Introduction
Activating mutations of EGFR and KRAS
genes are characteristic mutations, or so-called ‘driver
mutations’, of lung adenocarcinomas (1–3).
Approximately 80% of patients with EGFR mutations
(mEGFR) respond efficiently to treatment with EGFR-tyrosine
kinase inhibitors (TKIs) (1,2), but KRAS mutations (mKRAS)
are considered to predict resistance to EGFR-TKI therapy (4). Over the past decade, other ‘driver
mutations’ in ALK (5),
HER2 (6), and BRAF
(7) have been found in lung
adenocarcinomas, although mEGFR and mKRAS remain the
most frequent ‘driver mutations’ in lung adenocarcinomas (8). Significantly, mEGFR and
mKRAS are mutually exclusive and exhibit a characteristic
association with clinical factors, particularly ethnicity;
mEGFR are frequently observed in Asian individuals, women
and never-smokers, but mKRAS are frequently observed in
Caucasian individuals, men and smokers (9,10).
A copy number gain (CNG) is another mechanism of
oncogenic activation (11). A
large-scale project to characterize copy number alterations in
primary lung adenocarcinomas confirmed that EGFR and
KRAS loci were significantly recurrent events when using a
high-resolution genome-wide approach (12). A recent systematic review and
meta-analysis revealed that EGFR CNGs (gEGFR) were
associated with responsiveness and improved survival outcomes in
patients with non-small cell lung cancer (NSCLC) who were treated
with EGFR-TKIs (13,14). Although gEGFR are reportedly
frequent among never-smokers with NSCLC whose samples are collected
in Western countries (15,16), ethnic differences in the frequency of
gEGFR have not been intensively investigated. The frequency
of KRAS CNGs (gKRAS) is reportedly low (7–11%) in
lung adenocarcinoma (17–19) and the association between gKRAS
and clinical factors has been controversial. Of particular note is
the fact that gEGFR and gKRAS occur significantly
frequently in mEGFR and mKRAS cases, respectively
(15,17–20).
These lines of evidence suggest that a significant
mutually exclusive association between EGFR and KRAS
alterations is present in lung adenocarcinomas. In addition, there
is already a great deal of information about ethnicity and
mEGFR and mKRAS, but much less about the CNGs of
these genes. Only a modest number of studies have analyzed
mutations and CNGs in the same study and linked it with ethnicity.
To the best of our knowledge, the concordant association between
all four genetic alterations and ethnicity has not been extensively
investigated using an adequate statistical method. The present
study evaluated the impact of ethnic differences on the frequencies
of mutations and CNGs of the EGFR and KRAS genes in
lung adenocarcinomas, while considering gender and the smoking
status, using a polynomial logistic regression model.
Materials and methods
Tumor samples
We have previously determined the mutational status
and copy number of the EGFR and KRAS genes in
resected NSCLC samples (17). Among
these samples, the present study restudied 302 surgically resected
lung adenocarcinomas with complete information on mutational status
and copy number of the EGFR and KRAS genes, and
clinical information such as gender, smoking status and ethnicity.
Genomic DNA extracted from frozen tissues was obtained from four
countries: Japan [n=148; Okayama University, Okayama, Japan (n=73)
and Chiba University, Chiba, Japan (n=75)], the United States
(n=87), Australia (n=22) or Canada (n=45)]. All Japanese cases were
of Asian individuals; the 87 cases from the United States consisted
of 2 Asian, 4 African-American, 4 Hispanic and 77 Caucasian
individuals; the 22 Australian cases consisted of 1 Asian and 21
Caucasian individuals; and the 45 Canadian cases consisted of 15
Asian and 30 Caucasian individuals. For this study, the definition
of non-Asian individuals (n=136) consisted of Caucasian (n=128),
African-American (n=4) and Mexican-American (n=4) individuals. The
characteristics of the 302 cases are presented in Table I. Females and never-smokers occurred
significantly more frequently in the Asian group than in the
non-Asian group. Study permission was granted by the Institutional
Review Board of Okayama University (permission ref. Genome 173) and
written informed consent was obtained from all patients at each
collection site.
 | Table I.Patient characteristics and genetic
alterations in Asian and non-Asian groups. |
Table I.
Patient characteristics and genetic
alterations in Asian and non-Asian groups.
|
|
| Asian (n=166) | Non-Asian
(n=136) |
|
|---|
|
|
|
|
|
|
|---|
| Subsets | Total, n | n | % | n | % | P-value |
|---|
| Gender |
|
|
|
|
|
|
|
Female | 143 | 70 | 42.2 | 73 | 53.7 | 0.049 |
|
Male | 159 | 96 | 57.8 | 63 | 46.3 |
|
| Smoking status |
|
|
|
|
|
|
|
Never | 115 | 75 | 45.2 | 40 | 29.4 | 0.006 |
|
Ever | 187 | 91 | 54.8 | 96 | 70.6 |
|
| Stage |
|
|
|
|
|
|
| I | 188 | 109 | 65.7 | 79 | 58.1 | NS* |
| II | 35 | 15 |
9.0 | 20 | 14.7 |
|
|
III | 61 | 35 | 21.1 | 26 | 19.1 |
|
| IV | 13 | 3 |
1.8 | 10 |
7.4 |
|
| No
data |
5 |
4 |
2.4 |
1 |
0.7 |
|
| EGFR
mutation |
|
|
|
|
|
|
|
Mutation | 77 | 63 | 38.0 | 14 | 10.3 | <0.0001 |
|
Wild | 225 | 103 | 62.0 | 122 | 89.7 |
|
| EGFR
CNG |
|
|
|
|
|
|
|
CNG | 104 | 76 | 45.8 | 28 | 20.6 | <0.0001 |
| No
gain | 198 | 90 | 54.2 | 108 | 79.4 |
|
| Any EGFR
alterations |
|
|
|
|
|
|
|
Mutation or CNG | 143 | 107 | 64.5 | 36 | 26.5 | <0.0001 |
| None of
EGFR | 159 | 59 | 35.5 | 100 | 73.5 |
|
| KRAS
mutation |
|
|
|
|
|
|
|
Mutation | 69 | 22 | 13.3 | 47 | 34.6 | <0.0001 |
|
Wild | 233 | 144 | 86.7 | 89 | 65.4 |
|
| KRAS
CNG |
|
|
|
|
|
|
|
CNG | 17 | 11 |
6.6 | 6 |
4.4 | NS |
| No
gain | 285 | 155 | 93.4 | 130 | 95.6 |
|
| Any KRAS
alterations |
|
|
|
|
|
|
|
Mutation or CNG | 78 | 28 | 16.9 | 50 | 36.8 | 0.0001 |
| None of
KRAS | 224 | 138 | 83.1 | 86 | 63.2 |
|
Detection of gene mutations by direct
sequencing
The mutational status of exons 18 to 21 of the
EGFR gene and exon 2 of the KRAS gene was determined
by direct sequencing, as previously described (17,21,22).
Briefly, genomic DNA was amplified by conventional PCR using the
conditions stated in Table II. The
PCR products were incubated with exonuclease I and shrimp alkaline
phosphatase (GE Healthcare Life Sciences, Piscataway, NJ, USA), and
sequenced using the ABI PRISM® BigDye™ Terminator Cycle Sequencing
kit (PerkinElmer, Inc., Foster City, CA, USA). All sequence
variants were confirmed by sequencing the products of independent
polymerase chain reaction (PCR) in each direction.
 | Table II.Conditions for direct PCR sequencing
and quantitative PCR of gene copy number. |
Table II.
Conditions for direct PCR sequencing
and quantitative PCR of gene copy number.
| Gene | Primer sequence, 5′
to 3′ | Amplicons, bp | Tm, °C | Cycles, n |
|---|
| Direct
sequencing |
|
|
|
|
|
KRAS, exon 2 | F:
GTATTAACCTTATGTGTGACA | 222 | 55 | 37 |
|
| R:
GTCCTGCACCAGTAATATGC |
|
|
|
|
EGFR, exon 18 | F:
AGCATGGTGAGGGCTGAGGTGAC | 263 | 65 | 35 |
|
| R:
ATATACAGCTTGCAAGGACTCTGG |
|
|
|
|
EGFR, exon 19 | F:
CCAGATCACTGGGCAGCATGTGGCACC | 265 | 65 | 35 |
|
| R:
AGCAGGGTCTAGAGCAGAGCAGCTGCC |
|
|
|
|
EGFR, exon 20 | F:
GATCGCATTCATGCGTCTTCACC | 362 | 65 | 35 |
|
| R:
TTGCTATCCCAGGAGCGCAGACC |
|
|
|
|
EGFR, exon 21 | F:
TCAGAGCCTGGCATGAACATGACCCTG | 297 | 65 | 35 |
|
| R:
GGTCCCTGGTGTCAGGAAAATGCTGG |
|
|
|
| Gene copy
number |
|
KRAS | F:
CACCCTAGACAAGCAGCCAATA | – | 60 | 45 |
|
| R:
AAGCCCTGCCGCAAAAA |
|
|
|
|
EGFR | F:
CAAGGCCATGGAATCTGTCA | – | 60 | 45 |
|
| R:
CTGGAATGAGGTGGAGGAACA |
|
|
|
|
LINE-1 | F:
AAAGCCGCTCAACTACATGG | – | 60 | 45 |
|
| R:
TGCTTTGAATGCGTCCCAGAG |
|
|
|
Validation of gene copy number
alteration by quantitative (q)PCR assay
gEGFR and gKRAS were determined by
qPCR assay using Power SYBR® Green PCR Master Mix (Applied
Biosystems Life Technologies, Foster City, CA, USA), as previously
reported (17,22). LINE-1 was used as a reference
gene for all copy number analyses. The PCR conditions of each gene
are provided in Table II, and gene
dosages of EGFR, KRAS and LINE-1 were
calculated using the standard curve method. The relative copy
number of each sample was determined to compare the ratio of the
target gene and LINE-1 in each sample with the ratio in
human genomic DNA (EMD Millipore, Billerica, MA, USA) as a diploid
control. Based on our previous studies (17,22), CNG
was defined as values >3.
Statistical analysis
The primary endpoint of the present cross-sectional
study was to examine the ethnic differences (Asian vs. non-Asian)
in EGFR and KRAS alterations (mutations and CNGs) in
lung adenocarcinoma. To assess this, polynomial logistic regression
models adjusted for gender (female vs. male) and smoking status
(never vs. ever) were applied without any type of
EGFR/KRAS alteration as a reference group.
Cross-sectional odds ratios (ORs) and 95% confidence intervals
(CIs) were applied as a measure of association. Each OR indicates
how many times cases of Asian ethnicity are more likely to harbor
the specified pattern of alteration of EGFR/KRAS than
cases of non-Asian ethnicity. Fisher's exact test was used for
comparing the baseline characteristics of the Asian and non-Asian
groups. Exact 95% CIs were estimated with prevalence of each
combination of alteration. P<0.05 was defined as a threshold of
statistical significance. All the statistical analyses were
executed by STATA version 11 (StataCorp LP, College Station, TX,
USA).
Results
Mutations and CNGs of EGFR or KRAS and
clinical factors
mEGFR (EGFR mutation independent of
EGFR CNG), gEGFR (EGFR CNG independent of
EGFR mutation), mKRAS and gKRAS were present
in 26% (n=77), 34% (n=104), 23% (n=69) and 6% (n=17) of the 302
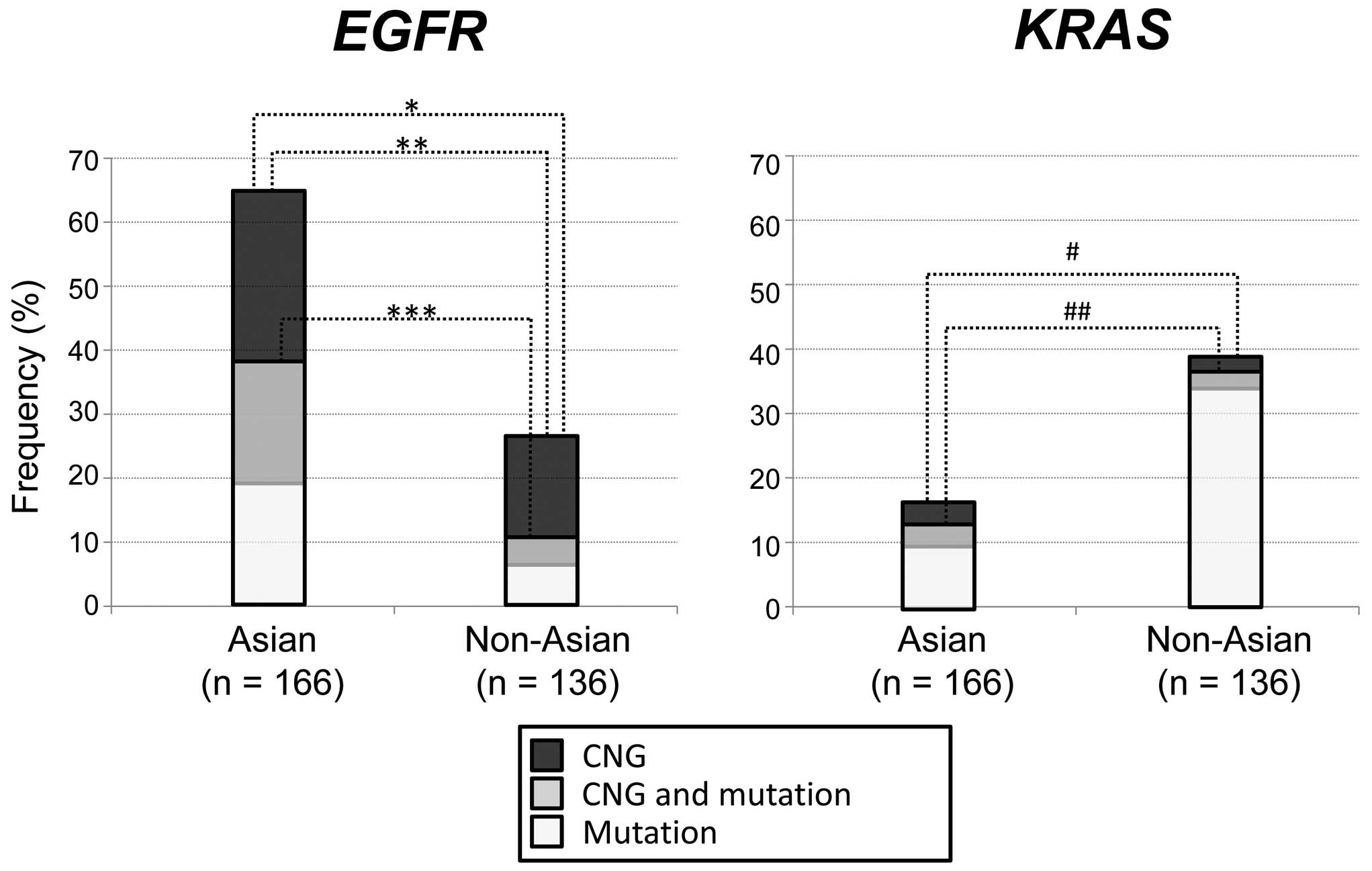
cases, respectively. mEGFR, gEGFR, mKRAS and
gKRAS were present in 38, 46, 13 and 7% of Asian individuals
(n=166) and 10, 21, 35 and 4% of non-Asian individuals (n=136),
respectively, indicating that CNGs were more frequently present
than mutations in EGFR but not in KRAS between the
two ethnic groups (Table I; Fig. 1). mEGFR (P<0.0001),
gEGFR (P<0.0001) or any EGFR alteration
(mEGFR or gEGFR; P<0.0001) were significantly more
frequent in Asian compared with non-Asian individuals. By contrast,
mKRAS (P<0.0001) and any KRAS alteration
(mKRAS or gKRAS; P=0.0001) were significantly more
frequent in non-Asian compared with Asian individuals (Table I; Fig.
1). With regard to other clinical factors, the never smoking
status was significantly associated with mEGFR (P<0.0001)
and gEGFR (P=0.046), whereas the presence of a smoking
history was significantly associated with mKRAS
(P<0.0001; Table III). The
female gender was a significant factor that was associated with
mEGFR (P=0.0005).
 | Table III.Association between EGFR and
KRAS alterations and characteristics in 302 lung
adenocarcinomas. |
Table III.
Association between EGFR and
KRAS alterations and characteristics in 302 lung
adenocarcinomas.
|
|
| mEGFR
(n=77) | gEGFR
(n=104) | mKRAS
(n=69) | gKRAS
(n=17) |
|---|
|
|
|
|
|
|
|
|---|
| Subsets | Total, n | n | % | P-value | n | % | P-value | n | % | P-value | n | % | P-value |
|---|
| Gender |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Female | 143 | 50 | 35.0 | 0.0005 | 42 | 29.4 | NS | 32 | 22.4 | NS | 4 |
2.8 | 0.048 |
|
Male | 159 | 27 | 17.0 |
| 62 | 39.0 |
| 37 | 23.3 |
| 13 |
8.2 |
|
| Ethnicity |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Asian | 166 | 63 | 38.0 | <0.0001 | 76 | 45.8 | <0.0001 | 22 | 13.3 | <0.0001 | 11 |
6.6 | NS |
|
Non-Asian | 136 | 14 | 10.3 |
| 28 | 20.6 |
| 47 | 34.6 |
| 6 |
4.4 |
|
| Smoking status |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Never | 115 | 57 | 49.6 | <0.0001 | 48 | 41.7 | 0.046 | 12 | 10.4 | <0.0001 | 4 |
3.5 | NS |
|
Smoker | 187 | 20 | 10.2 |
| 56 | 29.9 |
| 57 | 30.5 |
| 13 |
7.0 |
|
| Stage |
|
|
|
|
|
|
|
|
|
|
|
|
|
| I | 188 | 50 | 26.6 | NSa | 65 | 34.6 | NSa | 48 | 25.5 | NS | 11 |
5.9 | NSa |
| II | 35 | 11 | 31.4 |
| 12 | 34.3 |
| 6 | 17.1 |
| 0 |
0.0 |
|
|
III | 61 | 13 | 21.3 |
| 20 | 32.8 |
| 8 | 13.1 |
| 4 |
6.6 |
|
| IV | 13 | 2 | 15.4 |
| 4 | 30.8 |
| 4 | 30.8 |
| 1 |
7.7 |
|
| No
data |
5 | 1 | 20.0 |
| 3 | 60.0 |
| 3 | 60.0 |
| 1 | 20.0 |
|
Inter-association between mutations
and CNGs of EGFR and KRAS is retained between the two ethnic
groups
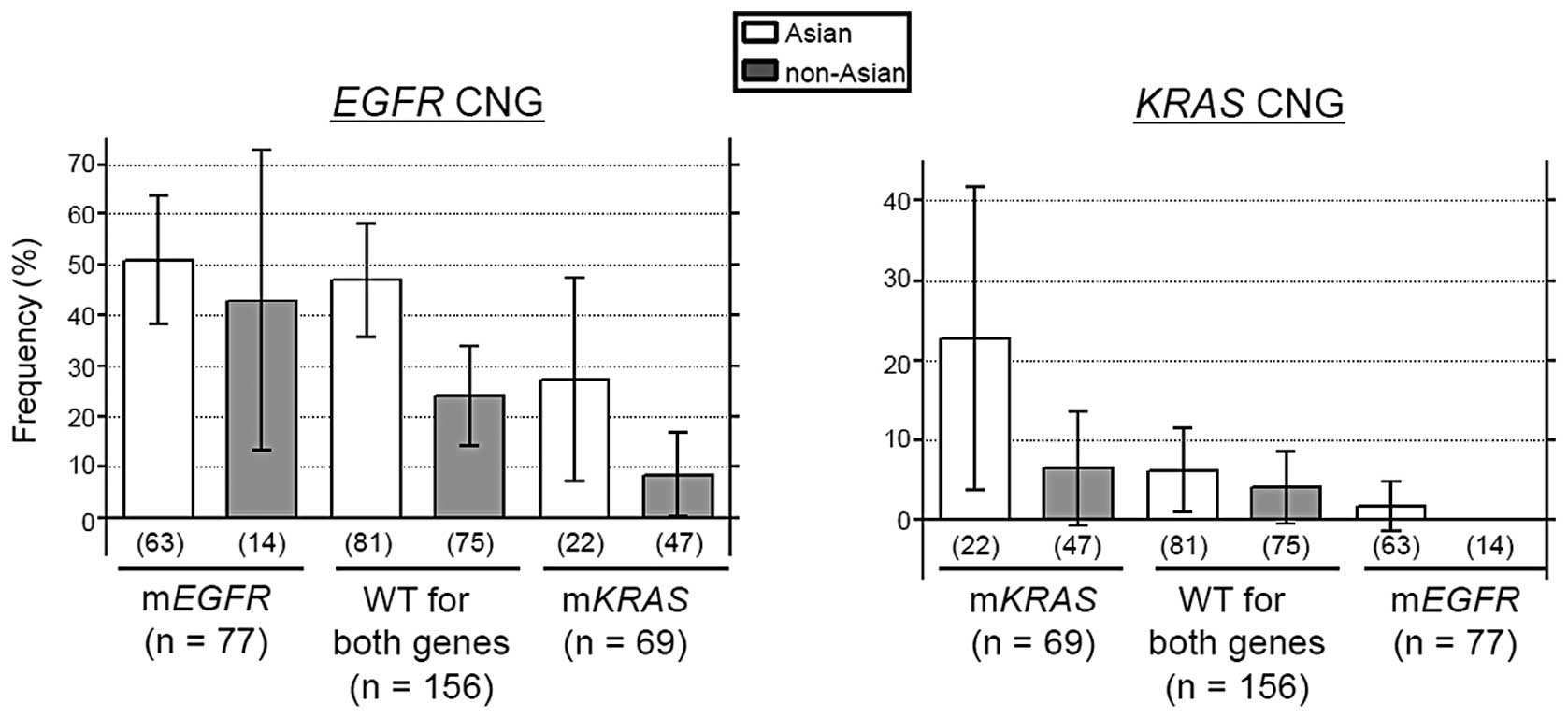
The present study evaluated the effect of ethnic
difference on the inter-association between mutations and CNGs of
the EGFR and KRAS genes by categorizing 302 cases
into three groups according to mutational status: i) mEGFR
(n=77), ii) mKRAS (n=69) and iii) wild-type for EGFR
and KRAS (n=156). gEGFR (Asian individuals, P=0.338;
non-Asian individuals, P=0.041) and gKRAS (Asian
individuals, P=0.007; non-Asian individuals, P=0.124) occurred
significantly more frequently in their respective mutant cases
(Fig. 2). Between the Asian and
non-Asian individuals, the frequencies of gEGFR and
gKRAS were lowest in the mKRAS and mEGFR
groups, respectively (Fig. 2).
mEGFR and mKRAS were completely mutually exclusive in
the two ethnic groups and any EGFR alterations (either
mEGFR or gEGFR) were almost exclusive with any
KRAS alterations (either mKRAS or gKRAS)
between the two ethnic groups (P=0.016 in Asians and P=0.004 in
non-Asians). These findings suggested that the inter-association
between the mutation and CNG of an identical gene and between
alterations of the EGFR and KRAS genes were retained
in the two ethnic groups.
Ethnic differences of EGFR and KRAS
alterations with respect to gender and smoking status
As gender and smoking status were other significant
factors associated with the frequency of the mutations and CNGs of
the EGFR and KRAS genes (Table III) and as the proportions of
females and never-smokers were significantly biased toward the
Asian group, a polynomial logistic regression model was performed
with adjustments for gender and smoking status; patients without
any EGFR/KRAS alterations were used as the reference
group (Table IV). According to the
polynomial logistic regression models, it was confirmed that all
types of EGFR alterations [mEGFR (P<0.001), gEGFR
(P=0.005), mgEGFR (mutation and CNG of EGFR gene;
P<0.001), any EGFR (P<0.001), mEGFR alone
(mEGFR without EGFR CNG; P<0.001) and gEGFR
alone (gEGFR without EGFR mutation; P=0.017)] were
significantly more frequent among Asian individuals compared with
among non-Asian individuals (ORs, 2.36–6.67). KRAS
alterations occurred less frequently among Asian individuals than
among non-Asian individuals, although a statistical significance
was not detected using the polynomial model.
 | Table IV.Odds ratios for Asian individuals
harboring EGFR or KRAS alterations. |
Table IV.
Odds ratios for Asian individuals
harboring EGFR or KRAS alterations.
| Genetic alterations
(EGFR/KRAS) | Asian, n (%) | Non-Asian, n
(%) | OR (95% CI) | P-value |
|---|
| None/None | 43 (25.9) | 56 (41.2) | 1.00
(Reference) |
|
| Mut alone/None | 31 (18.7) | 8 (5.9) | 4.94
(1.95–12.6) | <0.001 |
| Mut+CNG/None | 31 (18.7) | 6 (4.4) | 6.67
(2.48–17.9) | <0.001 |
| CNG alone/None | 33 (19.9) | 16 (11.8) | 2.43
(1.17–5.07) | 0.017 |
| None/Mut alone | 16 (9.6) | 40 (29.4) | 0.56
(0.27–1.16) | 0.118 |
| None/Mut+CNG | 0 (0.0) | 3 (2.2) | NE | NE |
| None/CNG alone | 0 (0.0) | 1 (0.7) | NE | NE |
| Mut+CNG/CNG | 1 (0.6) | 0 (0.0) | NE | NE |
| CNG/Mut | 1 (0.6) | 4 (2.9) | 0.34
(0.04–3.25) | 0.35 |
| CNG/Mut+CNG | 5 (3.0) | 0 (0.0) | NE | NE |
| CNG/CNG | 5 (3.0) | 2 (1.5) | 2.92
(0.53–16.1) | 0.219 |
| Mut or
CNG/None | 95 (57.2) | 30 (22.1) | 4.24
(2.29–7.86) | <0.001 |
| None/Mut or
CNG | 16 (9.6) | 44 (32.4) | 0.51
(0.25–1.03) | 0.059 |
| Mut/None | 62 (37.4) | 14 (10.3) | 4.76
(2.30–9.83) | <0.001 |
| CNG/None | 64 (38.6) | 22 (16.2) | 2.36
(1.30–4.28) | 0.005 |
| None/Mut | 16 (9.6) | 43 (31.6) | 0.53
(0.26–1.08) | 0.080 |
| None/CNG | 0 (0.0) | 3 (2.9) | NE | NE |
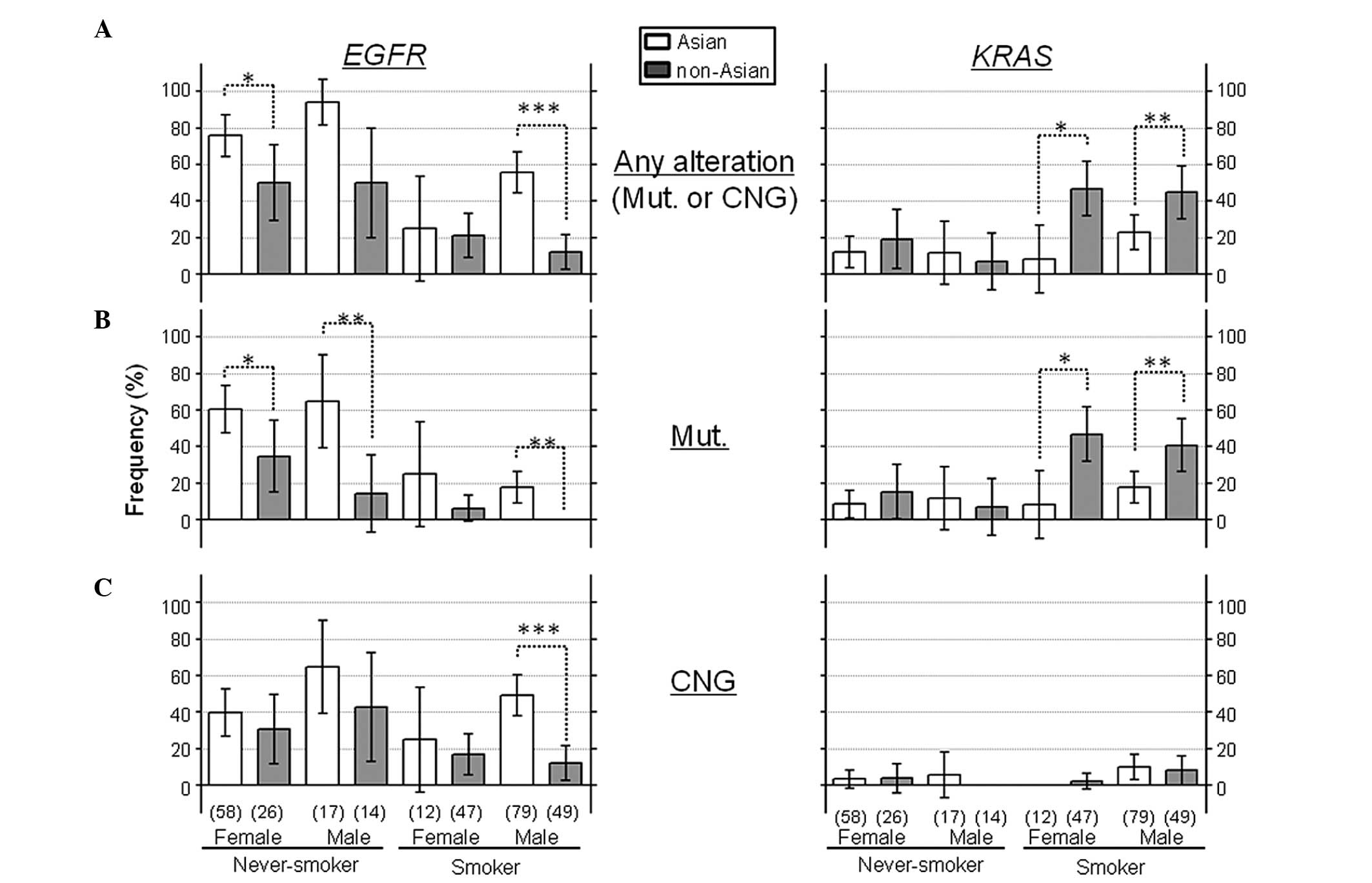
Additionally, the 302 cases were subcategorized into
8 groups according to gender, smoking status and ethnicity: i)
Asian never-smoker, female (A-NS-F; n=58), ii) Asian never-smoker,
male (A-NS-M; n=17), iii) Asian smoker, female (A-SM-F; n=12), iv)
Asian smoker, male (A-SM-M; n=79), v) non-Asian never-smoker,
female (NA-NS-F; n=26), vi) non-Asian never-smoker, female
(NA-NS-M; n=14), vii) non-Asian smoker, female (NA-SM-F; n=47), and
viii) non-Asian smoker, male (NA-SM-M; n=49). Each group was
compared, noting the effect of ethnicity on the frequencies of
these genetic alterations (Fig. 3).
mEGFR and gEGFR occurred more frequently in every
Asian group than in their corresponding non-Asian group,
independent of gender and smoking status, whereas mKRAS
occurred more frequently in non-Asians than in Asians among the
smoker groups, but this ethnic difference was not observed among
the never-smoker groups (Fig. 3).
Discussion
The present study investigated the impact of ethnic
differences on the genetic alterations of EGFR and/or
KRAS genes, and found that ethnic differences were
associated with the frequencies of these genetic alterations,
particularly EGFR alterations, even when gender and smoking
status were taken into consideration. Ethnic differences in the
frequencies of molecular alterations have been described in several
studies. We previously reported that the frequencies of the
aberrant methylation of CpG islands in certain tumor-suppressor
genes, such as MGMT and GSTP1, were different between
non-Asian populations (American and Australian cases) and Asian
populations (Japanese and Taiwanese cases) (23). A recent study that evaluated CNGs in
lung adenocarcinomas using a common high-resolution single
nucleotide polymorphism microarray, also reported that discrete
differences in copy number aberrations was present between
East-Asian and Western European individuals (chromosome 16p CNGs in
East-Asian individuals, and chromosome 19p losses in Western
European individuals) (24).
Oncogenes can be activated by mutations, CNGs and/or
translocations (11). Any one of
these genetic alterations is able to activate oncogenes, but
interactions among these alterations can occur. In fact, the
EGFR and KRAS genes are known to be activated by
activating mutations and CNGs, and the inter-association between
these genetic alterations have been investigated in previous
studies (15,17,18,25).
Although these studies have not revealed ethnic differences for
these genetic alterations, it has been reported that i) the
EGFR gene is more dominantly activated by CNGs than by
mutations, while the KRAS gene is more activated by
mutations than by CNGs; ii) mEGFR and mKRAS are
mutually exclusive; and iii) gEGFR and gKRAS occur
significantly more frequently among their respective mutant cases.
As a result of these findings, EGFR alterations
(mEGFR and/or gEGFR) may be almost exclusive of
KRAS alterations, as confirmed in the present study. This
study added novel insights into the inter-association between these
genetic alterations and ethnicity. The inter-association between
mutations and CNGs of the same gene, and between alterations of the
EGFR and KRAS genes, were similar in the Asian and
non-Asian groups: i.e., in each ethnic group, gEGFR and
gKRAS were significantly frequent among the respective
mutant cases, and EGFR alterations (mEGFR and/or
gEGFR) were exclusive of KRAS alterations. This fact
strongly suggests that the inter-association between CNG and
mutations in each gene is retained in Asian and non-Asian
ethnicities.
In the present study, DNA samples were collected
from four countries with mixed populations of different
ethnicities, such as Japanese, other Asian (non-Japanese),
Caucasian, African-American and Mexican-American. A total of 4
African-Americans were included in the non-Asian group, as
African-Americans have been reported to show similar frequencies of
mEGFR and mKRAS to Caucasians (26–28), and
it was confirmed that none of the 4 African-Americans harbored
mEGFR and that 2 harbored mKRAS. The mutations and
CNGs in 12 Asian patients with lung adenocarcinomas whose DNA
samples were obtained from Western countries were also determined.
It was confirmed that EGFR alterations were more frequent
than KRAS alterations among these Asians samples (data not
shown), as previously reported (29).
In addition, Asian patients with NSCLC who immigrated to Canada
from Asian countries reportedly showed a preferential response to
EGFR-TKI treatment (30). These lines
of evidence suggest that ethnic differences in the molecular
spectra of EGFR and KRAS are not affected by
environmental factors, and that ethnicity is an important factor
determining the molecular spectrum of lung adenocarcinoma.
In conclusion, the EGFR and KRAS
profiles in lung adenocarcinoma differ between Asian and non-Asian
populations, suggesting that ethnicity affects the molecular
characteristics of lung adenocarcinoma.
Acknowledgements
The authors would like to thank Dr. Makoto Suzuki
(Department of Thoracic Surgery, Graduate School of Medical
Sciences, Kumamoto University, Kumomoto, Japan) for providing
clinical DNA samples of Japanese patients from Chiba University.
This study was partly supported by the National Cancer Center
Research and Development Fund (grant no. 22700916), and a Lung
Cancer SPORE grant (P50 CA70907).
References
|
1
|
Paez JG, Jänne PA, Lee JC, et al: EGFR
mutations in lung cancer: Correlation with clinical response to
gefitinib therapy. Science. 304:1497–1500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lynch TJ, Bell DW, Sordella R, et al:
Activating mutations in the epidermal growth factor receptor
underlying responsiveness of non-small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rodenhuis S, van de Wetering ML, Mooi WJ,
Evers SG, van Zandwijk N and Bos JL: Mutational activation of the
K-ras oncogene. A possible pathogenetic factor in adenocarcinoma of
the lung. N Engl J Med. 317:929–935. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pao W, Wang TY, Riely GJ, et al: KRAS
mutations and primary resistance of lung adenocarcinomas to
gefitinib or erlotinib. PLoS Med. 2:e172005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Soda M, Choi YL, Enomoto M, et al:
Identification of the transforming EML4-ALK fusion gene in
non-small-cell lung cancer. Nature. 448:561–566. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stephens P, Hunter C, Bignell G, et al:
Lung cancer: Intragenic ERBB2 kinase mutations in tumours. Nature.
431:525–526. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Naoki K, Chen TH, Richards WG, Sugarbaker
DJ and Meyerson M: Missense mutations of the BRAF gene in human
lung adenocarcinoma. Cancer Res. 62:7001–7003. 2002.PubMed/NCBI
|
|
8
|
Pao W and Girard N: New driver mutations
in non-small-cell lung cancer. Lancet Oncol. 12:175–180. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shigematsu H and Gazdar AF: Somatic
mutations of epidermal growth factor receptor signaling pathway in
lung cancers. Int J Cancer. 118:257–262. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Suda K, Tomizawa K and Mitsudomi T:
Biological and clinical significance of KRAS mutations in lung
cancer: An oncogenic driver that contrasts with EGFR mutation.
Cancer Metastasis Rev. 29:49–60. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vogelstein B and Kinzler KW: Cancer genes
and the pathways they control. Nat Med. 10:789–799. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Weir BA, Woo MS, Getz G, et al:
Characterizing the cancer genome in lung adenocarcinoma. Nature.
450:893–898. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dahabreh IJ, Linardou H, Kosmidis P,
Bafaloukos D and Murray S: EGFR gene copy number as a predictive
biomarker for patients receiving tyrosine kinase inhibitor
treatment: a systematic review and meta-analysis in non-small-cell
lung cancer. Ann Oncol. 22:545–552. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dahabreh IJ, Linardou H, Siannis F,
Kosmidis P, Bafaloukos D and Murray S: Somatic EGFR mutation and
gene copy gain as predictive biomarkers for response to tyrosine
kinase inhibitors in non-small cell lung cancer. Clin Cancer Res.
16:291–303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cappuzzo F, Hirsch FR, Rossi E, et al:
Epidermal growth factor receptor gene and protein and gefitinib
sensitivity in non-small-cell lung cancer. J Natl Cancer Inst.
97:643–655. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hirsch FR, Varella-Garcia M, Cappuzzo F,
et al: Combination of EGFR gene copy number and protein expression
predicts outcome for advanced non-small-cell lung cancer patients
treated with gefitinib. Ann Oncol. 18:752–760. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Soh J, Okumura N, Lockwood WW, et al:
Oncogene mutations, copy number gains and mutant allele specific
imbalance (MASI) frequently occur together in tumor cells. PLoS
ONE. 4:e74642009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sasaki H, Hikosaka Y, Kawano O, Moriyama
S, Yano M and Fujii Y: Evaluation of Kras gene mutation and copy
number gain in non-small cell lung cancer. J Thorac Oncol. 6:15–20.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wagner PL, Perner S, Rickman DS, et al: In
situ evidence of KRAS amplification and association with increased
p21 levels in non-small cell lung carcinoma. Am J Clin Pathol.
132:500–505. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gandhi J, Zhang J, Xie Y, et al:
Alterations in genes of the EGFR signaling pathway and their
relationship to EGFR tyrosine kinase inhibitor sensitivity in lung
cancer cell lines. PLoS One. 4:e45762009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shigematsu H, Lin L, Takahashi T, et al:
Clinical and biological features associated with epidermal growth
factor receptor gene mutations in lung cancers. J Natl Cancer Inst.
97:339–346. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yamamoto H, Shigematsu H, Nomura M, et al:
PIK3CA mutations and copy number gains in human lung cancers.
Cancer Res. 68:6913–6921. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Toyooka S, Maruyama R, Toyooka KO, et al:
Smoke exposure, histologic type and geography-related differences
in the methylation profiles of non-small cell lung cancer. Int J
Cancer. 103:153–160. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Broet P, Dalmasso C, Tan EH, et al:
Genomic profiles specific to patient ethnicity in lung
adenocarcinoma. Clin Cancer Res. 17:3542–3550. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ichihara S, Toyooka S, Fujiwara Y, et al:
The impact of epidermal growth factor receptor gene status on
gefitinib-treated Japanese patients with non-small-cell lung
cancer. Int J Cancer. 120:1239–1247. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cote ML, Haddad R, Edwards DJ, et al:
Frequency and type of epidermal growth factor receptor mutations in
African Americans with non-small cell lung cancer. J Thorac Oncol.
6:627–630. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Reinersman JM, Johnson ML, Riely GJ, et
al: Frequency of EGFR and KRAS mutations in lung adenocarcinomas in
African Americans. J Thorac Oncol. 6:28–31. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Leidner RS, Fu P, Clifford B, et al:
Genetic abnormalities of the EGFR pathway in African American
patients with non-small-cell lung cancer. J Clin Oncol.
27:5620–5626. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tsao AS, Tang XM, Sabloff B, et al:
Clinicopathologic characteristics of the EGFR gene mutation in
non-small cell lung cancer. J Thorac Oncol. 1:231–239.
2006.PubMed/NCBI
|
|
30
|
Ho C, Murray N, Laskin J, Melosky B,
Anderson H and Bebb G: Asian ethnicity and adenocarcinoma histology
continues to predict response to gefitinib in patients treated for
advanced non-small cell carcinoma of the lung in North America.
Lung Cancer. 49:225–231. 2005. View Article : Google Scholar : PubMed/NCBI
|