Introduction
Colorectal cancer (CRC) is one of the most common
cancers worldwide, with more than one million newly diagnosed cases
reported annually. In total, ~700,000 CRC-associated mortalities
occurred worldwide in 2012, accounting for 8% of all cancer
mortalities, and making CRC the fourth most common cause of
cancer-associated mortality (1). The
standard treatment for patients with unresectable metastases
includes chemotherapy regimens based on irinotecan, oxaliplatin,
fluoropyrimidines, anti-vascular endothelial growth factor
(anti-VEGF) therapy (bevacizumab) and anti-epidermal growth factor
receptor (anti-EGFR) therapy, such as panitumumab and cetuximab.
These treatment options are efficient in a small percentage of
patients, and it is important to identify specific biomarkers to
determine the patients that are likely to benefit from anti-EGFR
therapy (2–4).
EGFR triggers a downstream signalling cascade
through, for example, the Kirsten rat sarcoma viral oncogene
homolog-serine/threonine-protein kinase B-Raf and the
phosphatidylinositol 3-kinase (PI3K), catalytic subunit
α-phosphatase and tensin homolog-Akt pathways, which regulate cell
proliferation, survival, apoptosis resistance, invasion and
migration (2,3,5–8). The PIK3CA gene encodes the p110α
subunit of PI3K α and belongs to class IA of the PI3Ks. The PI3K α
protein is composed of regulatory subunit p85, which mediates
anchorage to EGFR-specific docking sites, and catalytic subunit
p110, which generates a second messenger that is responsible for
the activation of Akt in response to the activation of growth
factors from various ligands. These ligands include epidermal
growth factor (EGF) or VEGF. Somatic mutations in cancer cells only
occur in PIK3CA and PI3KR1, which encodes the p85 α
subunit (3,6,9). These
mutations are concentrated in two key regions of the PIK3CA
gene, consisting of the helical domain of exon 9 and the kinase
domain of exon 20 (7,8,10).
Activating mutations in PIK3CA are detected in 7–32%
patients, with G>A transversions in exon 9 being the most
commonly observed configuration, which may coincide with
KRAS and BRAF mutations. Tumours with the
PIK3CA gene mutation are characterised by a predominant
proximal colonic location (10,11) and by
the frequent presence of mucinous differentiation (10).
Mutations of EGFR-dependent signalling molecules
confer resistance to EGFR-specific antibody therapy. KRAS
mutation is the first molecular marker of response to EGFR
inhibitors (11). It has been
hypothesised that the PIK3CA gene mutation may also affect
the response to anti-EGFR therapy in patients with metastatic CRC
(12,13). Certain studies indicate that
PIK3CA exon 20 mutations negatively affect the response
rate, disease control rate, progression-free survival (PFS) time
and overall survival (OS) time, whilst PIK3CA exon 9
mutations demonstrate no significant effect on objective response
(3,4).
The aim of the present study was to evaluate the
importance of mutation in the PIK3CA gene as a prognostic
factor in CRC. Additionally, the frequency of PIK3CA
mutations in patients with CRC and the incidence of mutations in
particular exons were examined. The association between the
PIK3CA gene mutation and mutations in other downstream
effectors of the EGFR signalling pathway was also analysed, in
addition to the association between the PIK3CA gene mutation
and various clinical or pathological features.
Materials and methods
Patient characteristics
Based on the database of the Military Institute of
Medicine (Warsaw, Poland), 156 patients that were consecutively
diagnosed with CRC were identified. The patients had been treated
with palliative chemotherapy at the Oncology Department of the
Military Institute of Medicine between 2006 and 2010. The inclusion
criteria were as follows: Confirmed histopathological diagnosis of
CRC; aged >18 years; presence of measurable lesions, determined
by Response Evaluation Criteria in Solid Tumours, version 1.1
(14); adequate haematological
parameters, consisting of a neutrophil count of
≥1.5×109, platelet count of ≥100×109/l and
haemoglobin count of ≥9.0 g/dl; adequate biochemical parameters,
comprising a bilirubin level <2 × upper limit of normal (ULN);
an aspartate transaminase (AST); alanine transaminase (ALT) level
<2.5 × ULN; a glomerular filtration rate (GFR) of >50 ml/min;
and, in premenopausal women, an absence of pregnancy. The exclusion
criteria were as follows: Renal insufficiency, demonstrated by a
GFR <50 ml/min; hepatic insufficiency, demonstrated by AST and
ALT levels >2.5 × ULN; and severe concomitant disease, such as
unstable cardiac angina. This study was approved by the ethics
commmittee of the Military Institute of Medicine and written
informed consent was obtained from all patients.
Histopathological examination of
tumour specimens
Primary tumour specimens were collected from CRC
patients. Formalin-fixed paraffin embedded (FFPE) tissue blocks
were cut into serial 5 µm-thick sections for haematoxylin and eosin
staining. The presence of tumour tissue was verified by an
experienced pathologist. Subsequently, tissue samples from at least
three serial sections were macrodissected to ensure that the
specimens contained ≥80% tumour cells.
DNA extraction
DNA from FFPE colorectal tumour tissues was isolated
from 10–30-µm thick sections subsequent to macrodissection,
resulting in the selection of specimens containing ≥80% tumour
cells. Tumour samples were extracted with xylene and ethanol to
remove paraffin, and placed in 1% SDS/proteinase K (10 mg/ml) at
56°C overnight. DNA was isolated using the NucliSENS easyMag
platform (bioMérieux, Marcy l'Etoile, France) for automated nucleic
acid extraction.
KRAS, BRAF and PIK3CA mutation
analysis
The detection of mutations in codons 12 and 13 of
exon 1 of the KRAS gene and exons 11 and 15 of the
BRAF gene was conducted using a previously described method
(15). The analysis of mutations in
exons 9 and 20 of the PIK3CA gene was performed by direct
sequencing, as described by Samuels et al (16) and Li et al (17), with a number of modifications. The
primers for exon 9 were designed to avoid amplification of
homologous sequences located at the chromosome 22q11.2 cat-eye
syndrome region and on chromosome 16. Genomic DNA obtained from
tumour samples was amplified by polymerase chain reaction (PCR)
using the following primers: Forward strand exon (FSE)9,
5′-TTGCTTTTTCTGTAAATCATCTGTG-3′; Reverse strand exon (RSE)9, for
exon 9 of PIK3CA, 5′-CTGCTTTATTTATTCCAATAGGTATG-3′; FSE20,
5′-ACATCATTTGCTCCAAACTGA-3′, RSE20, for exon 20 of PIK3CA,
5′-CATAACATGAAATTGCGCATT-3′. PCR was conducted in a total volume of
10 µl, containing 2 µl of the extracted genomic DNA, using 10X PCR
buffer, 1.5 mmol/l MgCl2, 0.2 µmol/l of each primer, 0.1
mmol/l of deoxynucleoside triphosphate, and 1 unit of Taq DNA
polymerase (Eurx Ltd., Gdańsk, Poland). PCR conditions were as
follows: 95°C for 10 min; 45 cycles of 95°C for 30 sec, 59°C for 30
sec and 72°C for 30 sec; and finally 7 min at 72°C. The
amplification products were purified using the DNA Gel-Out kit (DNA
Gdańsk, Gdynia, Poland). Automated sequencing was conducted using
the BigDye® Terminator v3.1 Cycle Sequencing Kit (Life
Technologies, Warsaw, Poland). Sequencing reactions were purified
using the ExTerminator kit (DNA Gdańsk), and analysed on a 3500
Genetic Analyzer sequencer (Life Technologies). A wild-type control
DNA sample without PIK3CA mutation and a known mutation
sample (substitution 1633 G>A, E545K within exon 9 and
substitution 3140 A>G, H1047R within exon 20) were included in
the experiment. The mutation was confirmed by sequencing at least
two independent PCR products.
Statistical analysis
The OS time was defined as the time elapsed between
the commencement of the first line of palliative chemotherapy, and
the date of mortality or of the final follow-up, and was estimated
according to the Kaplan-Meier method. The cut-off date for the
present analysis was December 2013.
The χ2 test was used to investigate the
association between variables in the two treatment groups with
respect to baseline characteristics. The log-rank test was
performed in the Kaplan-Meier survival analyses to assess
differences between the groups with regard to OS time. P<0.05
was considered to indicate a statistically significant difference.
Multivariate analyses of OS time were performed by Cox
proportional-hazard regression using the forward stepwise method,
all variables determined to be significant in the univariate
analysis were included in the multivariate analysis. Analyses were
performed using the statistical package Statistica, version 7.0
(Statsoft, Inc., Tulsa, OK, USA).
Results
Patient characteristics
The characteristics of included patients are
summarised in Table I. The cohort
comprised 100 women and 56 men, with a median age of 67 years. The
majority of patients (91%) underwent primary tumour resection. The
primary tumour was located in the colon in 67 patients (42.9%), and
in the sigmoid colon or in the rectum in 89 patients (57.1%).
Metastases were located in the liver in 77 patients (70.6% of
patients with metastases), the lungs in 21 patients (19.3% of
patients with metastases), and other organs in 72 patients (66.0%
of patients with metastases). Lymph node metastasis was also
identified in 50.6% of patients. The majority of patients had a
good performance status (Karnofsky status of 80–100).
 | Table I.Characteristics of the patients with
colorectal cancer enrolled in the present study. |
Table I.
Characteristics of the patients with
colorectal cancer enrolled in the present study.
| Characteristic | Value |
|---|
| Total, n | 156 |
| Age in years,
median (range) | 67 (25–85) |
| Gender, n (%) |
|
|
Female | 100 (64.9) |
|
Male | 56
(35.9) |
| KRAS status,
n (%) |
|
|
Mutation | 44
(28.2) |
| Codon
12 | 41
(26.3) |
| Codon
13 | 3
(1.9) |
|
Wild-type | 112 (71.8) |
| BRAF status,
n (%) |
|
|
Mutation | 12 (7.7) |
|
Wild-type | 144 (92.3) |
| PIK3CA
status, n (%) |
|
|
Mutation | 15 (9.6) |
| Codon
9 | 7
(4.5) |
| Codon
20 | 8
(5.1) |
|
Wild-type | 141 (90.4) |
| Primary tumour
localisation, n (%) |
|
|
Colon | 67
(42.9) |
|
Sigmoid/rectum | 89
(57.1) |
| Localisation of
metastases (n=109), n (%) |
|
|
Liver | 77
(70.6) |
|
Lungs | 21
(19.3) |
| Other
localisations | 72
(66.0) |
| Karnofsky
performance status, n (%) |
|
|
100 | 79
(50.6) |
| 90 | 63
(40.4) |
| 80 | 12 (7.7) |
| 70 | 2
(1.3) |
| Histological
differentiation grade, n (%) |
|
|
High/moderate | 126 (80.8) |
|
Low/unknown | 30
(19.2) |
| Histological type,
n (%) |
|
|
Mucinous | 7
(4.5) |
|
Mixed | 44
(28.2) |
|
Cylindocellular | 3
(1.9) |
|
Tubular | 70
(44.9) |
|
Unclassified | 32
(20.5) |
| Previous adjuvant
chemotherapy, n (%) | 55
(35.3) |
| Lymph node status,
n (%) |
|
| N0 | 35
(22.4) |
| N1 | 42
(26.9) |
|
N2a | 20
(12.8) |
|
N2b | 17
(10.9) |
| Nx | 42
(27.0) |
| Invasive extent, n
(%) |
|
| Tx | 10 (6.4) |
| T1 | 1
(0.6) |
| T2 | 17
(10.9) |
| T3 | 105 (67.3) |
| T4 | 23
(14.7) |
KRAS, BRAF, PIK3CA gene mutation
status
KRAS mutations were present in 44 patients
(28.2%), of whom 41 patients (93.2%) possessed mutations in codon
12, and three patients (6.8%) possessed mutations in codon 13.
BRAF mutations were present in 12 patients (7.7%).
PIK3CA mutations were present in 15 patients (9.6%), of whom
seven (46.7%) had mutations in codon 9, and eight (53.3%) had
mutations in codon 20. Mutation in the PIK3CA gene was
detected in six patients who had KRAS gene mutations (40% of
PIK3CA mutated tumours) and in one patient with BRAF
mutations (6.6% of PIK3CA mutated tumours).
Clinicopathological variables and
PIK3CA gene mutation status
The evaluation of PIK3CA mutation status
relative to clinicopathological variables is summarised in Table II. PIK3CA gene mutations were
present in 15 patients (9.6%). An increased incidence of
PIK3CA gene mutations was detected in patients with involved
lymph nodes, with low-grade or unknown histological differentiation
of the tumour, and with tubular cancer. PIK3CA gene
mutations were also frequently present in patients with advanced
disease, T stage III or IV. However, the association between these
variables and PIK3CA status was not statistically
significant.
 | Table II.Comparison of PIK3CA gene
mutation status and clinicopathological variables (n=156). |
Table II.
Comparison of PIK3CA gene
mutation status and clinicopathological variables (n=156).
|
| PIK3CA
status |
|
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
variable | Wild-type | Mutation | Statistical
test | P-value |
|---|
| Patients, n | 141 | 15 |
|
|
| Age, years |
|
| 1026.0a | 0.8521 |
|
Median | 67 | 69 |
|
|
|
Range | 25–85 | 37–79 |
|
|
| Gender, n |
|
| 0.25c | 0.6165 |
|
Male | 52 | 4 |
|
|
|
Female | 89 | 11 |
|
|
| Histological
differentiation grade, n |
|
| 2.70c | 0.1003 |
|
High/moderate | 30 | 0 |
|
|
|
Low/unknown | 111 | 15 |
|
|
| Primary tumour
localisation, n |
|
| 0.07b | 0.7914 |
|
Sigmoid/rectum | 79 | 9 |
|
|
|
Colon | 61 | 6 |
|
|
| Karnofsky
performance status, n |
|
| 0.65c | 0.4214 |
|
≤80 | 14 | 0 |
|
|
|
>80 | 127 | 15 |
|
|
| Primary tumour
size, n |
|
| 0.04c | 0.8445 |
|
T1/T2 | 17 | 1 |
|
|
|
T3/T4 | 124 | 14 |
|
|
| Lymph node
involvement grade, n |
|
| 2.92b | 0.0873 |
| N0 | 29 | 6 |
|
|
| N
positive | 112 | 9 |
|
|
| Histological type,
n |
|
| 0.02b | 0.8835 |
|
Tubular | 63 | 7 |
|
|
|
Other | 78 | 8 |
|
|
| BRAF status,
n |
|
| 0.12c | 0.7242 |
|
Wild-type | 130 | 14 |
|
|
|
Mutation | 11 | 1 |
|
|
| KRAS status,
n |
|
| 1.13b | 0.2872 |
|
Wild-type | 103 | 9 |
|
|
|
Mutation | 38 | 6 |
|
|
Prognostic significance of PIK3CA gene
mutation status
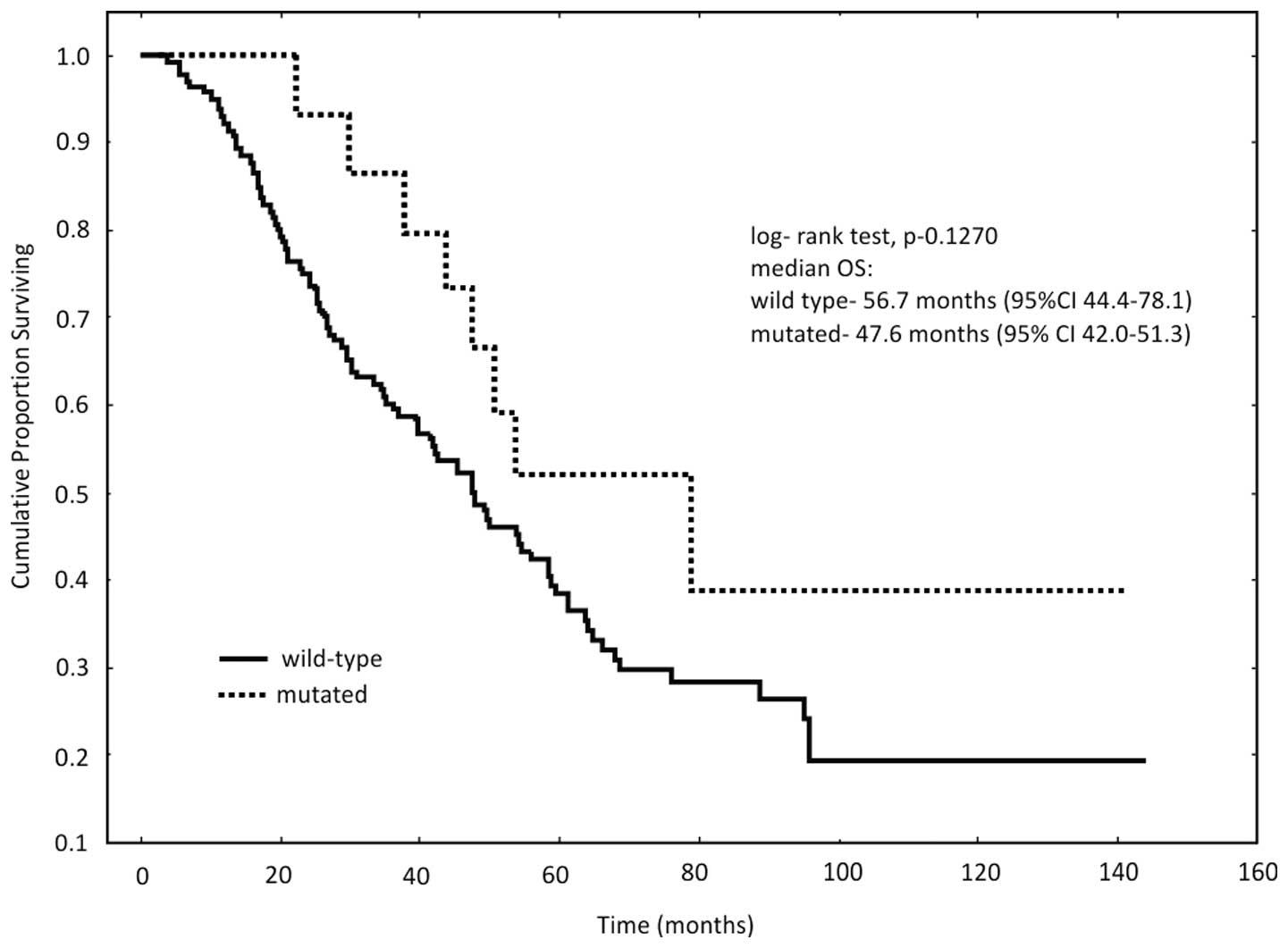
No significant difference in OS rate was identified
between patients with PIK3CA mutations and those with
wild-type PIK3CA genes (P=0.1270; Fig. 1). However, patients with PIK3CA
mutations tended to demonstrate a decreased OS rate. The median OS
in patients with wild-type PIK3CA genes was 56.7 months,
compared with 47.6 months in patients presenting with
mutations.
Clinical and pathological variables
identified by univariate analysis as potential prognostic factors
for OS rate
The results of the univariate analysis are
summarised in Table III. Univariate
analysis of the present patient cohort identified that gender and
lymph node involvement acted as prognostic factors that influenced
OS rate, as female patients survived for 57.5 months, compared with
39.3 months for male patients (P=0.0111, and the median OS in
patients without lymph node metastases was 61.4 months, compared
with 45.4 months in patients presenting with metastases (P=0.0122).
The OS rate associated with other clinical parameters, consisting
of age, primary tumour localisation, Karnofsky performance status,
histological type, histological differentiation grade, primary
tumour size and gene mutation status, did not differ significantly
between groups.
 | Table III.Univariate analysis of OS rate
(log-rank test). |
Table III.
Univariate analysis of OS rate
(log-rank test).
| Clinical
parameter | n | Median OS,
months | P-value |
|---|
| Age, years |
|
| 0.9269 |
|
<70 | 102 | 59.2 |
|
|
≥70 | 54 | 45.4 |
|
| Gender |
|
| 0.0111a |
|
Male | 56 | 39.3 |
|
|
Female | 100 | 57.5 |
|
| Primary tumour
localisation |
|
| 0.9432 |
|
Sigmoid/rectum | 67 | 44.6 |
|
|
Colon | 89 | 49.6 |
|
| Karnofsky
performance status |
|
| 0.6373 |
|
≤80 | 14 | 20.9 |
|
|
>80 | 142 | 49.4 |
|
| Lymph node
involvement |
|
| 0.0122a |
|
Present | 121 | 45.4 |
|
|
Absent | 35 | 61.4 |
|
| Histological
type |
|
| 0.9808 |
|
Tubular | 86 | 48.8 |
|
|
Other | 70 | 47.8 |
|
| Histological
differentiation grade |
|
| 0.1331 |
|
High/moderate | 126 | 52.4 |
|
|
Low/unknown | 30 | 29.3 |
|
| Primary tumour
size |
|
| 0.1280 |
|
T1/T2 | 18 | 60.1 |
|
|
T3/T4 | 138 | 47.4 |
|
| PIK3CA
status |
|
| 0.1271 |
|
Mutation | 15 | 56.7 |
|
|
Wild-type | 141 | 47.6 |
|
| KRAS
status |
|
| 0.7740 |
|
Mutation | 44 | 47.9 |
|
|
Wild-type | 112 | 48.8 |
|
| BRAF
status |
|
| 0.6398 |
|
Mutation | 12 | 22.7 |
|
|
Wild-type | 144 | 49.4 |
|
Clinical and pathological variables
identified by multivariate analysis as potential prognostic factors
for OS rate
The results of the multivariate analysis are
summarised in Table IV. Multivariate
analysis identified that lymph node involvement grade [hazard ratio
(HR), 1.68; P=0.0467] and male gender (HR, 1.57; P=0.0249) were
adverse prognostic factors for OS rates. KRAS, BRAF
and PIK3CA gene mutation status was not found to
significantly affect OS rate in this analysis.
 | Table IV.Multivariate analysis of overall
survival rate. |
Table IV.
Multivariate analysis of overall
survival rate.
| Clinical parameter
comparison | Multivariate
analysis, HR (95% CI) | P-value |
|---|
| Lymph node
involvement | 1.68
(1.01–2.82) |
0.0467 |
| Gender | 1.57
(1.06–2.32) |
0.0249 |
| PIK3CA
status | NS | >0.05 |
| KRAS
status | NS | >0.05 |
| BRAF
status | NS | >0.05 |
Discussion
The treatment of cancer is increasingly based on
targeted therapy, including morphological identification of tumour
histology, tumour staging and identification of target pathways and
molecules. It has been established that KRAS mutation is a
marker of resistance to anti-EGFR therapy in patients with CRC
(4,18–21).
Despite the exclusion of patients with KRAS-mutant tumours,
anti-EGFR treatment fails in numerous patients with CRC. A number
of studies have demonstrated a negative correlation between
PIK3CA mutations and clinical outcomes (3,4,8,10,12,22).
The aim of the current study was to evaluate the
incidence of PIK3CA gene mutation in patients with CRC at
all stages, and also to determine the association between mutation
of the PIK3CA gene and mutations in other downstream
effectors of the EGFR signalling pathway. Additionally, the
incidence of mutations in exon 9 and exon 20 of the PIK3CA
gene were examined. The present study also evaluated the role of
the PIK3CA gene mutation and the select clinical and
pathological variables of these tumours as potential prognostic
factors.
Activating mutations in the PIK3CA gene are
identified in 7–32% of CRC patients. In the present study,
PIK3CA mutations were detected in 9.6% of CRCs, which is
consistent with previously published data (3,7,8,10–12,18,23,24).
However, in contrast to a number of earlier studies (3,10,11,18,23–25),
the present analysis identified a similar frequency of mutations in
exons 9 and 20. Overall, 46.7% of patients possessed mutations in
codon 9, while 53.3% possessed mutations in codon 20.
Pentheroudakis et al (25)
detected the PIK3CA mutation in exon 9 in 54% of cases, and
in exon 20 in 13.5% of cases. Double PIK3CA gene mutations
in exons 9 or 20 were not detected in the present cohort, in
contrast to the studies conducted by Rock et al (18) and Sartore-Bianchi et al
(26). In the present study, mutation
in the PIK3CA gene coincided with KRAS gene mutations
in six patients, comprising 40% of PIK3CA mutated tumours,
and with BRAF mutations in one patient, comprising 6.6% of
PIK3CA mutated tumours. Similar associations have been
reported in previous studies (10,12,24,26).
Tumours with PIK3CA mutations were
characterised by a predominantly distal colonic location, the
frequent presence of tubular differentiation and a low grade of
histological differentiation. These results are in contrast with
previously published studies, with certain studies identifying no
clinical features that were associated with PIK3CA gene
mutations (7,22), while others identified an association
between PIK3CA gene mutation and tumour mucinous
differentiation and proximal colon location (10,24).
Notably, patients with more advanced disease, T stage III or IV, or
those demonstrating the involvement of lymph nodes presented with
an increased rate of PIK3CA gene mutations. Similar
associations have been previously reported (7,27). The
difference in the clinicopathological characteristics of the tumour
between the mutation statuses of particular exons was not estimated
due to the small number of tumours demonstrating PIK3CA
mutations in the present patient population.
In the present study, the results of the univariate
and multivariate analyses into the role of clinical and
pathological variables revealed a positive, statistically
significant association between female gender and uninvolved lymph
nodes on the overall patient survival.
The present study did not confirm a prognostic role
for PIK3CA mutation status in CRC patients, in contrast to
the results obtained by Rosty et al (10) and Therkildsen et al (28), who observed a shorter survival time in
patients with PIK3CA mutations. The current results are
consistent with findings reported by Cathomas (24), Zhu et al (7) and Karapetis et al (29), that PIK3CA exhibited no
prognostic impact. Ogino et al (30) also reported that tumour PIK3CA
mutation status is not associated with stage III colon cancer
prognosis. Compared with carriers of wild-type PIK3CA,
patients with a PIK3CA-mutated tumour had a shorter OS rate.
However, this trend was not statistically significant. Similar data
have been reported in previous studies (11,18,26).
Studies have reported differences between exon 9 and 20 mutations
with regard to their effects on PFS and OS, noting that
PIK3CA exon 20 mutations were significantly associated with
poorer PFS and OS (3,4). The biological effects of mutations in
exons 9 and 20 of the PIK3CA gene were not compared in the
present study due to the small number of patients with mutant
PIK3CA. It has previously been reported that the coexistence
of PIK3CA exon 9 and 20 mutations is associated with poor
prognosis in CRC patients (31).
The results of the present study indicate that
aberrations in PIK3CA did not contribute significant
prognostic information. The role of the PIK3CA mutation
status remains unclear; therefore future prospective multi-centre
trials involving CRC patients are essential in order to fully
assess the clinical relevance of the PIK3CA mutation
status.
In summary, activating mutations in the
PIK3CA gene were present in 9.6% of colorectal carcinomas,
and coincided with mutations in other downstream effectors of the
EGFR signalling pathway. The results from this analysis of CRC
patients of all disease stages indicates that the PIK3CA
mutation status is not a prognostic factor in these patients. In
addition, there is no statistically significant association between
PIK3CA mutation and clinicopathological factors.
References
|
1
|
Ferlay J, Soerjomataram I, Ervik M,
Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and
Bray F: GLOBOCAN 2012: Estimated Cancer Incidence, Mortality and
Prevalence Worldwide in 2012. http://globocan.iarc.frApril 30–2014
|
|
2
|
Baldus SE, Schaefer KL, Engers R, Hartleb
D, Stoecklein NH and Gabbert HE: Prevalence and heterogeneity of
KRAS, BRAF, and PIK3CA mutations in primary
colorectal adenocarcinomas and their corresponding metastases. Clin
Cancer Res. 16:790–799. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Custodio A and Feliu J: Prognostic and
predictive biomarkers for epidermal growth factor receptor-targeted
therapy in colorectal cancer: Beyond KRAS mutations. Crit
Rev Oncol Hematol. 85:45–81. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang ZY, Wu XY, Huang YF, Di MY, Zheng DY,
Chen JZ, Ding H, Mao C and Tang JL: Promising biomarkers for
predicting the outcomes of patients with KRAS wild-type
metastatic colorectal cancer treated with anti-epidermal growth
factor receptor monoclonal antibodies: A systematic review with
meta-analysis. Int J Cancer. 133:1914–1925. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Soeda H, Shimodaira H, Watanabe M, Suzuki
T, Gamoh M, Mori T, Komine K, Iwama N, Kato S and Ishioka C:
Clinical usefulness of KRAS, BRAF, and PIK3CA
mutations as predictive markers of cetuximab efficacy in
irinotecan- and oxaliplatin-refractory Japanese patients with
metastatic colorectal cancer. Int J Clin Oncol. 18:670–677. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
German S, Aslam HM, Saleem S, Raees A,
Anum T, Alvi AA and Haseeb A: Carcinogenesis of PIK3CA. Hered
Cancer Clin Pract. 11:52013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu K, Yan H, Wang R, Zhu H, Meng X, Xu X,
Dou X and Chen D: Mutations of KRAS and PIK3CA as
independent predictors of distant metastases in colorectal cancer.
Med Oncol. 31:162014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stintzing S and Lenz HJ: A small cog in a
big wheel: PIK3CA mutations in colorectal cancer. J Natl Cancer
Inst. 105:1775–1776. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ikenoue T, Kanai F, Hikiba Y, et al:
Functional analysis of PIK3CA gene mutations in human colorectal
cancer. Cancer Res. 65:4562–4567. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rosty C, Young JP, Walsh MD, et al: PIK3CA
activating mutation in colorectal carcinoma: Associations with
molecular features and survival. PLoS ONE. 8:e654792013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shen Y, Wang J, Han X, Yang H, Wang S, Lin
D and Shi Y: Effectors of epidermal growth factor receptor pathway:
The genetic profiling of KRAS, BRAF, PIK3CA, NRAS
mutations in colorectal cancer characteristics and personalized
medicine. PLoS ONE. 8:122013.
|
|
12
|
Soeda H, Shimodaira H, Watanabe M, Suzuki
T, Gamo M, Takahashi M, Komine K, Kato S and Ishioka C: KRAS
mutation in patients with metastatic colorectal cancer does not
preclude benefit from oxaliplatin-or irinotecan-based treatment.
Mol Clin Oncol. 2:356–362. 2014.PubMed/NCBI
|
|
13
|
Huang L, Liu Z, Deng D, Tan A, Liao M, Mo
Z and Yang X: Anti-epidermal growth factor receptor monoclonal
antibody-based therapy for metastatic colorectal cancer: A
meta-analysis of the effect of PIK3CA mutations in KRAS
wild-type patients. Arch Med Sci. 10:1–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Eisenhauer EA, Therasse P, Bogaerts J, et
al: New response evaluation criteria in solid tumours: Revised
RECIST guideline (version 1.1). Eur J Cancer. 45:228–247. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stec R, Bodnar L, Charkiewicz R, et al:
K-Ras gene mutation status as a prognostic and predictive factor in
patients with colorectal cancer undergoing irinotecan- or
oxaliplatin-based chemotherapy. Cancer Biol Ther. 13:1235–1243.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Samuels Y, Wang Z, Bardelli A, et al: High
frequency of mutations of the PIK3CA gene in human cancers.
Science. 304:5542004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li VS, Wong CW, Chan TL, Chan AS, Zhao W,
Chu KM, So S, Chen X, Yuen ST and Leung SY: Mutations of PIK3CA in
gastric adenocarcinoma. BMC Cancer. 5:292005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
De Roock W, Claes B, Bernasconi D, et al:
Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on
the efficacy of cetuximab plus chemotherapy in
chemotherapy-refractory metastatic colorectal cancer: A
retrospective consortium analysis. Lancet Oncol. 11:753–762. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Razis E, Pentheroudakis G, Rigakos G, et
al: EGFR gene gain and PTEN protein expression are favorable
prognostic factors in patients with KRAS wild-type
metastatic colorectal cancer treated with cetuximab. J Cancer Res
Clin Oncol. 140:737–748. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tejpar S and Piessevaux H: Personalized
medicine in metastatic colorectal cancer treated with
anti-epidermal growth factor receptor agents: A future opportunity?
Asia Pac J Clin Oncol. 10:2–10. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Domagała P, Hybiak J, Sulżyc-Bielicka V,
Cybulski C, Ryś J and Domagała W: KRAS mutation testing in
colorectal cancer as an example of the pathologist's role in
personalized targeted therapy: A practical approach. Pol J Pathol.
63:145–164. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ogino S, Nosho K, Kirkner GJ, et al:
PIK3CA mutation is associated with poor prognosis among patients
with curatively resected colon cancer. J Clin Oncol. 27:1477–1484.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Herreros-Villanueva M, Gomez-Manero N,
Muñiz P, García-Girón C and Coma del Corral MJ: PIK3CA mutations in
KRAS and BRAF wild type colorectal cancer patients. A
study of Spanish population. Mol Biol Rep. 38:1347–1351. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cathomas G: PIK3CA in Colorectal Cancer.
Front Oncol. 4:352014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pentheroudakis G, Kotoula V, De Roock W,
et al: Biomarkers of benefit from cetuximab-based therapy in
metastatic colorectal cancer: Interaction of EGFR ligand expression
with RAS/RAF, PIK3CA genotypes. BMC Cancer. 13:492013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sartore-Bianchi A, Martini M, Molinari F,
et al: PIK3CA mutations in colorectal cancer are associated with
clinical resistance to EGFR-targeted monoclonal antibodies. Cancer
Res. 69:1851–1857. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Prenen H, De Schutter J, Jacobs B, De
Roock W, Biesmans B, Claes B, Lambrechts D, Van Cutsem E and Tejpar
S: PIK3CA mutations are not a major determinant of resistance to
the epidermal growth factor receptor inhibitor cetuximab in
metastatic colorectal cancer. Clin Cancer Res. 15:3184–3188. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Therkildsen C, Bergmann TK,
Henrichsen-Schnack T, Ladelund S and Nilbert M: The predictive
value of KRAS, NRAS, BRAF, PIK3CA and PTEN for
anti-EGFR treatment in metastatic colorectal cancer: A systematic
review and meta-analysis. Acta Oncol. 53:852–864. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Karapetis CS, Jonker D, Daneshmand M, et
al: NCIC Clinical Trials Group and the Australasian
Gastro-Intestinal Trials Group: PIK3CA, BRAF, and PTEN
status and benefit from cetuximab in the treatment of advanced
colorectal cancer—results from NCIC CTG/AGITG CO.17. Clin Cancer
Res. 20:744–753. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ogino S, Liao X, Imamura Y, et al:
Alliance for Clinical Trials in Oncology: Predictive and prognostic
analysis of PIK3CA mutation in stage III colon cancer intergroup
trial. J Natl Cancer Inst. 105:1789–1798. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liao X, Morikawa T, Lochhead P, et al:
Prognostic role of PIK3CA mutation in colorectal cancer: Cohort
study and literature review. Clin Cancer Res. 18:2257–2268. 2012.
View Article : Google Scholar : PubMed/NCBI
|