Introduction
Ribavirin
(1-β-D-ribofuranosy-1,2,4-triazole-3-carboxamide) was developed as
an antiviral agent against RNA and DNA viruses and its function was
first described in 1972 by Sidwell et al (1). Ribavirin, in combination with
interferon (IFN), is one of the standard treatment
strategies for hepatitis C virus (HCV) infections (2). Ribavirin was expected to be clinically
advantageous in the treatment of various viral infections, however,
to date the clinical utilization of ribavirin has been limited to
the treatment of HCV infections.
Recently, interest in the efficacy of ribavirin for
the treatment of tumors has increased due to two reasons. Firstly,
ribavirin inhibits inosine-5′-monophosphate dehydrogenase (IMPDH)
(3). IMPDH is a key enzyme in
guanosine triphosphate synthesis, and IMPDH expression levels and
activity are elevated in certain types of tumor. Furthermore, IMPDH
is also associated with the proliferation and transformation of
malignant tumor cells (4).
Therefore, an IMPDH inhibitor may be a candidate for antitumor
chemotherapy. Secondly, ribavirin inhibits the eukaryotic
translation initiation factor 4E (eIF4E) (5,6). eIF4E
has two major functions in gene expression: Messenger (m)RNA
translation and mRNA export (7).
The eIF4E protein is present in the nucleus and the cytoplasm; in
the nucleus, eIF4E facilitates the export of a subset of specific
growth-promoting mRNAs. In the cytoplasm, eIF4E recruits mRNA with
highly structured 5′-untranslated regions to the ribosome to
promote translation. Therefore, eIF4E, which is overexpressed in
~30% of human cancers (5,8), may have oncogenic potential.
The antitumor efficacy of ribavirin has been
reported in breast cancer and leukemia (5,9),
however, to the best of our knowledge it has yet to be reported in
malignant glioma. The aim of the present investigation was to
evaluate the antitumor efficacy of ribavirin on malignant glioma
cells, to identify novel predictive genes for ribavirin sensitivity
through the evaluation of gene expression profiles and to assess
the influence of ribavirin on the cell cycle of malignant glioma
cells.
Materials and methods
Cell lines and cell culture
Human malignant glioma cells of the A-172, AM-38,
T98G, U-251MG and YH-13 cell lines were obtained from Health
Science Research Resources Bank (Osaka, Japan), and the U-87MG and
U-138MG cell lines were purchased from the American Type Culture
Collection (Manassas, VA, USA). All of the cell lines were cultured
in Dulbecco’s modified Eagle’s medium (Nissui Pharmaceutical,
Tokyo, Japan) supplemented with 10% fetal bovine serum (Life
Technologies, Grand Island, NY, USA) in a standard humidified
incubator at 37°C with an atmosphere of 5% CO2.
Cell culture growth with ribavirin
Malignant glioma cell proliferation was evaluated
using a Z1 Coulter Counter® (Beckman Coulter, Brea, CA,
USA) to count the cell growth in 24-well plates (Iwaki, Chiba,
Japan). Each well was seeded with 1×104 cells and
cultured for 24 h prior to ribavirin treatment to allow adherence
of the cells to the plate. The culture medium was replenished with
fresh medium containing ribavirin (0.1–1,000 μM), and the cells
were cultured for 72 h. The proliferated cells were trypsinized
with trypsin-EDTA solution (Invitrogen Life Technologies, San
Diego, CA, USA) and counted using the Z1 Coulter
Counter®. The cell culture growth experiments were
repeated a minimum of seven times at each concentration. The half
maximal inhibitory concentration (IC50), with regard to
the growth of the malignant glioma cell culture, was determined
from the concentration of ribavirin required for 50% growth
inhibition in comparison to untreated control cells.
RNA preparation and hybridization
Total RNA was extracted from the malignant glioma
cells using an RNeasy® mini kit (Qiagen, Valencia, CA,
USA) and quantified by spectrophotometry (Jasco V-550
spectrophotometer; Jasco Interntational Co., Ltd., Tokyo, Japan).
Double-stranded complementary (c)DNA was generated from the total
RNA (5 μg) using a One-Cycle cDNA Synthesis kit (Affymetrix, Inc.,
Santa Clara, CA, USA). Biotinylated cRNA was then synthesized from
the cDNA in an in vitro transcription reaction (IVT) by
employing an IVT labeling kit (Affymetrix, Inc.). The biotinylated
cRNA (10 μg) was fragmented and hybridized to a DNA oligonucleotide
expression array (Human Genome U133A 2.0 Array; Affymetrix, Inc.)
containing >22,277 probe sets for ~14,500 human genes (certain
genes are represented on the array by multiple probe sets). The
hybridized probe array was washed and stained with a
streptavidin-phycoerythrin conjugate (Molecular Probes Life
Technologies, Carlsbad, CA, USA) using a GeneChip®
Fluidics Station 450 (Affymetrix, Inc.) according to the
manufacturer’s instructions, as previously described (10,11).
Identification of discriminatory genes
for ribavirin sensitivity
The probe array was scanned with a confocal laser
scanner (GeneChip® Scanner 3000; Affymetrix, Inc.) and
analyzed using GeneChip® Operating Software 1.1
(Affymetrix, Inc.), allowing the gene expression levels to be
calculated from the signal intensities. All of the genes
represented on the GeneChip® were globally normalized
and scaled to the signal intensity to provide a gene expression
value. To avoid contributions from artificial sources of variation
in the experimentally measured expression patterns, each cell line
was grown in three independent cultures and the entire process was
conducted independently on mRNA that was extracted from each
culture. The expression array analysis for each cell line was then
run in triplicate.
Cell cycle distribution analysis
The cells were plated at 2×105 cells per
25-cm2 flask (Iwaki) and incubated for 24 h to allow
attachment of the cells to the flask. The culture medium was then
replenished with fresh medium containing 10 μM ribavirin and the
cells were harvested using trypsin-EDTA solution at 0, 4, 8 and 24
h. The cells were rinsed with phosphate-buffered saline, fixed
using ice-cold 70% ethanol, incubated with 200 μg/ml RNase A (Roche
Diagnostics, Basel, Switzerland) for 30 min at room temperature and
stained with 2 μg/ml propidium iodide (PI) solution (Miltenyi
Biotec, Inc., San Diego, CA, USA). The fluorescence was measured by
flow cytometry, using the BD FACSCalibur™ flow cytometer (BD
Biosciences, Franklin Lakes, NJ, USA), at a fluorescence wavelength
of 610 nm. The resulting DNA histogram was analyzed using FlowJo
software (BioLegend, Inc., San Diego, CA, USA).
Statistical analysis
To identify the discriminative genes for ribavirin
sensitivity, the Pearson’s correlation test was performed to
evaluate the association between the IC50 of ribavirin
and the gene expression level of each gene (Microsoft Office Excel
2007, Redmond, WA, USA).
Results
Antitumor efficacy of ribavirin
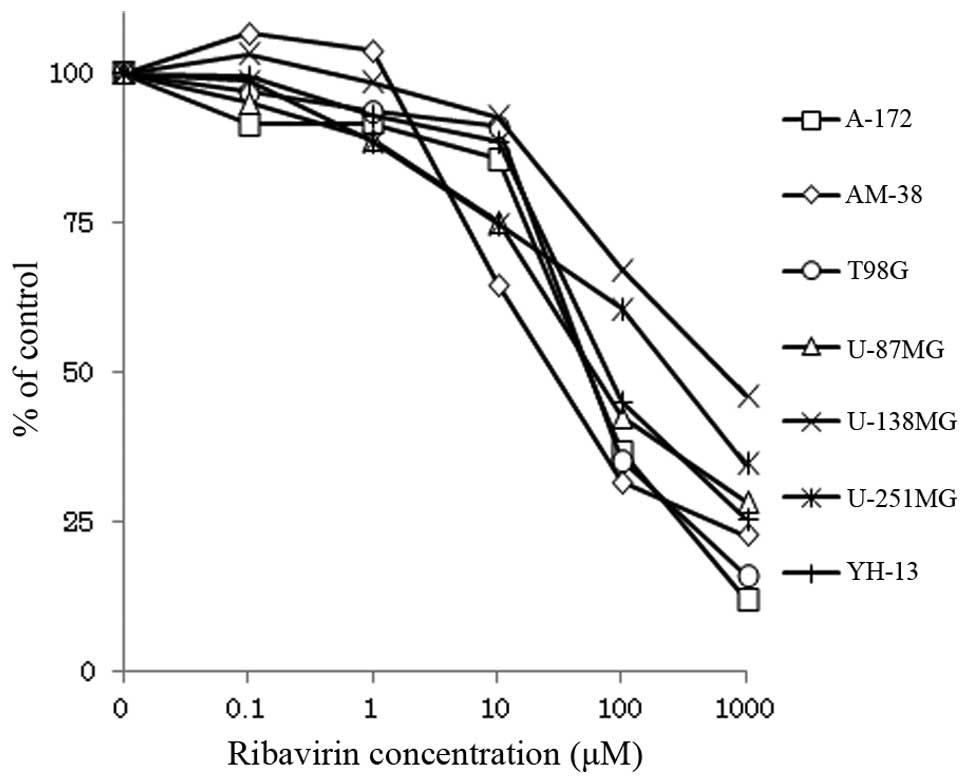
Seven malignant glioma cell lines were treated with
0.1–1,000 μM ribavirin and cultured for 72 h. The proliferated
cells were trypsinized and counted using a Z1 Coulter
Counter®. As demonstrated by Fig. 1, the growth of all of the cell lines
was inhibited by ribavirin in a dose-dependent manner, although the
sensitivity of each cell line to ribavirin varied. The
IC50 of ribavirin for five of the malignant glioma cell
lines (A-172, AM-38, T98G, U-87MG and YH-13) was <100 μM,
whereas, the IC50 for the other two cell lines (U-138MG
and U-251MG) was >250 μM (Table
I).
 | Table IIC50 of ribavirin for
malignant glioma cell lines. |
Table I
IC50 of ribavirin for
malignant glioma cell lines.
| Cell line | IC50,
μM |
|---|
| A-172 | 53.6 |
| AM-38 | 27.9 |
| T98G | 55.0 |
| U-87MG | 59.7 |
| U-138MG | 664.2 |
| U-251MG | 257.7 |
| YH-13 | 76.9 |
Identification of discriminatory genes
for ribavirin sensitivity
Of the 22,277 probe sets, genes that were not
expressed were omitted, therefore, 16,913 probe sets were available
for subsequent analysis. The expression profiling data for the
malignant glioma cell lines were consistent with data from previous
reports (10,11). Various genes that were expressed in
the seven malignant glioma cell lines included in the present study
were observed to be upregulated or downregulated relative to
ribavirin sensitivity. The negative and positive correlation values
of the association between gene expression level and ribavirin
sensitivity are indicated in Table
II by the Pearson’s correlation coefficient and the
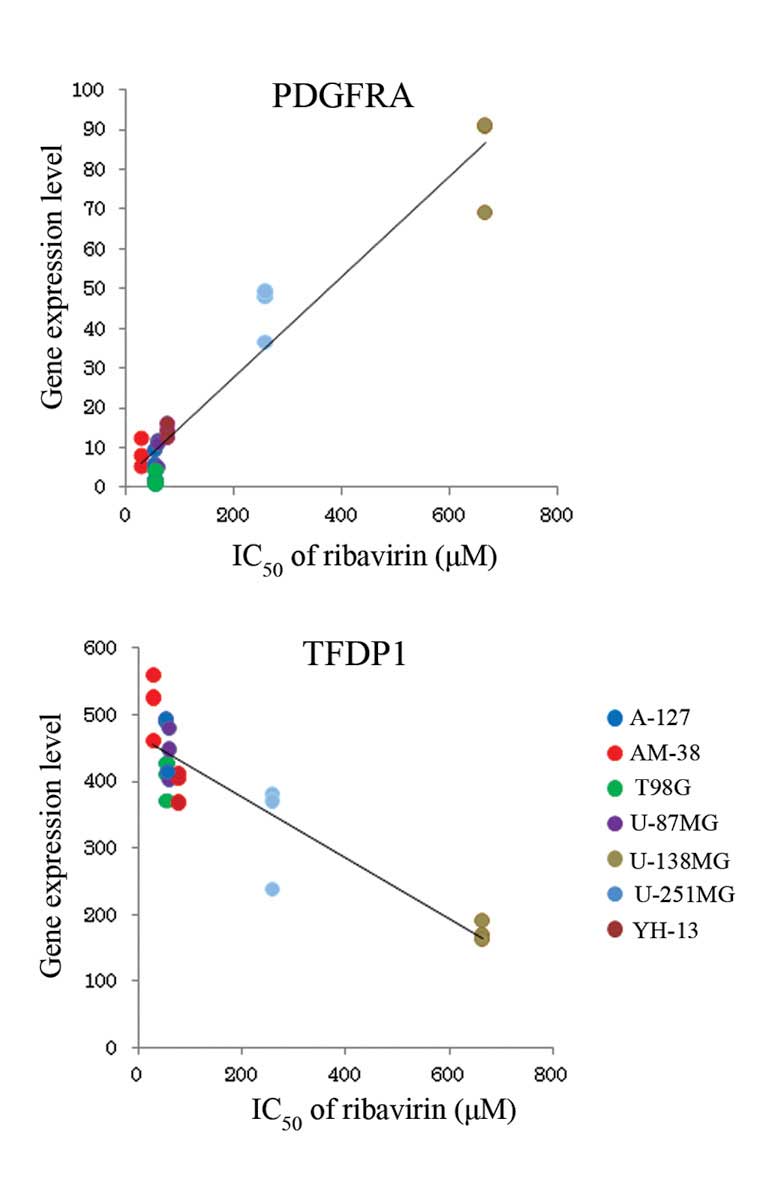
corresponding P-value. The gene expression levels of PDGFRA
and TFDP1 indicate the greatest positive and negative
correlation with ribavirin sensitivity, respectively (Fig. 2).
 | Table IIDifferentially expressed genes
associated with ribavirin sensitivity. |
Table II
Differentially expressed genes
associated with ribavirin sensitivity.
| A, Positive
differentially expressed genes |
|---|
|
|---|
| Symbol | Gene title | Correlation | P-value |
|---|
| PDGFRA | Platelet-derived
growth factor receptor, α polypeptide 0.968 | <0.0001 | |
| ZFP36L2 | Zinc finger protein
36, C3H type-like 2 | 0.951 | <0.0001 |
| UBL3 | Ubiquitin-like 3 | 0.944 | <0.0001 |
| HSPB2 | Heat shock 27 kDa
protein 2 | 0.940 | <0.0001 |
| HERC2P2,
HERC2P3 | Hect domain and RLD 2
pseudogene 2; Hect domain and RLD 2 pseudogene 3 | 0.940 | <0.0001 |
| MEOX2 | Mesenchyme homeobox
2 | 0.933 | <0.0001 |
| SDC2 | Syndecan 2 | 0.929 | <0.0001 |
| POSTN | Periostin, osteoblast
specific factor | 0.921 | <0.0001 |
| PLCB1 | Phospholipase C, β 1
(phosphoinositide-specific) | 0.920 | <0.0001 |
| KCNJ8 | Potassium
inwardly-rectifying channel, subfamily J, member 8 | 0.918 | <0.0001 |
| MAP4K4 | Mitogen-activated
protein kinase kinase kinase kinase 4 | 0.917 | <0.0001 |
| DOK5 | Docking protein
5 | 0.916 | <0.0001 |
| STS | Steroid sulfatase
(microsomal), isozyme S | 0.913 | <0.0001 |
| HAS2 | Hyaluronan synthase
2 | 0.912 | <0.0001 |
| AP1G1 | Adaptor-related
protein complex 1, γ 1 subunit | 0.911 | <0.0001 |
|
| B, Negative
differentially expressed genes |
|
| Symbol | Gene title | Correlation | P-value |
|
| TFDP1 | Transcription factor
Dp-1 | −0.894 | <0.0001 |
| OSBPL9 | Oxysterol binding
protein-like 9 | −0.806 | <0.0001 |
| PLAA | Phospholipase
A2-activating protein | −0.796 | <0.0001 |
| BTF3 | Basic transcription
factor 3 | −0.795 | <0.0001 |
| PPWD1 | Peptidylprolyl
isomerase domain and WD repeat containing 1 | −0.792 | <0.0001 |
| LARS | Leucyl-tRNA
synthetase | −0.792 | <0.0001 |
| NDUFA8 | NADH dehydrogenase
(ubiquinone) 1 α subcomplex, 8, 19 kDa | −0.790 | <0.0001 |
| ERAP2 | Endoplasmic reticulum
aminopeptidase 2 | −0.787 | <0.0001 |
| PTPN4 | Protein tyrosine
phosphatase, non-receptor type 4 (megakaryocyte) | −0.784 | <0.0001 |
| HMGCR |
3-hydroxy-3-methylglutaryl-CoA
reductase | −0.781 | <0.0001 |
| TAF9 | TAF9 RNA polymerase
II, TBP-associated factor, 32 kDa | −0.777 | <0.0001 |
| KLF5 | Kruppel-like factor
5 (intestinal) | −0.774 | <0.0001 |
| XRCC4 | X-ray repair
complementing defective repair in Chinese hamster cells 4 | −0.769 | <0.0001 |
| MSH3 | MutS homolog 3 | −0.762 | <0.0001 |
| FBL | Fibrillarin | −0.753 | <0.0001 |
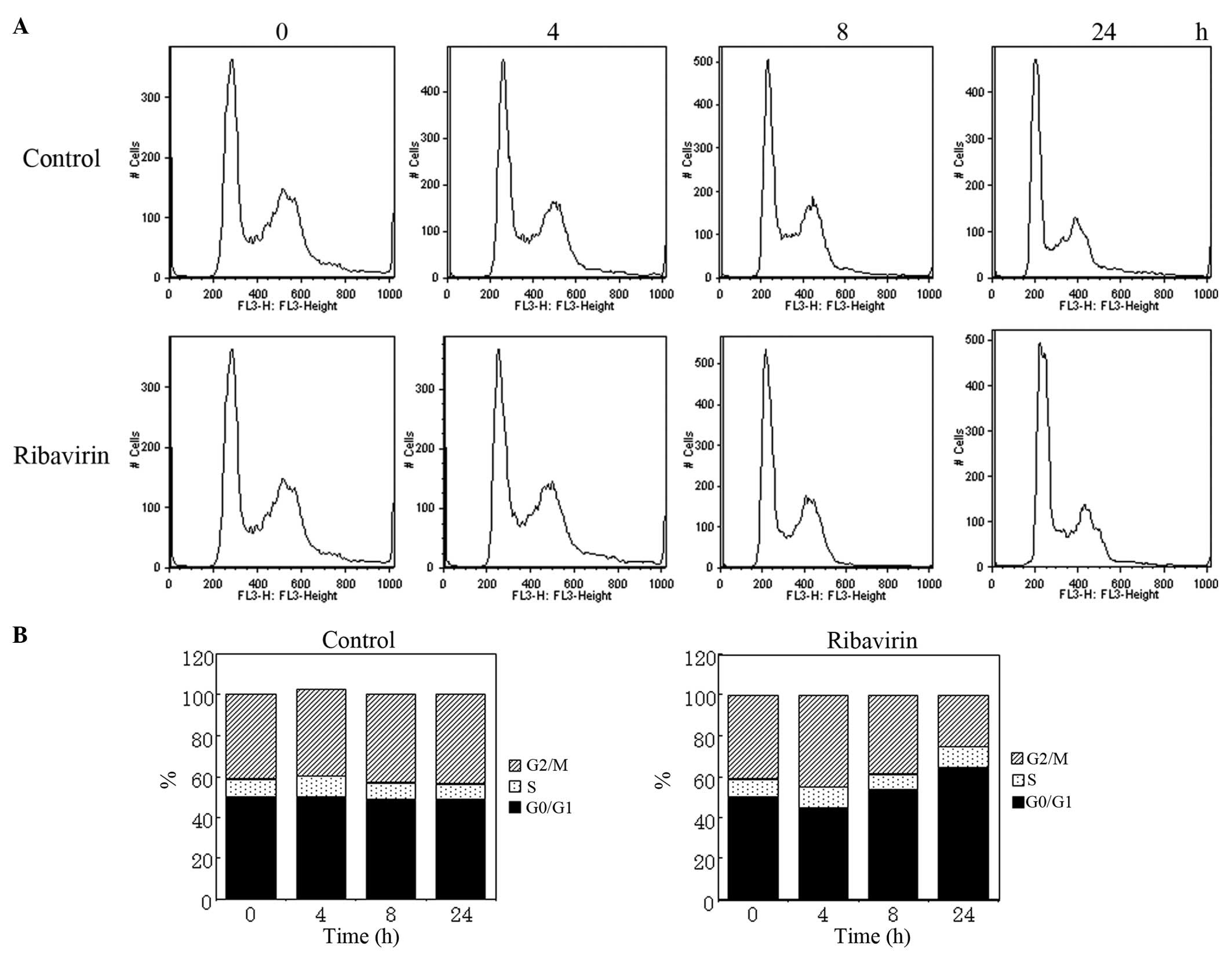
Cell cycle distribution analysis
To clarify the antitumor efficacy of ribavirin on
malignant glioma cells, alterations in the cell cycle distribution
were examined using U-87MG cells treated with ribavirin.
Unsynchronized cells were treated with 10 μM ribavirin for 4, 8 and
24 h. The harvested cells were fixed using ethanol, treated with
RNase A and stained with PI for analysis by fluorescence-activated
cell sorting (FACS; Fig. 3A).
Histograms presenting the FACS data (Fig. 3B) demonstrate an increase in the
population of cells in the G0/G1 phase following treatment with
ribavirin, with time-lapse indicating that the antitumor efficacy
of ribavirin results from the accumulation of cells in the G0/G1
phase.
Discussion
To the best our knowledge, the current study is the
first to demonstrate that ribavirin inhibits the growth of
malignant glioma cells. Ribavirin was initially developed as an
antiviral agent against RNA and DNA viruses and is predominantly
used, in combination with IFN, as a treatment strategy for HCV
infection (2). Although ribavirin
was expected to be developed as a therapeutic treatment for various
other viral infections, its clinical use has thus far been
restricted to the treatment of HCV infections. Interest in the
antitumor efficacy of ribavirin has been increasing due to its
ability to inhibit IMPDH and eIF4E (1,6).
Furthermore, the antitumor efficacy of ribavirin has been reported
in the treatment of breast cancer and leukemia (5,9).
The antitumor efficacy of ribavirin on malignant
glioma cell lines was measured and it was revealed that the growth
of these cell lines was inhibited by ribavirin in a dose-dependent
manner. Of the seven malignant glioma cell lines investigated in
the current study, the IC50 of five of these cell lines
(A-172, AM-38, T98G, U-87MG and YH-13) was <100 μM and the
IC50 of the other two glioma cell lines (U-138MG and
U-251MG) was >250 μM. Based on these data, the malignant glioma
cell lines can be divided into ribavirin-effective and
ribavirin-resistant groups. The genes that were positively and
negatively correlated with ribavirin sensitivity are listed in
Table II. To the best of our
knowledge, none of the genes identified in the current study were
previously expected to be associated with ribavirin or chemotherapy
sensitivity. TFDP1 has been associated with the cell cycle
and seven genes (PDGFRA, ZEP36L2, STS,
TFDP1, HAS2, FBL and KLF5) have
previously been associated with cell proliferation. In the present
study, PDGFRA demonstrated the greatest positive correlation
between gene expression level and the IC50 of ribavirin.
Platelet-derived growth factor (PDGF) is a major mitogen for glial
cells and connective tissue, and is key in the development of the
central nervous system, wound healing, inflammation and neoplasia
(12). PDGF receptor α is the cell
surface receptor of PDGF. PDGFRA mRNA overexpression was
detected in the low- and high-grade astrocytomas (13) and PDGFRA amplification is
typical in the signaling pathway that results in the development of
secondary glioblastoma (14).
PDGFRA amplification is important for subgroup classification of
malignant gliomas. To the best our knowledge, PDGFRA amplification
and overexpression has not been reported, with regard to its
association with chemotherapy sensitivity. However, although the
association between rebavirin and PDGFRA amplification is unclear,
we hypothesize that PDGFRA expression may be involved in the
antitumor efficacy of ribavirin. Although it has been reported that
ribavirin has an effect on the types of breast cancer that
overexpress eIF4E, eIF4E was not extracted and analyzed in the
present study as eIF4E is upregulated in all high-grade astrocytic
tumors (15) and hence all
malignant glioma cell lines. Furthermore, IMPDH was not included in
the present study as it was reported that ribavirin inhibits
leukemic cell proliferation through multiple signaling pathways
(16), therefore, ribavirin may act
on malignant glioma cells via mechanisms that do not involve eIF4E
or IMPDH.
Finally, a cell cycle analysis of the harvested
U-87MG cells, which were treated with ribavirin revealed that cells
accumulated at the G0/G1 boundary; however, the sub-G1 population,
which indicates apoptotic cells, did not increase. Vallée et
al (17) reported that, in a
melanoma cell line, the cell cycle was blocked in the G0/G1 phase
by treatment with 100 μM ribavirin. The results of the present
study also indicate that ribavirin is important in the inhibition
of cell growth in malignant glioma by inducing G0/G1 arrest. To the
best of our knowledge, no studies have reported that ribavirin
alone is able to induce apoptosis in malignant glioma cells.
However, Schlosser et al (18) reported that the number of human
hepatoma apoptotic cells was increased by treatment with 50 μM
ribavirin plus α-IFN. This synergistic effect is also expected to
occur in malignant glioma cells treated with ribavirin combined
with IFN or temozolomide (TMZ).
The present standard postoperative treatment
strategy for malignant glioma is TMZ plus radiation and the median
treatment survival time is 14.6 months (19). The results of the current study
indicate that ribavirin may present as a novel agent for malignant
glioma chemotherapy and that PDGFRA expression levels may be
a significant marker of the antitumor efficacy of ribavirin against
malignant gliomas.
References
|
1
|
Sidwell RW, Huffman JH, Khare GP, Allen
LB, Witkowski JT and Robins RK: Broad-spectrum antiviral activity
of Virazole: 1-beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide.
Science. 177:705–706. 1972.
|
|
2
|
Fried MW, Shiffman ML, Reddy KR, et al:
Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus
infection. N Engl J Med. 347:975–982. 2002.
|
|
3
|
Yamada Y, Natsumeda Y and Weber G: Action
of the active metabolites of tiazofurin and ribavirin on purified
IMP dehydrogenase. Biochemistry. 27:2193–2196. 1988.
|
|
4
|
Jackson RC, Weber G and Morris HP: IMP
dehydrogenase, an enzyme linked with proliferation and malignancy.
Nature. 256:331–333. 1975.
|
|
5
|
Assouline S, Culjkovic B, Cocolakis E, et
al: Molecular targeting of the oncogene eIF4E in acute myeloid
leukemia (AML): a proof-of-principle clinical trial with ribavirin.
Blood. 114:257–260. 2009.
|
|
6
|
Kentsis A, Topisirovic I, Culjkovic B,
Shao L and Borden KL: Ribavirin suppresses eIF4E-mediated oncogenic
transformation by physical mimicry of the 7-methyl guanosine mRNA
cap. Proc Natl Acad Sci USA. 101:18105–18110. 2004.
|
|
7
|
Culjkovic B, Topisirovic I and Borden KL:
Controlling gene expression through RNA regulons: the role of the
eukaryotic translation initiation factor eIF4E. Cell Cycle.
6:65–69. 2007.
|
|
8
|
Borden KL and Culjkovic-Kraljacic B:
Ribavirin as an anti-cancer therapy: acute myeloid leukemia and
beyond? Leuk Lymphoma. 51:1805–1815. 2010.
|
|
9
|
Pettersson F, Yau C, Dobocan MC, et al:
Ribavirin treatment effects on breast cancers overexpressing eIF4E,
a biomarker with prognostic specificity for luminal B-type breast
cancer. Clin Cancer Res. 17:2874–2884. 2011.
|
|
10
|
Yoshino A, Ogino A, Yachi K, et al: Gene
expression profiling predicts response to temozolomide in malignant
gliomas. Int J Oncol. 36:1367–1377. 2010.
|
|
11
|
Yoshino A, Tashiro S, Ogino A, et al: Gene
expression profiles predicting the response to IFN-β and a
combination of temozolomide and IFN-β in malignant gliomas. Int J
Oncol. 39:529–542. 2011.
|
|
12
|
Heldin CH and Westermark B:
Platelet-derived growth factor: mechanism of action and possible in
vivo function. Cell Regul. 1:555–566. 1990.
|
|
13
|
Hermanson M, Funa K, Koopmann J, et al:
Association of loss of heterozygosity on chromosome 17p with high
platelet-derived growth factor alpha receptor expression in human
malignant gliomas. Cancer Res. 56:164–171. 1996.
|
|
14
|
Kleihues P, Burger PC and Aldape KD:
Glioblastoma. WHO Classification of Tumours of the Central Nervous
System. Louis DN, Ohgaki H, Wiestler OD and Cavenee WK: 4th
edition. IARC Press; Lyon, France: pp. 33–49. 2007
|
|
15
|
Gu X, Jones L, Lowery-Norberg M and Fowler
M: Expression of eukaryotic initiation factor 4E in astrocytic
tumors. Appl Immunohistochem Mol Morphol. 13:178–183. 2005.
|
|
16
|
Kökény S, Papp J, Weber G, Vaszkó T,
Carmona-Saez P and Oláh E: Ribavirin acts via multiple pathways in
inhibition of leukemic cell proliferation. Anticancer Res.
29:1971–1980. 2009.
|
|
17
|
Vallée S, Fouchier F, Braguer D, Marvaldi
J and Champion S: Ribavirin-induced resistance to heat shock,
inhibition of the Ras-Raf-1 pathway and arrest in G(1). Eur J
Pharmacol. 404:49–62. 2000.
|
|
18
|
Schlosser SF, Schuler M, Berg CP, et al:
Ribavirin and alpha interferon enhance death receptor-mediated
apoptosis and caspase activation in human hepatoma cells.
Antimicrob Agents Chemother. 47:1912–1921. 2003.
|
|
19
|
Stupp R, Mason WP, van den Bent MJ, et al;
European Organisation for Research and Treatment of Cancer Brain
Tumor and Radiotherapy Groups; National Cancer Institute of Canada
Clinical Trials Group. Radiotherapy plus concomitant and adjuvant
temozolomide for glioblastoma. N Engl J Med. 352:987–996. 2005.
|