Introduction
MicroRNAs (miRNAs), a class of small and non-coding
RNAs (length, 19–25 nucleotides), have the potential to regulate
~30% of human genes, which may attribute to the development of
cancer (1–3). miRNAs regulate gene expression by
binding to the 3′-untranslated region (3′-UTR) of the targeted
mRNAs. Numerous studies have indicated that abnormal expression of
miRNA is associated with the development and progression of various
types of human cancer (4–6). Although the biological function of
miRNAs remains unknown, specific miRNAs have functions similar to
tumor suppressors or oncogenes. Previous studies investigating the
expression levels of miRNAs in cancer have indicated their
significance and potential use in the classification of cancer or
as diagnostic indicators (5,7).
Breast cancer (BC) is one of the most common types
of carcinoma and results in a high female mortality rate worldwide
(1). In China, the number of
BC-associated mortality rate has increased markedly in recent years
(3,7). Although early detection of BC is
important in order to reduce the mortality rate of this disease,
the methods available for primary detection, including mammography,
ultrasonography and magnetic resonance imaging, may result in
misdiagnosis or missed diagnosis. The expression pattern of each
miRNA molecule varies in different cancer phenotypes (including BC)
and, thus, can be used in tumor classification, diagnosis, therapy
and prognosis (4). Over the past
decades, the association between miRNAs and human BC has been
extensively investigated (8–10).
Previous studies have demonstrated that the expressional changes of
a number of miRNAs are involved in BC development and progression
(2,5), while serum or plasma miRNA expression
levels were found to be different in BC patients and healthy
controls (3). Therefore, miRNAs
represent a novel approach to BC diagnosis.
The present study aimed to further investigate the
association between serum miRNA expression levels and the clinical
diagnosis of BC. The circulating let-7c levels were determined in
the serum of BC patients and healthy controls by performing
individual-based reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) assays, in accordance with previous
reports (2,3,11). In
addition, the present study investigated whether the serum let-7c
level can be used as a potential biomarker for BC diagnosis.
Materials and methods
Study subjects
The present study was conducted in the Inpatient
Department of Medical Oncology at the Affiliated Hospital of
Binzhou Medical University (Yantai, China) and performed in
accordance with the relevant guidelines of the Medical Ethics
Committee of Binzhou Medical University.
In total, 90 females with BC (age range, 27–79
years), who were pathologically diagnosed with BC for the first
time between June 1st 2010 and July 31st 2013, were included in the
present study. The patients had not been previously treated with
chemotherapy or postmenopausal hormone therapy. In addition, 64
healthy controls were recruited from individuals who visited the
Affiliated Hospital of Binzhou Medical University for physical
examination within the same time period and were not diagnosed with
a tumor or physical illness. Prior to enrollment in the present
study, written informed consent was obtained from each
individual.
Immunohistochemistry
Fresh BC and healthy tissues from patients who
underwent surgery at the Affiliated Hospital of Binzhou Medical
University were obtained at the time of surgery, and immediately
prepared for pathological diagnosis, immunohistochemistry or RNA
isolation. The cancer sample sections containing a minimum of 60%
cancer cells were used in the experiments. All patients provided
their written consent to participate in this study. Subsequently,
the tissues were fixed in 4% paraformaldehyde, embedded in paraffin
and cut into 5-μM sections. Subsequently, the sections were
deparaffinized in xylene, rehydrated and antigen retrieval was
performed by incubation in 10 mM citrate buffer, pH 6.0 at 95–100°C
for 10 min. This was followed by washing in phosphate-buffered
saline (PBS). To quench the endogenous peroxidase activity, the
slides were incubated in 3% hydrogen peroxide for 15 min at room
temperature. Next, the BC slides were incubated in 10% normal goat
serum [Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.,
Beijing, China] for 20 min at room temperature to prevent
non-specific staining. Subsequently, the slides were incubated at
4°C overnight with appropriate dilutions of the following primary
antibodies: Rabbit monoclonal anti-estrogen receptor (ER; 1:200);
and rabbit monoclonal anti-progesterone receptor (PR; 1:200;
Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.). Next, the
samples were incubated with horseradish peroxidase-conjugated goat
anti-rabbit secondary antibodies (1:5000, Beijing Zhongshan Golden
Bridge Biotechnology Co., Ltd.). The negative controls were
incubated in 1× PBS without antibody, following the same procedure.
The samples was visualized using an ABC kit (Beijing Zhongshan
Golden Bridge Biotechnology Co., Ltd.) and positive ER and PR
status was considered when nuclear staining was >10%. The
expression of ER and PR was examined under the Olympus BX51 AX-70
microscope (Olympus, Tokyo, Japan).
Sample collection
Whole blood samples (3 ml) were obtained from each
subject. The serum samples were separated by centrifugation at
2,650 × g for 10 min at room temperature and stored at −80°C prior
to analysis (11).
miRNA isolation from serum or tissue
samples
miRNA was extracted from the serum samples using the
mirVana™ miRNA isolation kit (Ambion Life Technologies, Carlsbad,
CA, USA) according to the manufacturer’s instructions. The BC
tissues were homogenized in a denaturing lysis solution and treated
with TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA,
USA) to extract total RNA, according to the manufacturer’s
instructions. Subsequently, the mirVana™ miRNA isolation kit was
used to obtain the miRNAs from 30–50 mg total RNA samples.
RT-qPCR
Poly(A) tails were added to the extracted miRNAs
using poly(A) polymerase (Ambion Life Technologies) and the
complementary (c)DNA molecules were synthesized using the
5′-AACATGTACAGTCCATGGATGd(T)30N(A, G, C or T)-3′ primer. RT-qPCR of
Let-7c was performed using the following primers: Forward,
5′-GGTTGAGGTAGTAGGTTGTATGGT-3′; and reverse,
5′-AACATGTACAGTCCATGGATG-3′. Each RT-qPCR reaction mixture
contained 0.5 μl cDNA, 7.5 μl sterile water, 1 μl forward primer, 1
μl reverse primer and 10 μl of SYBR® Premix Ex Taq™
(Takara Biotechnology Co., Ltd., Dalian, China) and was performed
using the Rotor-Gene 3000 system (Corbett Life Science, Mortlake,
Australia) as follows: Initial denaturation at 95°C for 5 min,
followed by 40 cycles of denaturation at 95°C for 20 sec, annealing
at 60°C for 20 sec and extension at 72°C for 30 sec. As described
in previous studies, 5S ribosomal RNA was used as the reference
control (12,13). All the experiments were performed in
triplicate.
Statistical analysis
The experimental data were initially analyzed for
normal distribution and variance homogeneity using the Shapiro-Wilk
test and F-test, respectively. Normal distribution data are
represented as the mean ± standard deviation, and all the other
data are represented as median and quartiles. Differences in the
age, height or weight between the BC patients and healthy controls
were analyzed using the Student’s t-test. However, when the serum
expression levels of let-7c did not demonstrate normal
distribution, nonparametric tests were applied to analyze these
differences. In addition, continuous variables between the BC
patients and controls were analyzed by performing the Wilcoxon
rank-sum test. Statistical analyses were performed using
R© software (version 2.15.0; http://www.r-project.org). P<0.05 was considered to
indicate a statistically significant difference. Furthermore,
receiver operating characteristic (ROC) curves were generated using
the SPSS software (version 18.0; SPSS, Inc., Chicago, IL, USA) to
assess the diagnostic accuracy of each parameter. The area under
the ROC curve (AUC) was used to identify the optimal sensitivity
and specificity levels at which cancer patients can be
distinguished from healthy individuals.
Results
Clinical characteristics
The demographic and clinical characteristics of the
90 BC patients and 64 healthy controls that participated in the
present study are listed in Table
I. No statistically significant differences in the age, height
or weight were identified between the BC patients and healthy
controls. In addition, no clear statistically significant
differences in the menopausal status were identified between the BC
patients and healthy controls.
 | Table IDemographic and clinical
characteristics of the study samples. |
Table I
Demographic and clinical
characteristics of the study samples.
| Parameter | Healthy controls
(n=64) | Patients (n=90) | P-valuea |
|---|
| Age, yearsb | 43.781±15.831 | 47.900±9.882 | 0.068 |
| Weight, kgb | 62.638±8.844 | 65.408±8.238 | 0.090 |
| Height, cmb | 160.500±6.414 | 158.551±4.619 | 0.075 |
| Estrogen
receptor-positive/negative, n | - | 64/26 | - |
| Progesterone
receptor-positive/negative, n | - | 60/30 | - |
|
Premenopausal/postmenopausal, n | 35/29 | 48/42 | 0.998 |
| Median let-7c,
×103 copies/ml | 2.300 | 0.035 | <0.01 |
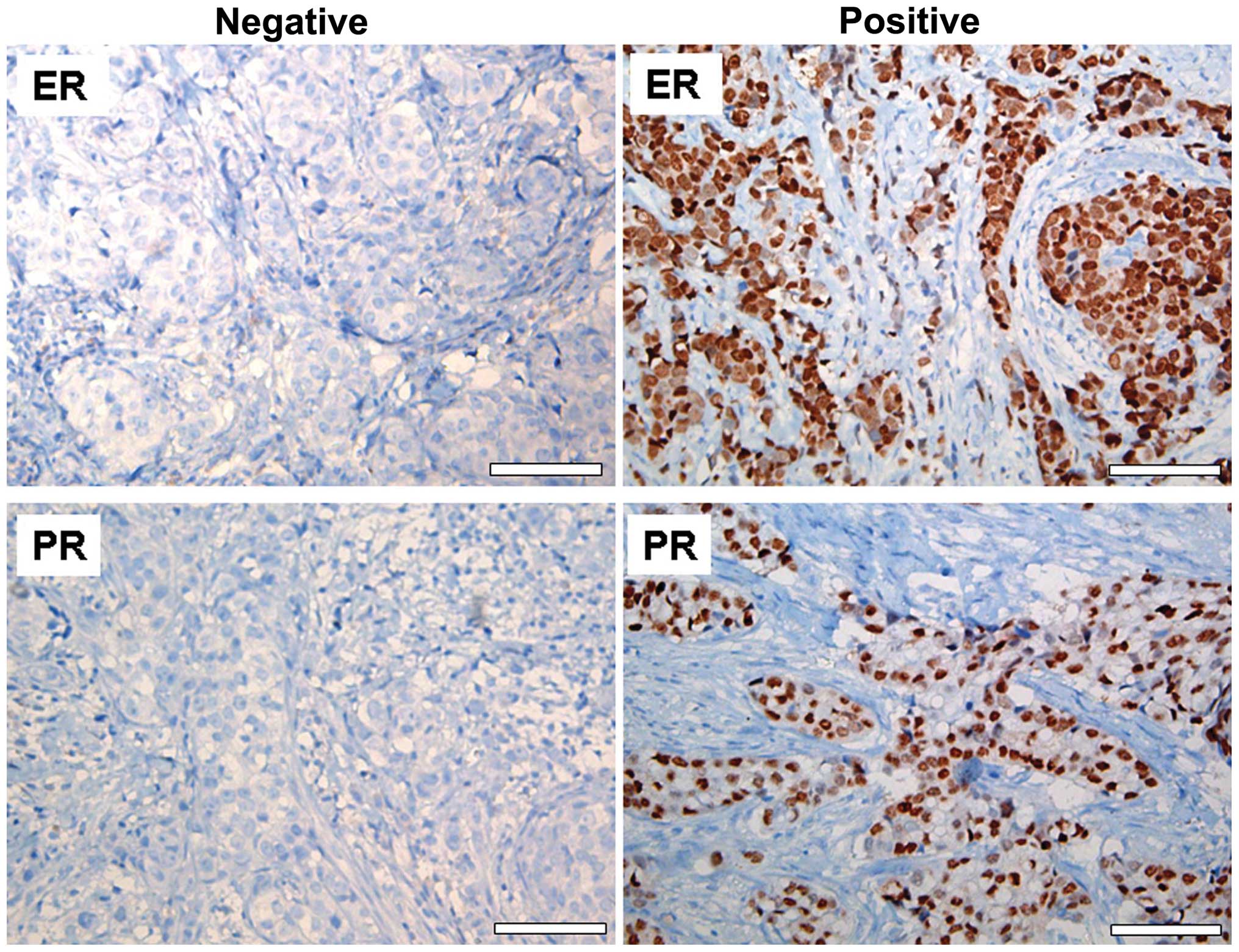
The presence of ER and PR in the BC tissue samples
was detected by immunohistochemical analysis. Nuclear staining of
>10% was considered to indicate positivity ER and PR status
(Fig. 1). Of the 90 BC patients, 64
patients were ER-positive (71.1%) and 60 were PR-positive
(66.6%).
Reduced let-7c expression levels in BC
tissues
Considerable evidence accumulated from a number of
previous studies has indicated that downregulation of let-7 family
miRNAs may be associated with a poor clinical outcome in BC
patients (14,15). Therefore, to investigate the role of
let-7c in BC, the present study analyzed the expression levels of
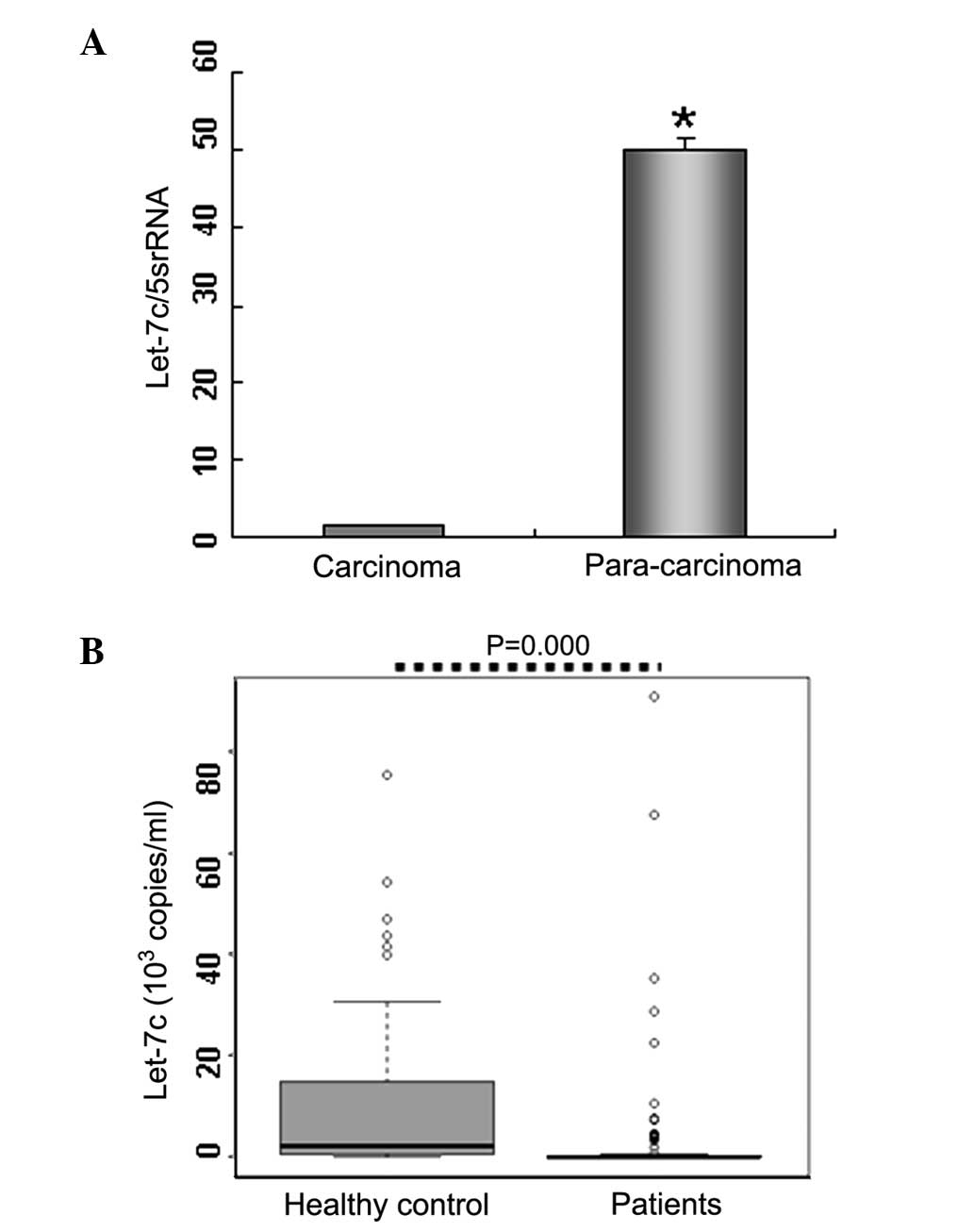
let-7c in BC tissues. The results indicated that let-7c expression
was markedly decreased (>20-fold lower) in BC tissues (n=4)
compared with paracarcinoma tissues (n=4; Fig. 2A), supporting the suppressive role
of let-7c in tumor proliferation.
Reduced serum let-7c expression levels in
BC patients
The serum expression levels of let-7c were detected
by performing qPCR analysis to investigate the potential role of
let-7c in the diagnosis of BC. The results of the present study
demonstrated that serum let-7c levels in BC patients
(0.035×103 copies/ml; n=90) were significantly lower
compared with the healthy controls (2.300×103 copies/ml;
n=64; P<0.01; Table I; Fig. 2B). These results were consistent
with the let-7c expression levels identified in the BC tissues
samples, indicating that let-7c may be an important factor in BC
diagnosis.
Correlation between ER/PR status and
circulating let-7c expression levels
ER and PR have been previously reported as important
factors associated with the etiology and therapeutic treatment
strategy of BC (16,17). Therefore, to investigate the
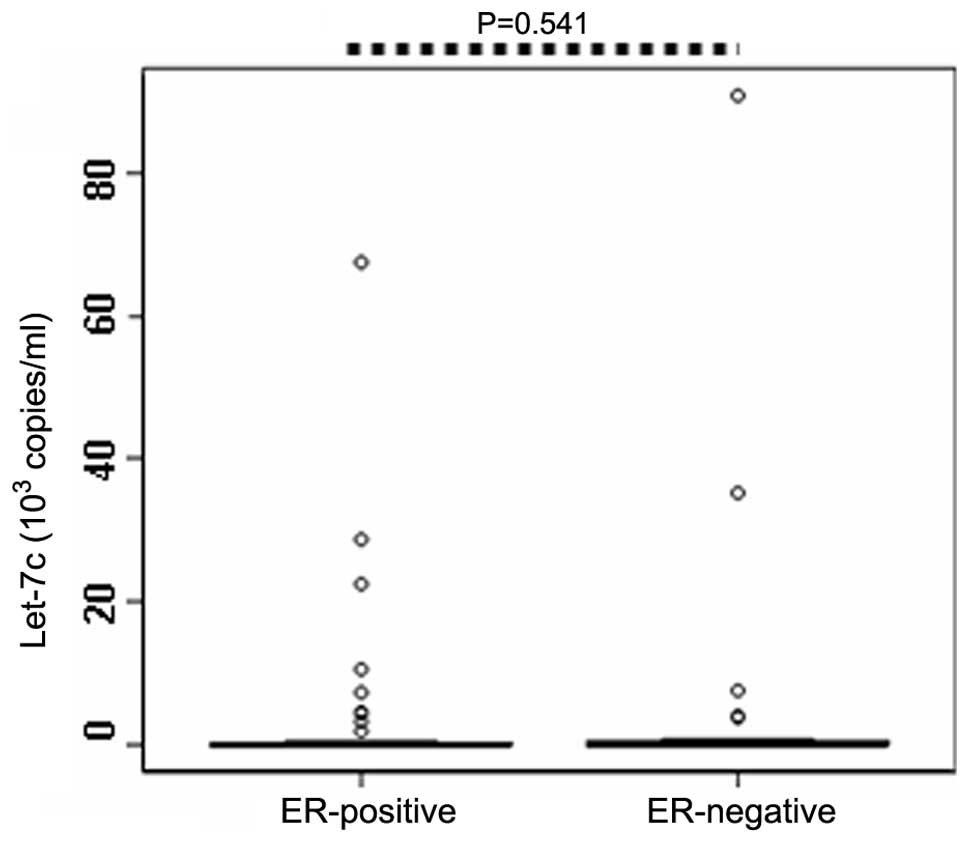
correlation between ER or PR status and the serum expression levels
of let-7c, let-7c expression was compared between ER- (or PR-)
positive and negative patients. The results demonstrated that serum
expression levels of let-7c in the ER-positive patients (n=64;
0.033×103 copies/ml) were not significantly different
compared with the ER-negative patients (n=26; 0.036×103
copies/ml; P=0.541; Table II;
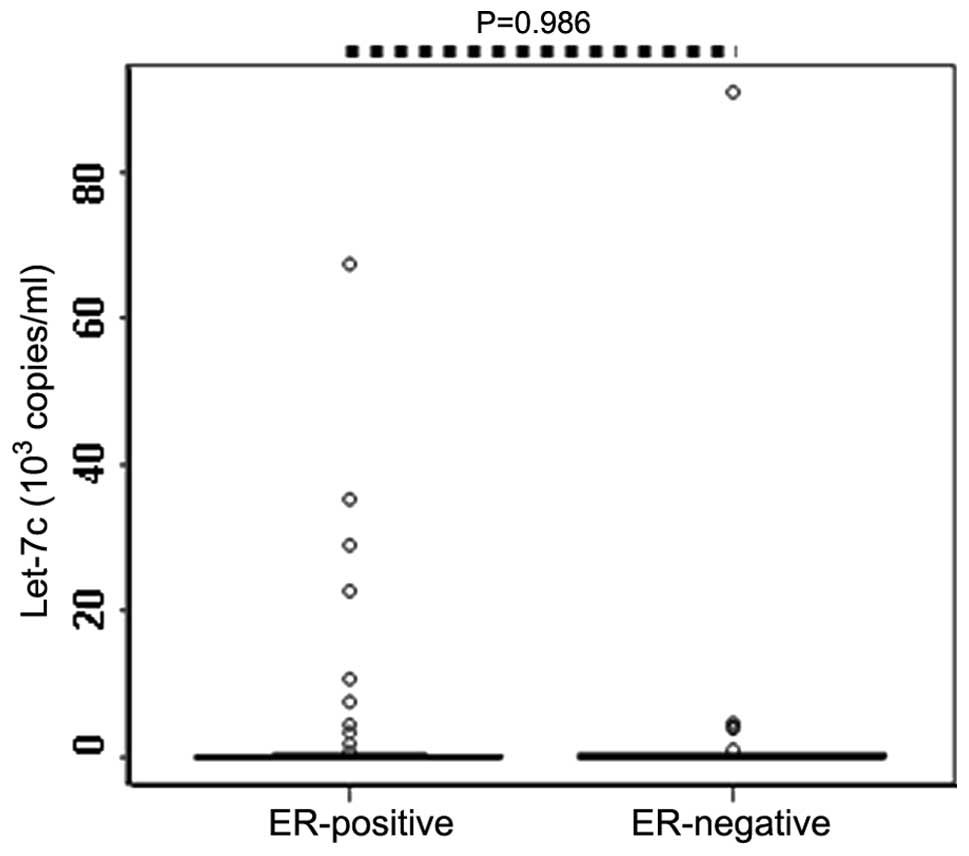
Fig. 3). Similarly, no
statistically significant difference was identified between the
serum levels of let-7c in the PR-positive (n=60) and PR-negative
patients (n=30; P=0.986; Table II;
Fig. 4).
 | Table IIAssociation of ER, PR and menopausal
status with let-7c. |
Table II
Association of ER, PR and menopausal
status with let-7c.
| Parameter | Median let-7c,
×103 copies/ml | P-valuea |
|---|
| ER | | 0.541 |
| Positive
(n=64) | 0.033 | |
| Negative
(n=26) | 0.036 | |
| PR | | 0.986 |
| Positive
(n=60) | 0.035 | |
| Negative
(n=30) | 0.033 | |
| Menopausal
status | | 0.040a |
| Premenopausal
(n=48) | 0.036 | |
| Postmenopausal
(n=42) | 0.032 | |
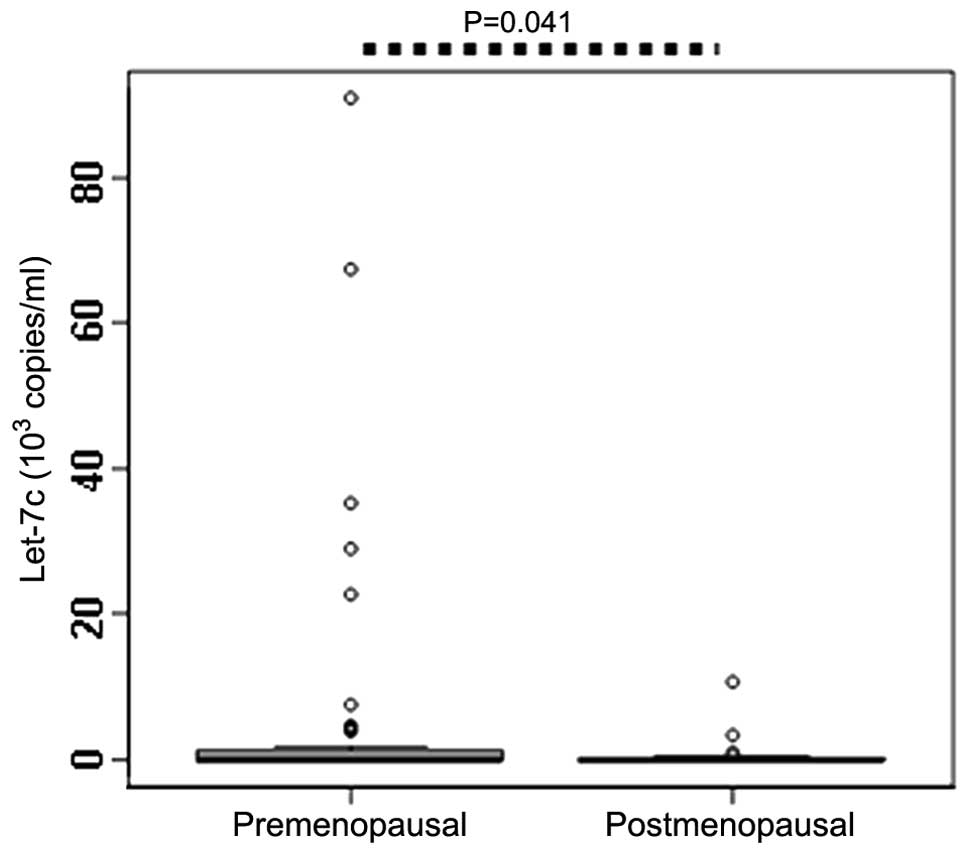
Correlation between menopausal status and
circulating let-7c expression levels
Since postmenopausal females with a high breast
density exhibit increased risk of developing BC (18), the serum let-7c expression levels
were compared between premenopausal and postmenopausal patients.
The results identified that the expression levels of let-7c in the
premenopausal patients (0.036×103 copies/ml) was
significantly higher compared with the postmenopausal patients
(0.032×103 copies/ml; P=0.040; Table II; Fig.
5), which indicates that the postmenopausal status may affect
the expression level of serum let-7c.
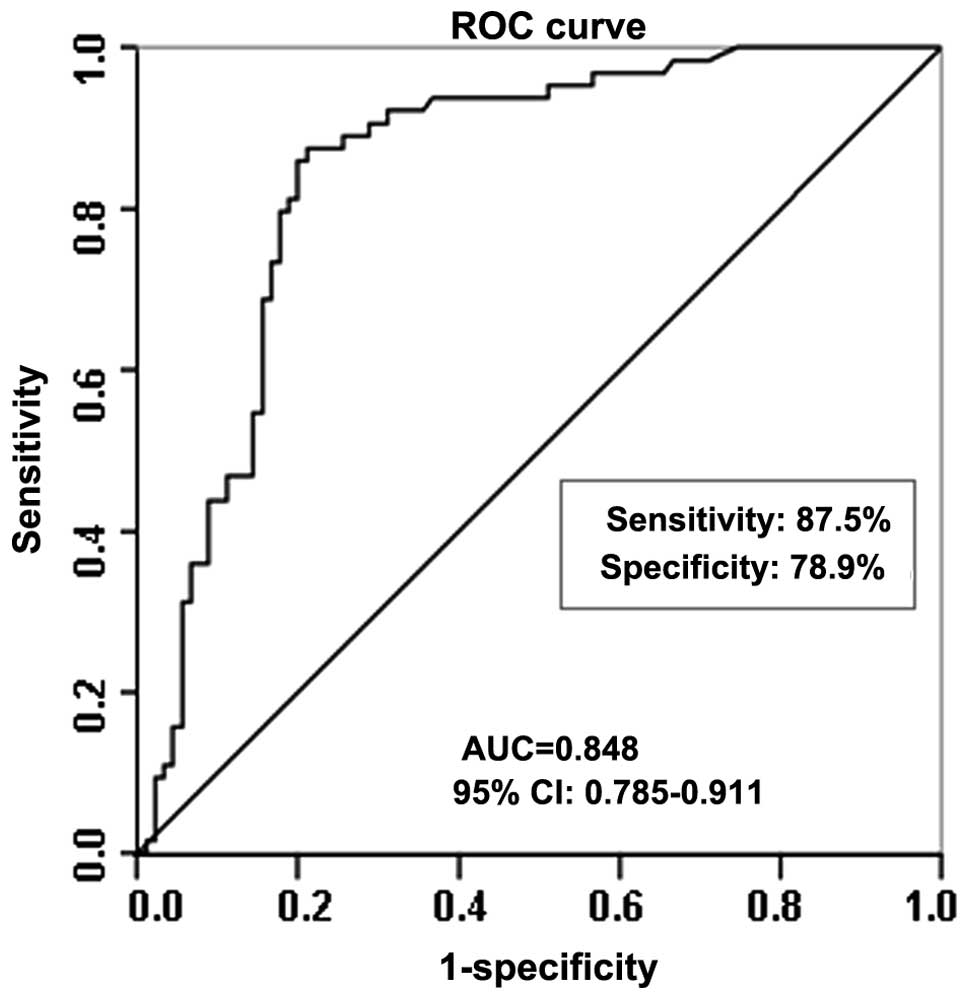
Diagnostic potential of let-7c expression
levels in BC
Statistical ROC analysis was used to investigate the
diagnostic potential of let-7c serum expression levels in BC
patients. The expression data for let-7c were plotted using the ROC
curves to identify a cut-off value that could distinguish breast
cancer patients from healthy controls. Using a cutoff level of
0.374×103 copies/ml, the serum expression levels of
let-7c presented sensitivity of 87.5% and specificity of 78.9% in
distinguishing the BC patients from the healthy controls, with an
AUC of 0.848 (95% confidence interval, 0.785–0.911; P<0.001;
Fig. 6).
Discussion
let-7 miRNAs are members of a highly conserved miRNA
family consisting of 12 genes (including let-7-a1, -a2, -a3, -b,
-c, -d, -e, -f1, -f2, -g, -i and miR-98), which are located on
eight different chromosomes (19).
Let-7 was initially described in Caenorhabditis elegans and
functionally conserved from lower invertebrates to higher mammals
(20). In the present study, a
comprehensive investigation of serum let-7c miRNA expression was
conducted in BC patients and healthy controls using RT-qPCR
(21). The expression levels of
let-7c were significantly decreased in the serum of the BC patients
compared with the healthy controls, which indicates that serum
let-7c may have a considerable diagnostic function in
differentiating BC patients from healthy controls.
The let-7 family members function as tumor
suppressors and have been associated with various target genes,
including Ras (20), high mobility
group AT-hook 2 (22,23) and B-cell lymphoma-extra large
(Bcl-xL) (24). let-7 expression is
downregulated in a number of malignancies. For instance, let-7 was
identified to be decreased in human hepatoma cells and tissues,
which are associated with enhanced expression of Bcl-xL (24). A previous study revealed that let-7c
was a downregulated epithelial miRNA and its functions were
delineated in unique cells derived from columnar cell hyperplasia
(25). Similarly, the present study
identified that let-7c expression was downregulated in BC tissues
compared with paracarcinoma tissues. Thus, the present and
aforementioned studies indicated the suppressive role of let-7
miRNAs in tumorigenesis.
A greater number of studies focusing on the function
of miRNAs have been conducted, particularly those investigating the
roles of circulating miRNAs in disease diagnosis (20). Circulating miRNAs isolated from the
serum or plasma are more stable compared with those isolated from
the blood (10). In addition,
circulating miRNAs are stable at room temperature and can survive
under the effects of RNase and DNase (1,11).
Thus, the expression patterns of circulating miRNAs may be used as
potential diagnostic indicators for a number of diseases, including
tumors, improving cancer diagnosis. To further investigate the role
of let-7c expression levels in the diagnosis of BC, the circulating
let-7c levels were compared between BC patients and healthy
controls. It was identified that let-7c expression was lower in the
serum of BC patients compared with the healthy controls.
Furthermore, at the optimal cut-off, the serum level of let-7c
exhibited sensitivity of 87.5% and specificity of 78.9% for
distinguishing BC patients from healthy controls.
The important role of pathological analysis in the
diagnosis of BC is biomarker testing, specifically the accurate
assessment of the ER and PR status of BC tissues (26,27).
Previously, significant associations have been reported between
tumor size (or the presence of distant metastases) in BC and
ER/PR-positive rate (or menopausal status) (28). Therefore, the association between
the expression level of let-7c and the ER/PR-positive rate, as well
as the menopausal status of the patients, was investigated in the
present study. The results indicated that 71.1% of primary BC
patients expressed ER, while 66.6% expressed PR. Subsequent
investigation into the association between ER (or PR)-positive
expression and serum expression levels of let-7c revealed that ER
(or PR)-positive expression did not affect the serum expression
levels of let-7c. However, let-7c expression in the premenopausal
patients was significantly higher compared with the postmenopausal
patients, indicating that the postmenopausal status may affect
let-7c expression levels. Moreover, Kerlikowske et al
reported that postmenopausal females with a high breast density
presented an increased risk of developing BC (18).
In conclusion, the present study identified that
let-7c expression was lower in BC tissues compared with
paracancerous tissues. Furthermore, the let-7c serum levels of the
BC patients were significantly lower compared with the levels of
the healthy controls and were affected by the menopausal status of
the patients. Although the results of the current study indicate
that serum let-7c expression levels may represent a novel
diagnostic biomarker for BC patients, well-designed studies with
larger sample sizes are required to further confirm the role of
let-7c in cancer diagnosis.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation (nos. 31371321, 31440061 and 81200601),
NCET-10-0919, the Taishan Scholars program of Shandong Province,
the Shandong Science and Technology Committee (no. ZR2012HQ035) and
the Foundation of Shandong Educational Committee (no. J11LC01).
Abbreviations:
|
miRNA
|
microRNA
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
3′-UTR
|
3′-untranslated region
|
|
BC
|
breast cancer
|
|
CI
|
confidence interval
|
|
ER
|
estrogen receptor
|
|
PR
|
progesterone receptor
|
|
ROC
|
receiver operating characteristic
|
|
AUC
|
area under the ROC curve
|
References
|
1
|
Asaga S, Kuo C, Nguyen T, et al: Direct
serum assay for microRNA-21 concentrations in early and advanced
breast cancer. Clin Chem. 57:84–91. 2011. View Article : Google Scholar
|
|
2
|
Heneghan HM, Miller N, Lowery AJ, et al:
MicroRNAs as novel biomarkers for breast cancer. J Oncol.
2009:9502012009.PubMed/NCBI
|
|
3
|
Roth C, Rack B, Müller V, et al:
Circulating microRNAs as blood-based markers for patients with
primary and metastatic breast cancer. Breast Cancer Res.
12:R902010. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis, function and role in cancer. Curr Genomics. 11:537–561.
2010. View Article : Google Scholar
|
|
5
|
Zhao B, Han H, Chen J, et al: MicroRNA
let-7c inhibits migration and invasion of human non-small cell lung
cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 342:43–51. 2014.
View Article : Google Scholar
|
|
6
|
Sempere LF, Freemantle S, Pitha-Rowe I, et
al: Expression profiling of mammalian microRNAs uncovers a subset
of brain-expressed microRNAs with possible roles in murine and
human neuronal differentiation. Genome Biol. 5:R132004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pelosi A, Careccia S, Lulli V, et al:
miRNA let-7c promotes granulocytic differentiation in acute myeloid
leukemia. Oncogene. 32:3648–3654. 2013. View Article : Google Scholar
|
|
8
|
Qian B, Katsaros D, Lu L, et al: High
miR-21 expression in breast cancer associated with poor
disease-free survival in early stage disease and high TGF-beta1.
Breast Cancer Res Treat. 117:131–140. 2009. View Article : Google Scholar
|
|
9
|
Wang PY, Sun YX, Zhang S, et al: Let-7c
inhibits A549 cell proliferation through oncogenic TRIB2 related
factors. FEBS Lett. 587:2675–2681. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu Q, Wang C, Lu Z, Guo L and Ge Q:
Analysis of serum genome-wide microRNAs for breast cancer
detection. Clin Chim Acta. 413:1058–1065. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen X, Ba Y, Ma L, et al:
Characterization of microRNAs in serum: a novel class of biomarkers
for diagnosis of cancer and other diseases. Cell Res. 18:997–1006.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang HH, Pang M, Dong W, et al: miR-511
induces the apoptosis of radioresistant lung adenocarcinoma cells
by triggering BAX. Oncol Rep. 31:1473–1479. 2014.PubMed/NCBI
|
|
13
|
Wang PY, Sun YX, Zhang S, et al: Let-7c
inhibits A549 cell proliferation through oncogenic TRIB2 related
factors. FEBS Lett. 587:2675–2681. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu X, Guo J, Zheng L, et al: The
heterochronic microRNA let-7 inhibits cell motility by regulating
the genes in the actin cytoskeleton pathway in breast cancer. Mol
Cancer Res. 11:240–250. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Piskounova E, Polytarchou C, Thornton JE,
et al: Lin28A and Lin28B inhibit let-7 microRNA biogenesis by
distinct mechanisms. Cell. 147:1066–1079. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cui X, Schiff R, Arpino G, Osborne CK and
Lee AV: Biology of progesterone receptor loss in breast cancer and
its implications for endocrine therapy. J Clin Oncol. 23:7721–7735.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Diaz NM: Laboratory testing for HER2/neu
in breast carcinoma: an evolving strategy to predict response to
targeted therapy. Cancer Control. 8:415–418. 2001.PubMed/NCBI
|
|
18
|
Kerlikowske K, Cook AJ, Buist DS, et al:
Breast cancer risk by breast density, menopause and postmenopausal
hormone therapy use. J Clin Oncol. 28:3830–3837. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qin B, Xiao B, Liang D, et al: MicroRNA
let-7c inhibits Bcl-xl expression and regulates ox-LDL-induced
endothelial apoptosis. BMB Rep. 45:464–469. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Johnson SM, Grosshans H, Shingara J, et
al: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jones CI, Zabolotskaya MV, King AJ, et al:
Identification of circulating microRNAs as diagnostic biomarkers
for use in multiple myeloma. Br J Cancer. 107:1987–1996. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sakurai M, Miki Y, Masuda M, et al: LIN28:
a regulator of tumor-suppressing activity of let-7 microRNA in
human breast cancer. J Steroid Biochem Mol Biol. 131:101–106. 2012.
View Article : Google Scholar
|
|
23
|
Zhu XM, Wu LJ, Xu J, Yang R and Wu FS:
Let-7c microRNA expression and clinical significance in
hepatocellular carcinoma. J Int Med Res. 39:2323–2329. 2011.
View Article : Google Scholar
|
|
24
|
Shimizu S, Takehara T, Hikita H, et al:
The let-7 family of microRNAs inhibits Bcl-xL expression and
potentiates sorafenib-induced apoptosis in human hepatocellular
carcinoma. J Hepatol. 52:698–704. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Björner S, Fitzpatrick PA, Li Y, et al:
Epithelial and stromal microRNA signatures of columnar cell
hyperplasia linking Let-7c to precancerous and cancerous breast
cancer cell proliferation. PLoS One. 9:e1050992014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Barnes DM and Hanby AM: Oestrogen and
progesterone receptors in breast cancer: past, present and future.
Histopathology. 38:271–274. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sofi GN, Sofi JN, Nadeem R, et al:
Estrogen receptor and progesterone receptor status in breast cancer
in relation to age, histological grade, size of lesion and lymph
node involvement. Asian Pac J Cancer Prev. 13:5047–5052. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Faheem M, Mahmood H, Khurram M, Qasim U
and Irfan J: Estrogen receptor, progesterone receptor and Her 2 Neu
positivity and its association with tumour characteristics and
menopausal status in a breast cancer cohort from northern Pakistan.
Ecancermedicalscience. 6:2832012.
|