Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignant neoplasms worldwide, with ~50% of new cases
estimated to occur in China (1).
Hepatitis B virus (HBV) or hepatitis C virus infection, ingestion
of food contaminated with aflatoxin B1 and alcohol consumption are
considered major risk factors for HCC development (1). HBV-related HCC development progresses
through three main, well-characterized stages that may occur over
several decades: HBV-related chronic hepatitis, liver cirrhosis and
HCC. Furthermore, over two-thirds of HCCs originate from
HBV-related liver cirrhosis (1,2).
Chromosomal abnormalities in HCC have been
well-documented, and comparative genomic hybridization (CGH) has
identified a consistent pattern of chromosomal losses and gains
associated with the development and progression of HCC (3,4).
Furthermore, a number of studies have demonstrated characteristic
chromosomal abnormalities in HCC by CGH and fluorescence in
situ hybridization (FISH) (3–5). The most
significant changes are partial or entire gains of chromosome arms
1q, 8q and 2q, and losses of 1p and 1q, 4q, 8p, 13q, 16q and 17p.
In one meta-analysis, using conventional CGH analysis with low
resolution (~2 Mb) from several studies, it was revealed that the
most prominent changes were gains of 1q (57.1%), 8q (46.6%), 6p
(23.3%) and 17q (22.2%), and losses of 8p (38%), 16q (35.9%), 4q
(34.3%), 17p (32.1%) and 13q (26.2%), and that losses of 4q, 13q
and 16q were associated with HBV infection. Using array CGH
analysis from four studies, it was revealed that loci with genomic
gains with a prevalence of >25% included 1q, 6p, 8q, 17q, 20p,
5p15.33 and 9q34.2–34.3, and loci with genomic loss with prevalence
of >25% comprised 4q, 6q, 8p, 9p, 13q, 14q, 16q and 17p, and
were associated with 31 classical molecular pathways, particularly
the antivirus immunological pathway (6).
Since the duration of HBV-related chronic hepatitis
and liver cirrhosis may be several decades (7), chromosomal abnormalities may occur and
accumulate in the cirrhotic liver during the progression from
regenerated nodules (RNs) to preneoplastic lesions, including
dysplastic nodules (DNs), and particularly high-grade DNs (HGDNs)
(8–10). However, less is known with regard to
genomic imbalances associated with the progression from cirrhosis
to HCC, and the association between DNs and HCC has remained to be
elucidated.
To investigate whether there is a genetic link
between DNs and HCC, genomic imbalances in HGDNs were screened and
analyzed by array-based CGH analysis, and the prevalence of the
most frequently identified genomic imbalances in HCC was
evaluated.
Material and methods
Tumor samples
For array CGH analysis, HGDNs were collected from
two male patients with HBV-related liver cirrhosis who were aged 56
and 62 years. In addition, 83 HCC cases were enrolled in the
present study, in order to determine the prevalence of the
identified loci in HCCs. The patients were diagnosed as follows: i)
With or without HBV infection, determined by positivity or
negativity for hepatitis B surface antigen; ii) tumor stage 1 or
>1 [corresponding to TNM stage I, T1N0M0, as classified by the
Union for International Cancer Control (11)]; iii) well, moderately or poorly
differentiated tumors, classified according to the World Health
Organization Classification of Tumors of the Digestive System
(12) (Table I).
 | Table I.Clinicopathological characteristics
of hepatocellular carcinoma patients. |
Table I.
Clinicopathological characteristics
of hepatocellular carcinoma patients.
|
| Informative
cases |
|---|
|
|---|
|
Characteristics | 5q13.2 | 8p23.1
(D8S503) | 8p23.1
(D8S1130) |
|---|
| Cases, n | 83 | 38 | 31 |
| Age, years |
|
|
|
|
Mean | 52.1 | 52.8 | 54.1 |
|
Range | 30–80 | 29–78 | 29–78 |
| Gender, n (%) |
|
|
|
|
Male | 72 (86.7) | 35 (92.1) | 27 (87.1) |
|
Female | 11 (13.3) | 3 (7.9) | 4 (12.9) |
| Tumor stage, n
(%) |
|
|
|
| 1 | 47 (56.6) | 23 (60.5) | 16 (51.6) |
|
>1 | 36 (43.4) | 15 (39.5) | 15 (48.4) |
| HBV infection, n
(%)a |
|
|
|
|
Positive | 70 (84.3) | 32 (84.2) | 26 (83.9) |
|
Negative | 9 (10.8) | 4 (10.5) | 3 (9.7) |
| Differentiation, n
(%) |
|
|
|
|
Well | 13 (15.7) | 7 (18.4) | 4 (12.9) |
|
Moderate | 43 (51.8) | 19 (50.0) | 16 (51.6) |
|
Poor | 27 (32.5) | 12 (31.6) | 11 (35.5) |
Samples were collected at the Department of
Pathology, Beijing Friendship Hospital, Capital Medical University
(Beijing, China) and the Minimally Invasive Hepatobiliary Cancer
Center, Beijing You-An Hospital, Capital Medical University
(Beijing, China), from January 2005 to April 2013. HGDNs and HCCs
were examined by three pathologists and diagnosed according to the
morphological criteria of RNs, DNs and HCC. Following surgical
resection of HCCs and HGDNs, the samples were fixed in 4%
formaldehyde and embedded in paraffin. Hematoxylin and eosin
(H&E) staining was applied to serial sections. Formaldehyde,
paraffin and H&E were purchased from Sangon Biotech (Shanghai,
China).
Genomic DNA from HCCs was extracted from paraffin
sections, as described previously, for conventional polymerase
chain reaction (PCR) analysis (13).
Extraction reagents were obtained from Sangon Biotech. In addition,
50 samples of genomic DNA were obtained from blood samples from
healthy individuals at the Liver Research Center, Beijing
Friendship Hospital, and used as normal controls.
Written informed consent was obtained from all
participants for the use of their clinical materials in research.
The study protocol was approved by the Medical Ethics Commission of
the Beijing Friendship Hospital, Capital Medical University.
Laser microdissection and DNA
extraction for array CGH analysis
Serial tissue sections of 10-µm thickness from each
cirrhotic liver sample were placed on an ultraviolet
light-absorbing membrane. HGDNs were obtained by laser
microdissection using an LMD6000 (Leica Microsystems Ltd., Wetzlar,
Germany). Following H&E staining, the slides were mounted on
the LMD6000 Microstat, and the selected nodules were dissected. The
dissectate (with the attached specimen) fell by gravity into the
caps of selected 0.5-ml microcentrifuge tubes filled with 40 µl
lysate buffer and 10 µl proteinase K (E.Z.N.A® FFPE DNA Kit, Omega
Bio-Tek, Inc., Norcross, GA, USA). The tubes were subsequently
incubated at 55°C in a water bath to allow digestion of the tissue
specimens. Following overnight digestion, genomic DNA was purified
according to the manufacturer's instructions and examined using a
BioSpec-nano Spectrophotometer (Shimadzu Biotech, Tokyo, Japan) at
a wavelength of 220–800 nm.
Array CGH
The genomic profile changes of paired DNA samples
were compared using a Roche NimbleGen 720K CGH microarray (Roche
NimbleGen Inc., Madison, WI, USA). Sample and gender-matched
reference DNA were labeled with Cy5-dUTP and Cy3-dUTP, respectively
using a NimbleGen Dual-Color DNA Labeling Kit (Roche Nimblegen
Inc., Madison, WI, USA). A genomic DNA purification module was used
to purify the labeled samples by precipitation with isopropanol
(Sigma-Aldrich) followed by washing with 80% ice-cold ethanol
(Sigma-Aldrich); these samples were then denatured at 95°C and
applied to microarrays. Following incubation at 65°C for 40 h, the
microarrays were washed prior to scanning with a GenePix 4200A
Scanner using GenePix 6.0 software (Molecular Devices, LLC,
Sunnyvale, Ca, USA). NimbleScan v2.4 software (Roche NimbleGen) was
used to normalize the raw data, which was subsequently processed
using Nexus software (Nexus Copy Number™, Biodiscovery, Hawthorne,
CA, USA) with default settings. The normalized data were also
processed with NimbleScan software using the segMNT algorithm at
the default settings (with minimum segment difference set to
0.1).
Analysis of loss of heterozygosity
(LOH) at 5q13.2
To screen for LOH at 5q13.2, differential PCR was
conducted for general transcription factor IIH subunit 2
(GTF2H2), a gene located at 5q13.2, with the GAPDH
sequence as a reference, as described previously (13,14). The
primer sequences were as follows: Forward, AGC AAA GCA CAC CTT
GAATG and reverse, ATG AAT ACA GCC AGG GCA AC, covering the 5′ end
of GTF2H2 gene (GTF2H2-1); forward, TAA CCT AGG CGT
GGG ATA GG and reverse, AAG TGA AAG CCA GCC ACA AC, covering the 3′
end of GTF2H2 gene (GTF2H2-2); and forward, AAC GTG TCA GTG
GTG GAC CTG and reverse, AGT GGG TGT CGC TGT TGA AGT for the
GAPDH sequence (Taijixingke Biotech, Beijing, China). PCR
was performed in a total volume of 20 µl, consisting of 2 µl DNA
solution (concentration, ~100 ng/µl), 10 µl GoTaq® PCR Mix (Applied
Biosystems Life Technologies, Beijing, China), and 0.8 µl primer
sets (1.25 µmol/l of each primer), in a Veriti™ 96-Well Thermal
Cycler (Applied Biosystems, Foster, CA, USA) with an initial
denaturing step at 95°C for 5 min followed by 30 cycles of
denaturation at 95°C for 50 sec, annealing at 61°C or 55°C for 60
sec, extension at 72°C for 60 sec, and a final extension at 72°C
for 3 min. Electrophoresis was performed on the PCR products on 8%
acrylamide gels (Sangon Biotech), and the gels were photographed
with a DC120 Zoom Digital Camera (Kodak, Rochester, NY, USA).
Gel-Pro analyzer 4.0 Image Analysis software (Media Cybernetics,
Rockville, MD, USA) was used to estimate the density of each PCR
fragment.
Quantitative PCR assays were performed for further
confirmation of cases with LOH at 5q13.2. PCR was performed in a
total volume of 20 µl, consisting of 1 µl DNA solution
(concentration, ~100 ng/µl), 10 µl SYBR® Green PCR Master Mix
(Applied Biosystems) and 2 µl primer sets (1.25 µmol/l of each
primer) in a 7500 Fast PCR cycler (Applied Biosystems) with an
initial denaturation step at 95°C for 10 min followed by 40 cycles
of denaturation at 95°C for 50 sec, then annealing at 60°C for 60
sec.
Ratios of relative signal intensity of tumor tissue
with that of adjacent non-tumor tissue of <0.67 were considered
as LOH, as described previously (14,15). Five
normal tissue samples obtained from Beijing You-An Hospital were
used as a reference when adjacent non-tumor tissue was unavailable.
Samples demonstrating LOH for GTF2H2-1 and GTF2H2-2
markers were considered to have LOH at 5q13.2.
Analysis of LOH at 8p23.1
DNA from matched tumor and corresponding non-tumor
liver tissue was analyzed for LOH at 8p23.1 by amplification of two
published microsatellite markers, D8S503 (forward, GGT TAC GAG TTT
TGT CCT TTG and reverse, GAA ACA AAC CAA TGT AGG AGTG) and D8S1130
(forward, GAA GAT TTG GCT CTG TTGGA and reverse, TGT CTT ACT GCT
ATA GCTT), which were selected from the Genome Database (http://www.gdb.org), as described previously (16,17). PCR
was performed in a total volume of 25 µl, consisting of 2 µl DNA
solution (concentration, ~100 ng/µl), 5 µl 5X Colorless GoTaq®
Flexi Buffer (Applied Biosystems), 2 µl 25 mM MgCl2, 0.5
µl 200 µM dNTP, 0.15 µl 5 U/µl GoTaq® Hot Start Polymerase, and 0.8
µl primer sets (1.25 µmol/l of each primer), in a Bio-Rad thermal
cycler (Bio-Rad Laboratories, Inc.) using the following program:
Initial denaturing step at 95°C for 5 min, followed by 30 cycles of
denaturation at 95°C for 50 sec, annealing at 59°C or 53°C for 60
sec, extension at 72°C for 60 sec and a final extension at 72°C for
3 min. The PCR products were subjected to electrophoresis on 8%
acrylamide gels, and images of the gels were captured with a DC120
Zoom Digital Camera (Kodak). The density of each PCR fragment was
estimated using Gel-Pro analyzer 4.0 Image Analysis software (Media
Cybernetics Ltd., Rockville, MD, USA).
Informative visual inspection was used to determine
whether the samples demonstrated LOH or no loss, or were
uninformative. LOH was defined as a loss of intensity of ≥50% in
one or more alleles in the tumor, compared with the identical
allele in the adjacent non-tumor tissue, as described previously
(16,17).
Statistical analysis
The χ2 and Fisher's exact tests were used
to determine associations between genomic imbalances and clinical
parameters using 23 SAS v9.2 software (SAS Institute Inc., Cary,
NC, USA). P<0.05 was considered to indicate a statistically
significant differences for all tests.
Results
Loss of 5q13.2 and 8p23.1 occur in
HGDNs of HBV-related liver cirrhosis
From each case of HBV-related liver cirrhosis, three
HGDNs were separated by laser microdissection and a total of six
HGDNs were subjected to analysis of genomic profile changes.
Array CGH analysis revealed that genomic imbalances
were randomly located in 14 chromosomal regions (Chr 1, 3, 4, 5, 7,
8, 9, 11, 14, 15, 16, 17, 19 and 22) in the HGDNs. These genomic
imbalances were primarily identified as genomic losses of small
segments, and were frequently observed at 1q21.1–21.2, 1q32.1,
4p16.1, 5q13.2, 6p22.1, 7q11.23, 8p23.1, 17q21.31–21.32 and
22q11.21, whereas genomic gains were commonly observed at 1q32.1,
11q13.4, 14q24.3 and 17p11.2 (Table
II). Of these identified genomic imbalances, losses at 5q13.2
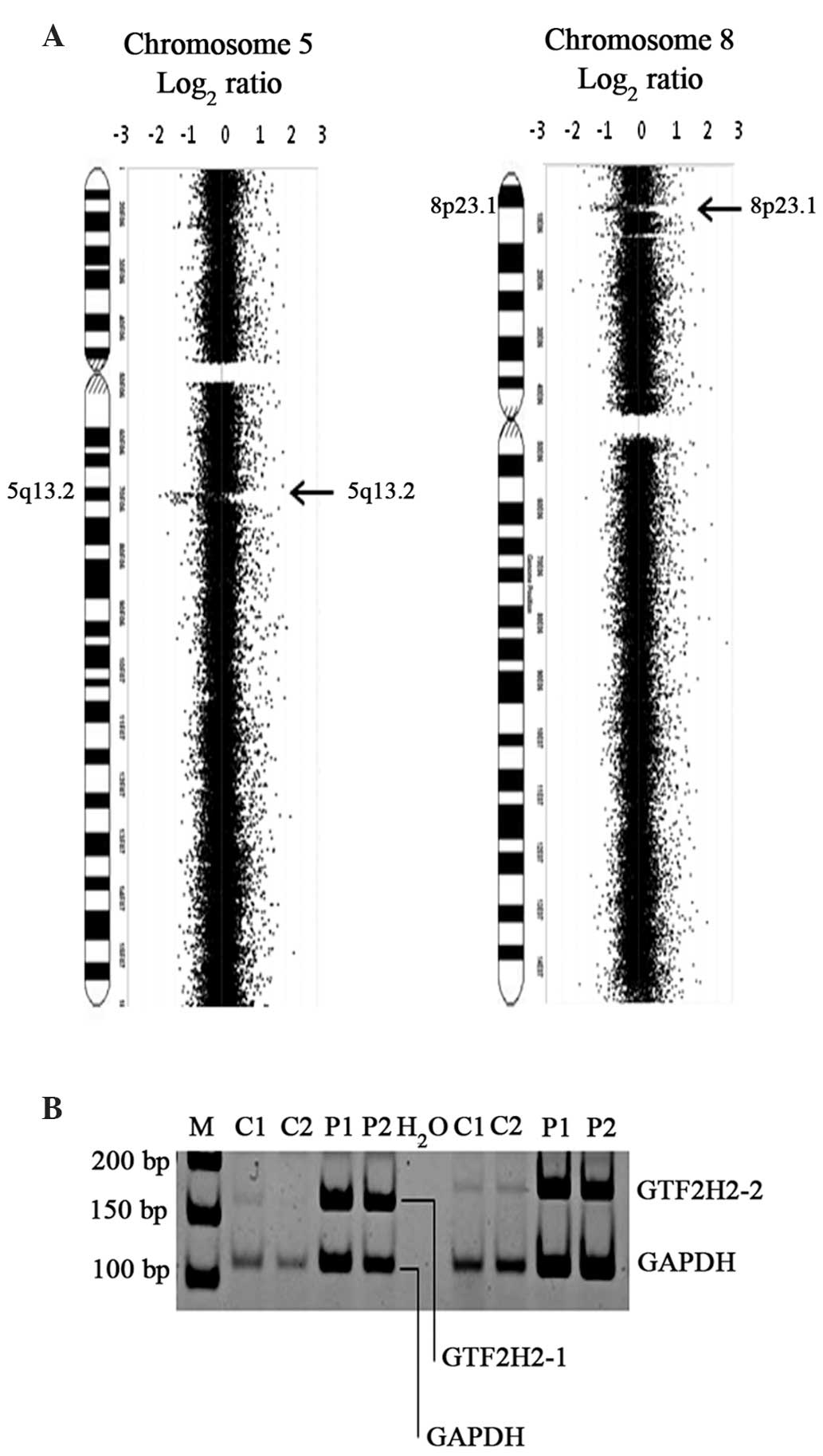
and 8p23.1 were observed most frequently (Fig. 1A). The deletion of GTF2H2 in
the 5q13.2 region, as revealed by PCR analysis, confirmed LOH at
5q13.2, indicating that the results obtained by array CGH analysis
were robust (Fig. 1B). In the HGDNs
of one case of HBV-related liver cirrhosis, loss of 16q22.1 was
identified, a region that harbors a tumor suppressor gene,
CDH1.
 | Table II.Array comparative genomic
hybridization analysis of loci with genomic imbalances identified
in high-grade dysplastic nodules of two hepatitis B virus-related
liver cirrhosis cases. |
Table II.
Array comparative genomic
hybridization analysis of loci with genomic imbalances identified
in high-grade dysplastic nodules of two hepatitis B virus-related
liver cirrhosis cases.
| Loci with genomic
imbalances | Position | Known harbored
genes |
|---|
| Loss |
|
|
|
1q21.1–21.2 |
145902910–148120004 | FCGR1A,
NBPF8, HIST2H4, et al |
|
1q32.1 |
204242050–204325021 | FAM72A,
SRGAP2 |
|
4p16.1 |
9009140–9351709 | DEFB-8,
DEFB-31, β-mannosyltransferase |
|
5q13.2 |
68941167–70587321 | GTF2H2,
OCLN, NAIP, et al |
|
6p22.1 |
26819195–27108243 | GUSBL1,
IMGL |
|
7q11.23 |
73787146–74897603 | GTF2I,
PMS2L5, NSUN5B, et al |
|
8p23.1 |
6907662–7903832 | SPAG11,
DEFB4, ZNF705B, et al |
|
17q21.31–21.32 |
41523026–42131252 | NSF,
ARL17P1, NBR2, et al |
|
22q11.21 |
17086780–17299469 | DGCR6,
PRODH, GGT2, et al |
| Gain |
|
|
|
1q32.1 |
199400460–199436053 | Titin isoform
N2-A |
|
11q13.4 |
78398022–78436027 | ARGEF17 |
|
14q24.3 |
77383793–77398912 | ADCK1 |
|
17p11.2 |
16899950–16930164 | MRIP |
Prevalence of LOH at 5q13.2 in
HCC
As no microsatellite marker has previously been
reported in the 5q13.2 region (position 68941167–70587321),
conventional LOH analysis could not be performed. Thus, in the
present study, two pairs of primers for PCR amplification of the
GTF2H2 gene located in this locus were used for the
examination of LOH at 5q13.2, using the GAPDH gene as a
control, as described in previous studies (13,14). Of
the 83 HCC cases investigated, 30 (36.1%) were found to have loss
of 5q13.2 (Fig. 2A and B); this
effect was not identified in any of the 50 healthy controls,
indicating that LOH at 5q13.2 occurred predominantly in tumors
(P<0.001).
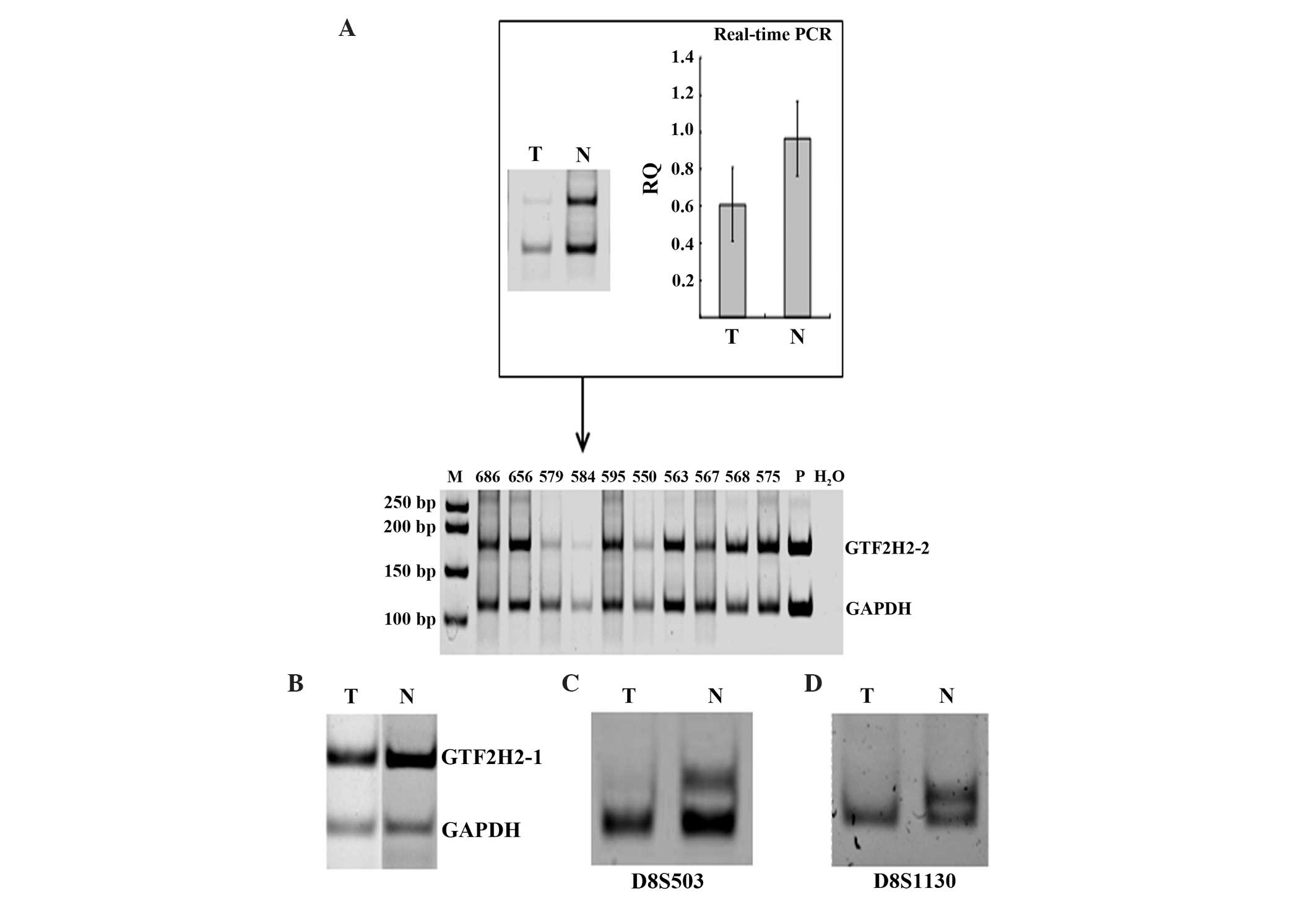 | Figure 2.Analysis of prevalence of LOH at
5q13.2 and 8p23.1 in HCC. (A) Analysis of LOH at 5q13.2 using
GTF2H2-2 as a marker. Differential PCR was performed on 10
HCC cases. Real-time PCR was performed on the genomic DNA of case
584, which had adjacent non-tumor tissue available, to confirm the
results identified by differential PCR. Cases 550, 579 and 584 were
identified as having LOH at 5q13.2. Lanes: P, normal DNA from
blood; M, 50 bp DNA ladder. RQ = 2−δCT, where δCT = CT
(target) - CT (reference). Values are presented as the mean ±
standard deviation. (B) Representative results for detection of LOH
5q13.2 with GTF2H2-1 marker, in a case of HCC (case 665). (C
and D) Representative results for identification of LOH at 8p23.1
in a case of HCC (case 678) showing the PCR products of the
microsatellite markers (C) D8S503 and (D) D8S1130. LOH at 5q13.2
occurred predominantly in tumors (P<0.001). T, tumor; N,
adjacent non-tumor tissue; PCR, polymerase chain reaction; RQ,
relative quantification; CT, threshold cycle; HCC, hepatocellular
carcinoma; LOH, loss of heterozygosity. |
Associations between the frequency of LOH at 5q13.2
and clinicopathological variables are summarized in Table III. No significant differences in
the frequency of LOH at 5q13.2 were identified between distinct
tumor stages, presence or absence of HBV infection, or varying
degrees of tumor differentiation.
 | Table III.Association between frequency of LOH
at 5q13.2 and clinical parameters of hepatocellular carcinoma. |
Table III.
Association between frequency of LOH
at 5q13.2 and clinical parameters of hepatocellular carcinoma.
|
|
| LOH 5q13.2 |
|
|
|---|
|
|---|
| Clinical
parameters | Patients, n | Cases, n | Percentage, % |
χ2-value | P-value |
|---|
| Tumor stage |
|
|
|
|
|
| 1 | 47 | 17 | 36.2 | 0.05 | 0.823 |
|
>1 | 36 | 13 | 36.1 |
|
|
| HBV
infectiona |
|
|
|
|
|
|
Negative | 9 | 5 | 55.6 | –b | 0.289 |
|
Positive | 70 | 25 | 35.7 |
|
|
|
Differentiation |
|
|
|
|
|
| Well or
moderate | 56 | 19 | 33.9 | 0.13 | 0.718 |
|
Poor | 27 | 11 | 40.7 |
|
|
Prevalence of LOH at 8p23.1 in
HCC
Two microsatellite markers, D8S503 and D8S1130, were
used for the examination of LOH at 8p23.1, as described previously
(15,16). Of the 83 HCC cases, 38 and 31
informative cases for D8S503 and D8S1130 were identified,
respectively. Of the 38 informative D8S503 cases, 26 were
identified as having LOH in D8S503 (68.4%; Fig. 2C), while of the 31 D8S1130 cases, 19
were identified with LOH in D8S1130 (61.29%; Fig. 2D).
Correlations between the frequency of LOH at 8p23.1
and clinicopathological variables are summarized in Table IV. No significant differences in the
frequency of LOH at 8p23.1 (D8S503/D8S1130) were identified between
tumor stages, presence or absence of HBV infection, or varying
degrees of tumor differentiation.
 | Table IV.Association between frequency of LOH
at 8p23.1 and clinical parameters of hepatocellular carcinoma. |
Table IV.
Association between frequency of LOH
at 8p23.1 and clinical parameters of hepatocellular carcinoma.
| A, LOH
8q23.1(D8S503) |
|---|
|
|---|
| Clinical
parameters | Patients, n
(n=38) | Cases, n (%) |
P-valueb |
|---|
| Tumor stage |
|
| 1.000 |
| 1 | 23 | 16 (69.6) |
|
|
>1 | 15 | 10 (66.7) |
|
| HBV
infectiona |
|
| 1.000 |
|
Positive | 32 | 22 (68.8) |
|
|
Negative | 4 | 3 (75.0) |
|
|
Differentiation |
|
| 0.461 |
| Well or
moderate | 26 | 19 (73.1) |
|
|
Poor | 12 | 7 (58.3) |
|
|
| B, LOH
8q23.1(D8S1130) |
|
| Clinical
parameters | Patients, n
(n=31) | Cases, n (%) |
P-valueb |
|
| Tumor stage |
|
| 0.715 |
| 1 | 16 | 9 (56.3) |
|
|
>1 | 15 | 10 (66.7) |
|
| HBV
infectiona |
|
| 0.269 |
|
Positive | 26 | 15 (57.7) |
|
|
Negative | 3 | 3 (100) |
|
|
Differentiation |
|
| 1.000 |
| Well or
moderate | 20 | 12 (60.0) |
|
|
Poor | 11 | 7 (63.6) |
|
Discussion
Recent studies have demonstrated that genomic loss
is important in the pathogenesis of HCC. Loss of 1p, 1q, 4q, 6q,
8p, 9p, 10q, 11q, 13q, 14q, 16q, 17p and 22q have been frequently
observed in HCC, and a series of tumor suppressor genes have been
identified in these regions, including PRDM5 (4q26),
TP53 (17p13.1), RB1 (13q14), CDH1 (16q22.1)
and P33ING1D1 (11q33.3-q34) (16,18,19).
Certain clinicopathological associations have been reported with
specific abnormalities: Loss of 4q has been associated with
enhanced α-fetoprotein levels, TP53 mutations (20), tumor size and vascular invasion
(21), while 9p and 6q losses have
been suggested to be independent predictors of poor prognosis of
HCC patients (22).
Therefore, little is known with regard to the impact
of genomic imbalances in HGDNs of liver cirrhosis. Gong et
al (23) reported that in one
nodule of altered hepatocytes (NAH) with small cellular change,
there were alterations in DNA copy number in four chromosomal
regions. Increases in DNA copy number were often detected at
1q25.2-q21.2, 8q and 19q13.43-q13.12, whereas decreases in DNA copy
number were frequently observed at 4p, 4q and 8p. By using a
microarray comprised of 2433 bacterial artificial chromosome clones
to analyze 15 atypical hepatocellular neoplasms (AHN), Kakar et
al (24) observed CGH
abnormalities in eight (53%) AHNs, including gains at 1q, 7p, 7q,
8p, 8q, 11p, 18p, 19p, 20p, 20q, 23p and 23q, and losses at 4q, 7q,
8p, 14q, 20q, 21q and X. However, owing to insufficient resolution
covering the human genome at 1.5 Mb, changes in small chromosomal
regions cannot be detected, and it remains difficult to explore
tumor-associated genes in large genomic areas. In the present
study, array CGH analyses were performed with a 720K array CGH
microarray covering the human genome at ~2 kb resolution. The
results revealed that genomic imbalances identified in HGDNs of
HBV-related liver cirrhosis were predominantly losses of small
segments, including loss of 4p16.1, 7q11.23 and 8p23.1, as well as
gain of 1q32.1, similar to the aforementioned findings in NAH or
AHN (23,24). Notably, certain chromosomal
aberrations in the HGDNs coincided with those frequently observed
in HCC, including loss of 1q21.1–21.2, 1q32.1, 8p23.1 and 22q11.21,
as well as gain of 1q32.1 (16,18,19).
However, loss of 4q, which was frequently observed in HCC in
previous studies, was not identified in the DNs of cirrhosis in the
present study; this is likely to be due to the insufficient number
of cases analyzed. It is notable that in one case of liver
cirrhosis, the loss of 16q22.1, which is frequently observed in HCC
and is associated with HBV infection, was identified in the HGDNs
in the present study. Thus, consistent with other studies, the
current results indicated that, as a precursor of HCC, HGDNs
contain genomic imbalances associated with the pathogenesis of
HBV-related HCC, and there may be genetic links between DN and
HCC.
LOH at 5q is a common chromosomal abnormality
amongst tumors of the digestive system. Based on the patterns of
LOH in all tumors in general, 11 distinct Chr5 regions were mapped
as LOH targets, including 5p15.3, 5p15.2-p15.1, 5p14.3-p13.2,
5q13.2-q23.3, 5q14.3-q15, 5q23.1-q31.1, 5q31.3-q33.1 and
5q33.1-q33.3 (25–27). Fewer genomic imbalances at Chr5 in HCC
have been reported, with the exception of a study by Ding et
al (25), which reported that
patients with non-cirrhotic HCC exhibited LOH primarily at 5q,
while patients with cirrhotic HCC had allelic loss at 5p.
Johannsdottir et al (27)
reported a high prevalence of LOH at 5q13.2-q23.3 in BRCA1-negative
breast tumors. In addition, microdeletions at 5q13.2 were reported
in a familial disease known as oculo-auriculo-vertebral spectrum,
and a correlation between 5q13.2 microdeletions and coarctation of
the aorta was hypothesized (28,29).
However, to the best of our knowledge, loss at 5q13.2 in HCC has
not previously been reported.
In the present study, loss at 5q13.2 was one of the
most frequently observed genomic imbalances identified in HGDNs,
with a prevalence of LOH at 5q13.2 in HCCs of up to 36.1%,
indicating that there may be a tumor suppressor gene at this loci.
The genomic loci of 5q13.2 encompasses the GTF2H2,
NAIP, and OCLN genes. GTF2H2, which encodes
the transcription factor IIH, is a signaling pathway gene and was
reported to be associated with the development of breast cancer
(30). NAIP encodes
baculoviral IAP repeat-containing protein 1 (BIRC1), which is
associated with apoptosis (29).
OCLN encodes occludin, which is an integral membrane protein
located at tight junctions and regulates the directional migration
of epithelial cells (31). However,
the present study identified no significant association between the
frequency of LOH at 5q13.2 and the clinical parameters of HCC.
Further study is required to explore the significance of LOH at
5q13.2 in the development of HCC.
Previous studies have reported that a loss of 8p is
the most common chromosomal alteration in a variety of types of
human cancer, and have postulated that one or several tumor
suppressor genes may lie within this region, including PINX1
in 8p23 and DLC1 in 8p22-p21.3 (32–34). Lu
et al (35) identified two
sites, 8p23.1 and 8p22, which potentially contain tumor suppressor
genes involved in human liver carcinogenesis. In the present study,
loss of 8p23.1 was another frequently observed locus with genomic
imbalances in HGDNs, similar to the results observed in previous
studies (23,24,32).
The present study also evaluated the prevalence of
LOH at 8p23.1 in HCCs, and showed its occurrence rate to be 61.29%
(D8S503) or 68.4% (D8S1130) in HCCs, similar to a previous study
that demonstrated the frequency of LOH at 8p23.2–21 to be 63% in
HCC in a Japanese population (35).
However, no significant mutation or absence of expression of the
genes at 8p23.2–21 has been identified (35), and the present study also found no
significant association between the frequency of LOH at 8p23.1 and
the clinicopathological characteristics of HCC. Although no
significant genetic alterations were detected in HCC at 8p23.1, the
results of the present study suggest that unknown genes in this
region may be significant in HCC, as suggested by a previous study
(35). Further studies are required
to explore the significance of LOH at 8p23.1 in the development of
HCC.
Although array CGH array analysis was only performed
on a small set of HGDNs, the present study demonstrated that, as
precursors of HCC, HGDNs contain genomic imbalances associated with
HCC pathogenesis, suggesting that chromosomal abnormalities may
occur and accumulate in preneoplastic lesions of liver cirrhosis.
Further studies are required to screen loci with genomic imbalances
in HGDNs in a greater number of cases, and to identify the critical
tumor suppressor genes in these loci to elucidate their roles in
the development of HCC from liver cirrhosis.
Acknowledgements
The authors would like to thank Dr Xiao-Yan Shi
(Department of Pathology, Beijing Friendship Hospital, Capital
Medical University) for his critical pathological review. This work
was supported by grants from the Wang Bao-En Liver Fibrosis
Foundation (grant no. 20100013), the Scientific Research Foundation
for Returned Overseas Chinese Scholars, State Education Ministry
(2011, No. 41) and a returning grant from the International Agency
for Research on Cancer (IARC, FEL/09/03). The Abstract from the
present study was presented as a poster in the 65th Annual Meeting
of the American Association for the Study of Liver Diseases: The
Liver Meeting 2014.
References
|
1
|
Theise ND, Chen CJ and Kew MC: Liver
cancerWorld Cancer Report 2014. Stewart B and Wild C: International
Agency for Research on Cancer; Lyon: pp. 577–593. 2014
|
|
2
|
Hsu YS, Chien RN, Yeh CT, et al: Long-term
outcome after spontaneous HBeAg seroconversion in patients with
chronic hepatitis B. Hepatology. 35:1522–1527. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim TM, Yim SH, Shin SH, et al: Clinical
implication of recurrent copy number alterations in hepatocellular
carcinoma and putative oncogenes in recurrent gains on 1q. Int J
Cancer. 123:2808–2815. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Patil MA, Gütgemann I, Zhang J, et al:
Array-based comparative genomic hybridization reveals recurrent
chromosomal aberrations and Jab1 as a potential target for 8q gain
in hepatocellular carcinoma. Carcinogenesis. 26:2050–2057. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Midorikawa Y, Tang W and Sugiyama Y:
High-resolution mapping of copy number aberrations and
identification of target genes in hepatocellular carcinoma. Biosci
Trends. 1:26–32. 2007.PubMed/NCBI
|
|
6
|
Guo X, Yanna, Ma X, et al: A meta-analysis
of array-CGH studies implicates antiviral immunity pathways in the
development of hepatocellular carcinoma. PLoS One. 6:e284042011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Han ZG: Recent progress in genomic
research of liver cancer. Sci China C Life Sci. 52:24–30. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
International Consensus Group for
Hepatocellular Neoplasia: Pathologic diagnosis of early
hepatocellular carcinoma: A report of the international consensus
group for hepatocellular neoplasia. Hepatology. 49:658–664. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
International Working Party: Terminology
of nodular hepatocellular lesions. Hepatology. 22:983–993. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Di Tommaso L, Sangiovanni A, Borzio M, et
al: Advanced precancerous lesions in the liver. Best Pract Res Clin
Gastroenterol. 27:269–284. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sobin LH and Wittekind CH: TNM
Classification of Malignant Tumours. 6th. Wiley; New York, NY: pp.
81–83. 2002
|
|
12
|
Theise ND, Curado MP, Franceschi S, et al:
Hepatocellular carcinomaWHO Classification of Tumors of the
Digestive System. Bosman FT, Carneiro F, Hruban RH and Theise ND:
International Agency for Research on Cancer; Lyon: pp. 205–216.
2010
|
|
13
|
Huang J, Grotzer MA, Watanabe T, et al:
Mutations in the Nijmegen breakage syndrome gene in
medulloblastomas. Clin Cancer Res. 14:4053–4058. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang J, Pang J, Watanabe T, Ng HK and
Ohgaki H: Whole genome amplification for array comparative genomic
hybridization using DNA extracted from formalin-fixed,
paraffin-embedded histological sections. J Mol Diagn. 11:109–116.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun FY, Wan DF, Qing WX, et al:
Determination of gene deletion in tumor tissue by semi-quantitative
polymerase chain reaction. Tumor. 20:135–137. 2000.(In
Chinese).
|
|
16
|
Tomlinson IP, Lambros MB and Roylance RR:
Loss of heterozygosity analysis: Practically and conceptually
flawed? Genes Chromosomes Cancer. 34:349–353. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou HL, Gong L, Zhang W, Du YX, Zhang JY
and Feng YM: Loss of heterozygosity on chromosomes 8 and 16 in
primary hepatocellular carcinoma. Xian Dai Zhong Liu Yi Xue.
20:1134–1138. 2012.(In Chinese).
|
|
18
|
Lau SH and Guan XY: Cytogenetic and
molecular genetic alterations in hepatocellular carcinoma. Acta
Pharmacol Sin. 26:659–665. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Niketeghad F, Decker HJ, Caselmann WH, et
al: Frequent genomic imbalances suggest commonly altered tumour
genes in human hepatocarcinogenesis. Br J Cancer. 85:697–704. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rashid A, Wang JS, Qian GS, Lu BX,
Hamilton SR and Groopman JD: Genetic alterations in hepatocellular
carcinomas: association between loss of chromosome 4q and p53 gene
mutations. Br J Cancer. 80:59–66. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zondervan PE, Wink J, Alers JC, et al:
Molecular cytogenetic evaluation of virus-associated and non-viral
hepatocellular carcinoma: analysis of 26 carcinomas and 12
concurrent dysplasias. J Pathol. 192:207–215. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Laurent-Puig P, Legoix P, Bluteau O, et
al: Genetic alterations associated with hepatocellular carcinomas
define distinct pathways of hepatocarcinogenesis. Gastroenterology.
120:1763–1773. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gong L, Li YH, Su Q, Chu X and Zhang W:
Clonality of nodular lesions in liver cirrhosis and chromosomal
abnormalities in monoclonal nodules of altered hepatocytes.
Histopathology. 56:589–599. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kakar S, Chen X, Ho C, et al: Chromosomal
abnormalities determined by comparative genomic hybridization are
helpful in the diagnosis of atypical hepatocellular neoplasms.
Histopathology. 55:197–205. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang F, Zhou C, Ling Y, et al: Allelic
analysis on chromosome 5 in sporadic colorectal cancer patients.
Zhonghua Zhong Liu Za Zhi. 24:458–460. 2002.(In Chinese).
PubMed/NCBI
|
|
26
|
Ding SF, Habib NA, Dooley J, et al: Loss
of constitutional heterozygosity on chromosome 5q in hepatocellular
carcinoma without cirrhosis. Br J Cancer. 64:1083–1087. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Johannsdottir HK, Jonsson G,
Johannesdottir G, et al: Chromosome 5 imbalance mapping in breast
tumors from BRCA1 and BRCA2 mutation carriers and sporadic breast
tumors. Int J Cancer. 119:1052–1060. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang XS, Xiao L, Li X, et al: Two
neighboring microdeletions of 5q13.2 in a child with
oculo-auriculo-vertebral spectrum. Eur J Med Genet. 53:153–158.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen CP, Lin CJ, Chen CY, et al: Maternal
transmission of interstitial microdeletion in 5q13.2 detected
during prenatal diagnosis of coarctation of the aorta. Taiwan J
Obstet Gynecol. 52:303–305. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Markaverich BM, Shoulars K and Rodriquez
MA: Luteolin regulation of estrogen signaling and cell cycle
pathway genes in MCF-7 human breast cancer cells. Int J Biomed Sci.
7:101–111. 2011.PubMed/NCBI
|
|
31
|
Du D, Xu F, Yu L, Zhang C, Lu X, Yuan H,
et al: The tight junction protein, occludin, regulates the
directional migration of epithelial cells. Dev Cell. 18:52–63.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kahng YS, Lee YS, Kim BK, Park WS, Lee JY
and Kang CS: Loss of heterozygosity of chromosome 8p and 11p in the
dysplastic nodule and hepatocellular carcinoma. J Gastroenterol
Hepatol. 18:430–436. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chan KL, Lee JM, Guan XY, Fan ST and Ng
IO: High-density allelotyping of chromosome 8p in hepatocellular
carcinoma and clinicopathologic correlation. Cancer. 94:3179–3185.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pineau P, Nagai H, Prigent S, et al:
Identification of three distinct regions of allelic deletions on
the short arm of chromosome 8 in hepatocellular carcinoma.
Oncogene. 18:3127–3134. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lu T, Hano H, Meng C, et al: Frequent loss
of heterozygosity in two distinct regions, 8p23.1 and 8p22, in
hepatocellular carcinoma. World J Gastroenterol. 13:1090–1097.
2007. View Article : Google Scholar : PubMed/NCBI
|