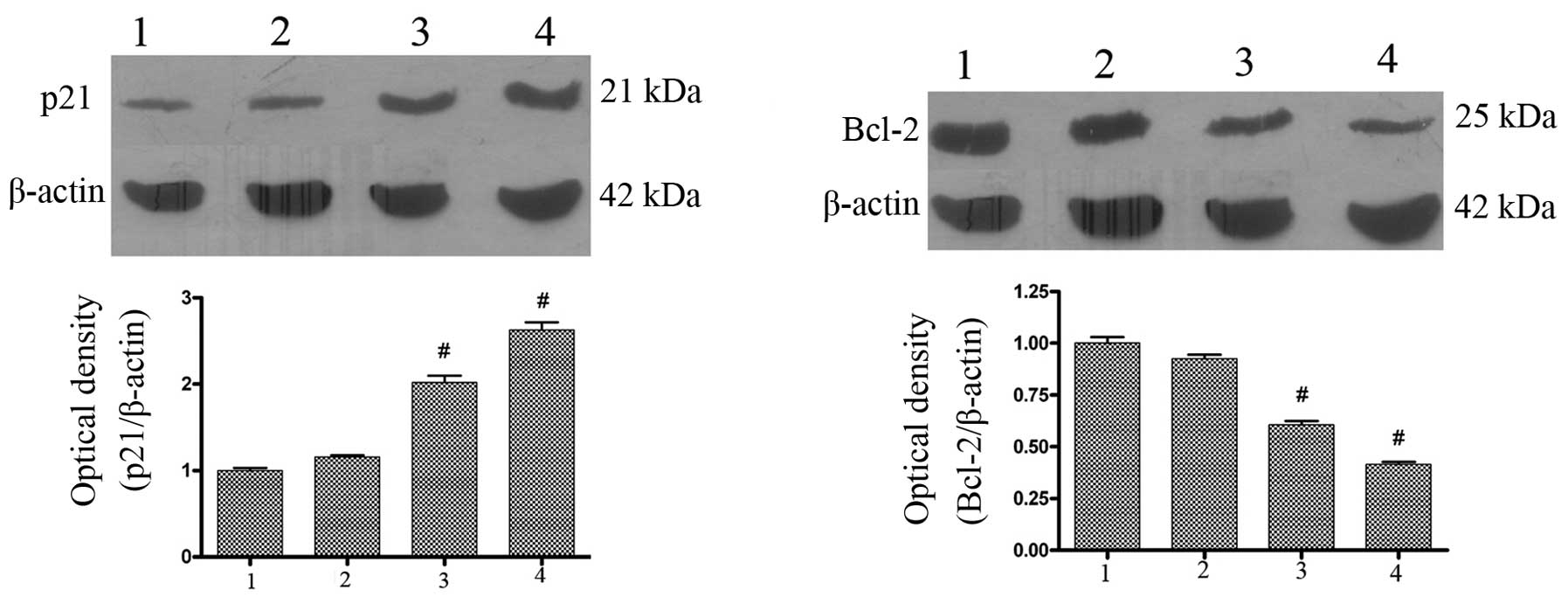 | Figure 6Effects of AdMax-pDC315-DRAM-EGFP
infection on Bcl-2 and p21 protein expression in SGC7901 cells. (A)
Effects of AdMax-pDC315-DRAMEGFP (MOI, 60) on Bcl-2 and p21 protein
expression. The SGC7901 cells were treated with
AdMax-pDC315-DRAM-EGFP (MOI, 60) and AdMax-pDC315-EGFP (MOI, 60)
for 12 or 24 h then harvested for extraction of total proteins.
AdMax-pDC315-DRAM-EGFP upregulated the expression of p21 and
downregulated the expression of Bcl-2 protein. 1, normal group; 2,
AdMax-pDC315-EGFP group; 3, AdMax-pDC315-DRAM-EGFP treatment for 12
h; 4, AdMax-pDC315-DRAM-EGFP treatment for 24 h. Statistical
comparisons were performed using Dunnett’s test (n=3). Values are
expressed as the mean ± standard deviation. #P<0.01,
compared with the control group. Cont, control; DRAM,
damage-regulated autophagy regulator; Bcl-2, B-cell lymphoma 2;
MOI, multiplicity of infection. |
Discussion
A number of studies have found that the baseline
levels of autophagy act as a tumor suppressor mechanism.
Nevertheless, stress-induced autophagy constitutes a major
pro-survival mechanism for tumors exposed to a hypoxic
microenvironment or to chemotherapeutic agents. Thus, autophagy
mediates either antitumor or pro-tumor functions (14,15),
and hence, is considered as a ‘double-edged sword’ in oncogenesis
and tumor progression (16).
The genetic inactivation of p53, the best-known
human oncosuppressor protein, has been observed in >50% of all
types of human cancer and mostly mediates tumor suppression, not
only by transactivating pro-apoptotic and cell cycle arresting
genes, but also by regulating autophagy. p53 mutations that
simultaneously abolish its pro-apoptotic and autophagy-inhibitory
functions behave as ‘multi-hit’ events, as opposed to ‘single-hit’
mutations that only affect the classical (pro-apoptotic and/or cell
cycle-arresting) functions of the p53 system (17,18).
Under genotoxic stress, p53 has been shown to
upregulate the transcription of DRAM. DRAM, a 238-amino acid
protein, which is highly conserved in higher eukaryotes, is
localized to the lysosomal membrane. Knockdown of DRAM expression
promoted survival following exposure to DNA-damage, and DRAM is
also required for p53-induced autophagy and cell death (5).
In the present study, the autophagic level is low in
the SGC7901 gastric cancer cell line; however, with the addition of
DRAM adenovirus the autophagy-specific marker, LC3, was upregulated
indicating an increased formation of autophagosomes induced by DRAM
infection. Beclin1, the mammalian ortholog of the yeast apg6/vps30
gene, plays a role in two fundamentally important cell biological
pathways, autophagy and apoptosis. Beclin1 is a major determinant
in the initiation of autophagy (18–21).
Beclin1 is monoallelically deleted in human breast
and ovarian cancers and is expressed at reduced levels in those
tumors (22,23). The findings of the present study
suggest that autophagy is induced by DRAM and its activation may
not contribute to the antitumor effects of DRAM. Moreover, DRAM
increased the expression of Beclin1, particularly the production of
p53. Bcl-2 and Bcl-xL are associated with the evolutionarily
conserved autophagy inducer, Beclin1, a haplo-insufficient tumor
suppressor (24), and inhibit
autophagy (25). The inhibition may
require Bcl-2 to localize on the endoplasmic reticulum (25,26)
and, notably, the BH3 domain of Beclin1 mediates their association
(27). In the present study, the
expression of Beclin1 was upregulated with the treatment of DRAM
and the expression of Bcl-2 was decreased, indicating that DRAM may
have decreased the expression of Bcl-2. The cell-cycle-regulating
protein, p21, is a cyclin-dependent kinase inhibitor coupled to a
wide variety of cell functions, including p53-dependent growth
suppression, cell cycle arrest following DNA damage, and the
inhibition and induction of apoptosis (28). In our study, following DRAM
infection, the expression of p53 was upregulated simultaneously to
the increase of p21.
In conclusion, the level of autophagy increased with
the addition of DRAM, which also induced proliferation of the
SGC7901 cells. The integrative effect of autophagy induced by DRAM
may have activated the proliferation of the SGC7901 cells. These
findings suggest that autophagy induces the survival of the SGC7901
tumor cell line. Further study should investigate the effects of
DRAM on primary culture gastric cancer cells collected from
patients with gastric cancer. This may provide novel treatment
strategies for patients with gastric cancer.
Acknowledgements
This study was supported by the Natural Science
Foundation of China (grant no. 81172348) and the Suzhou Science and
Technology Development Foundation (grant nos. 2010SYS201031 and
2011SYSD2011092).
References
|
1
|
Crew KD and Neugut AI: Epidemiology of
gastric cancer. World J Gastroenterol. 12:354–362. 2006.
|
|
2
|
Crighton D and Ryan KM: Splicing
DNA-damage responses to tumor cell death. Biochim Biophys Acta.
1705:3–15. 2004.
|
|
3
|
Ryan KM, Phillips AC and Vousden KH:
Regulation and function of the p53 tumor suppressor protein. Curr
Opin Cell Biol. 13:332–337. 2001.
|
|
4
|
Feng Z, Zhang H, Levine AJ and Jin S: The
coordinate regulation of the p53 and mTOR pathways in cells. Proc
Natl Acad Sci USA. 102:8204–8209. 2005.
|
|
5
|
Crighton D, Wilkinson S, O’Prey J, Syed N,
Smith P, Harrison PR, Gasco M, Garrone O, Crook T and Ryan KM:
DRAM, a p53-induced modulator of autophagy, is critical for
apoptosis. Cell. 126:121–134. 2006.
|
|
6
|
Klionsky DJ and Emr SD: Autophagy as a
regulated pathway of cellular degradation. Science. 290:1717–1721.
2000.
|
|
7
|
Kerley-Hamilton JS, Pike AM, Hutchinson
JA, Freemantle SJ and Spinella MJ: The direct p53 target gene,
FLJ11259/DRAM, is a member of a novel family of transmembrane
proteins. Biochim Biophys Acta. 1769:209–219. 2007.
|
|
8
|
Crighton D, O’Prey J, Bell HS and Ryan KM:
p73 regulates DRAM-independent autophagy that does not contribute
to programmed cell death. Cell Death Differ. 14:1071–1079.
2007.
|
|
9
|
Alesci S, Ramsey WJ, Bornstein SR,
Chrousos GP, Hornsby PJ, Benvenga S, Trimarchi F and
Ehrhart-Bornstein M: Adenoviral vectors can impair adrenocortical
steroidogenesis: clinical implications for natural infections and
gene therapy. Proc Natl Acad Sci USA. 99:7484–7489. 2002.
|
|
10
|
Niemann A, Takatsuki A and Elsässer HP:
The lysosomotropic agent monodansylcadaverine also acts as a
solvent polarity probe. J Histochem Cytochem. 48:251–258. 2000.
|
|
11
|
Ikeda K, Monden T, Kanoh T, Tsujie M,
Izawa H, Haba A, Ohnishi T, Sekimoto M, Tomita N, Shiozaki H and
Monden M: Extraction and analysis of diagnostically useful proteins
from formalin-fixed, paraffin-embedded tissue sections. J Histochem
Cytochem. 46:397–403
|
|
12
|
Biederbick A, Kern HF and Elsässer HP:
Monodansylcadaverine (MDC) is a specific in vivo marker for
autophagic vacuoles. Eur J Cell Biol. 66:3–14. 1995.
|
|
13
|
Yoshimori T: Autophagy: a regulated bulk
degradation process inside cells. Biochem Biophys Res Commun.
313:453–458. 2004.
|
|
14
|
Maiuri MC, Tasdemir E, Criollo A, Morselli
E, Vicencio JM, Carnuccio R and Kroemer G: Control of autophagy by
oncogenes and tumor suppressor genes. Cell Death Differ. 16:87–93.
2009.
|
|
15
|
Morselli E, Galluzzi L, Kepp O, Vicencio
JM, Criollo A, Maiuri MC and Kroemer G: Anti- and pro-tumor
functions of autophagy. Biochim Biophys Acta. 1793:1524–1532.
2009.
|
|
16
|
White E and DiPaola RS: The double-edged
sword of autophagy modulation in cancer. Clin Cancer Res.
15:5308–5316. 2009.
|
|
17
|
Soussi T: p53 alterations in human cancer:
more questions than answers. Oncogene. 26:2145–2156. 2007.
|
|
18
|
Soussi T and Lozano G: p53 mutation
heterogeneity in cancer. Biochem Biophys Res Commun. 331:834–842.
2005.
|
|
19
|
Liang XH, Jackson S, Seaman M, Brown K,
Kempkes B, Hibshoosh H and Levine B: Induction of autophagy and
inhibition of tumorigenesis by beclin 1. Nature. 402:672–676.
1999.
|
|
20
|
Yue Z, Jin S, Yang C, Levine AJ and Heintz
N: Beclin 1, an autophagy gene essential for early embryonic
development, is a haploinsufficient tumor suppressor. Proc Natl
Acad Sci USA. 100:15077–15082. 2003.
|
|
21
|
Zeng X, Overmeyer JH and Maltese WA:
Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase
complex in macroautophagy versus endocytosis and lysosomal enzyme
trafficking. J Cell Sci. 119:259–270. 2006.
|
|
22
|
Lum JJ, DeBerardinis RJ and Thompson CB:
Autophagy in metazoans: cell survival in the land of plenty
(review). Nat Rev Mol Cell Biol. 6:439–448. 2005.
|
|
23
|
Aita VM, Liang XH, Murty VV, Pincus DL, Yu
W, Cayanis E, Kalachikov S, Gilliam TC and Levine B: Cloning and
genomic organization of Beclin 1, a candidate tumor suppressor gene
on chromosome 17q21. Genomics. 59:59–65. 1999.
|
|
24
|
Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh
H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y,
Cattoretti G and Levine B: Promotion of tumorigenesis by
heterozygous disruption of the beclin 1 autophagy gene. J Clin
Invest. 112:1809–1820. 2003.
|
|
25
|
Pattingre S, Tassa A, Qu X, Garuti R,
Liang XH, Mizushima N, Packer M, Schneider MD and Levine B: Bcl-2
antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell.
122:927–939. 2005.
|
|
26
|
Hoyer-Hansen M, Bastholm L, Szyniarowski
P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N,
Elling F, Rizzuto R, Mathiasen IS and Jäättelä M: Control of
macroautophagy by calcium, calmodulindependent kinase kinase-beta,
and Bcl-2. Mol Cell. 25:193–205. 2007.
|
|
27
|
Oberstein A, Jeffrey PD and Shi Y: Crystal
structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a
novel BH3-only protein. J Biol Chem. 282:13123–13132. 2007.
|
|
28
|
Le NT and Richardson DR: Potent iron
chelators increase the mRNA levels of the universal
cyclin-dependent kinase inhibitor p21(CIP1/WAF1), but paradoxically
inhibit its translation: a potential mechanism of cell cycle
dysregulation. Carcinogenesis. 24:1045–1058. 2003.
|