Introduction
Lung cancer is the leading cause of cancer-related
mortality worldwide. In 2012, World Health Organization statistical
data reported that the numbers of global new cases and mortalities
from lung cancer were 1,824,701 and 1,589,800, respectively
(1). The majority of lung cancers are
diagnosed at an advanced stage, so the 5-year survival rate of lung
cancer patients is <15% (1). Based
on the histological type, lung cancers are divided into two
subtypes: Small cell lung cancer and non-small cell lung cancer
(NSCLC). NSCLC accounts for >85% cases, and can be further
classified into three main subtypes: Squamous cell carcinoma,
adenocarcinoma and large cell carcinoma (2–4).
Previous studies have identified a large quantity of
driver genes in lung carcinogenesis, genetic aberrations of which
can promote tumor cell proliferation, survival, angiogenesis,
invasion and metastasis, thus driving the development of lung
cancer (5). For example, epidermal
growth factor receptor (EGFR) mutations (6), EML4/ALK fusion genes (7), RAS mutations, BRAF mutations and
PI3K/mTOR mutations are frequently observed in lung cancer tissues,
and are predictive of response to targeted therapies (8).
The majority of lung cancers are sporadic, and
familial cases are extremely rare. Unlike sporadic cancer, germline
mutations may drive carcinogenesis in familial cases. For example,
functional mutations in BRCA1/2 genes may cause familial breast or
ovarian cancer. The family members who carry BRCA1/2 mutations have
a 70–80% possibility of developing cancers (9,10). Another
example in colorectal cancer demonstrates that DNA mismatch repair
gene MSH2 and MLH1 mutations carriers have a high risk of
developing colorectal cancer in their 40s (11,12).
To date, only three familial lung cancer cases have
been studied. In one study, it was reported that a lung squamous
cancer patient carried the EGFR R776H germline mutation, and that
the patient's daughter, also a lung cancer patient, carried the
same alteration (13). In another
study, 3/6 individuals in a Japanese family developed lung
adenocarcinoma. The germline EGFR V843I mutation was identified in
all the lung adenocarcinoma-affected members of this family
(14). In a study by Yamamoto et
al, it was found that mutations in HER2 may be the causative
variants of familial lung adenocarcinomas (15). The molecular mechanism of familial
lung cancer remains largely unknown.
The present study recruited a family of Chinese
descent in which multiple members were diagnosed with lung squamous
carcinoma. To find the causative mutations, whole exome sequencing
was conducted using a peripheral blood sample of one lung squamous
carcinoma patient, and certain variants were validated in more
samples. This study provides the basis for further studies, in
which further hereditary lung cancer members will be sequenced.
Materials and methods
DNA extraction
The selected family were from Northwest China,
Shaanxi province. All individuals were diagnosed with lung squamous
cell carcinoma and treated at the Department of Thoracic Surgery,
Second Affiliated Hospital, Medical School, Xi'an Jiaotong
University (Xi'an, China) in August 2011. Peripheral blood samples
were collected from 4 individuals, 3 of whom were lung cancer
patients, and 1 of whom had no malignant disease history. DNA was
extracted using a QIAamp DNA Mini kit (cat. no. 51106) according to
the manufacturer's instruction (Qiagen, Hilden, Germany).
Histological examination was performed by hematoxylin and eosin
staining of the bronchoscopic biopsies, needle aspiration biopsies
and surgical specimens.
The experiments were undertaken with the
understanding and written consent of each subject, and the study
conforms with The Code of Ethics of the World Medical Association
(Declaration of Helsinki). This study was performed in accordance
with the ethical standards of the Ethics Committee of the Second
Affiliated Hospital, Medical School, Xi'an Jiaotong University
(Xi'an, China).
Sanger sequencing
Specific primers were designed using Primer 5
(Premier Biosoft International, Palo Alto, CA, USA; Table I). The PCR materials included 0.25 mM
dNTPs, 1 unit of HotStarTaq (Qiagen), 1X HotStarTaq buffer (Qiagen)
and 0.4 µM primer. PCR conditions were 33 cycles of 95°C for 50
sec, 61°C for 40 sec and 72°C for 60 sec, following initial
denaturation at 95°C for 5 min. PCR products were purified using
SAP, and used as templates for the sequencing reaction using Big
Dye v3.1 (Applied Biosystems Life Technologies, Foster City, CA,
USA). Subsequent products were run on the ABI PRISM 3130×1 Genetic
Analyzer (Applied Biosystems Life Technologies). Electropherograms
were analyzed using Sequence Analysis Software version 5.2 (Applied
Biosystems Life Technologies).
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Primer | Sequence |
|---|
| EGFRrs2227983-F |
GTGACCCACTCTGTCTCCG |
| EGFRrs2227983-R |
GAAAACCCAAAACCTCCAA |
| NOTCH4-F |
GGCTTCCTGGGTGAGACG |
| NOTCH4-R |
TTGGTTGTGGGTAAGTGGATT |
| MAD1L18-F |
ATGACCAGAGCAGGACCAA |
| MAD1L1-R |
CACAGGGGAGGACAAACC |
| TNFRSF10A-F |
CTGATGAAATGGGTCAACAAA |
| TNFRSF10A-R |
TCCCGAGTAGCCGAGAATA |
| CDKN1Ars76-F |
TGTGAGGTAGATGGGAGCG |
| CDKN1Ars76-R |
GGGGAGGATTTGACGAGTG |
| CDKN1Ars18-F |
GTGTCTAATCTCCGCCGTGAC |
| CDKN1Ars18-R |
GCCTGCCTCCTCCCAACT |
| CDKN1Ars10-F |
GCGTCCTCTTCTTCTTGGC |
| CDKN1Ars10-R |
AAAGGAGAACACGGGATGAG |
| TP53rs1042522-F |
ACCTGTGGGAAGCGAAA |
| TP53rs1042522-R |
GGAAGGGACAGAAGATGACA |
| ATM-F |
AATCTCCTCCTTTCTGCTGC |
| ATM-R |
CCACTCTGCCAAGTATTATGCTAT |
| NBN-F |
GGTTACCTCAGTGCCATTTAC |
| NBN-R |
GCAGTGACCAAAGACCGA |
| XPC-F |
TGGCAGCGGCTCTGATTT |
| XPC-R |
GGCTCTGGTATGGTCTCAAGGTC |
| PDE4DIP-F |
CCCATGACTGAAGAGTTGCT |
| PDE4DIP-R |
CATCTCCCAAGTCCTACAAAG |
| CLTCL1-F |
CAGAAAGATGCTTACCACCC |
| CLTCL1-R |
GCTGCCTGATACTCACCGA |
| MLL3-F |
TTTCATCATCAGCAGACATAAGC |
| MLL3-R |
CACCCTACCTGTTTGGACC |
| ARID2-F |
TTCTTTGCTTTCCATCCTTG |
| ARID2-R |
CCTCCGTTTTACTCCTACCA |
| CLTCL1-F |
TTATTGTTGAAGATAGAAGCAGTT |
| CLTCL1-R |
AAAGTTAGGAGGCAGGTAGG |
| GNAS-F |
CGGAGCCTTCAGTGGTG |
| GNAS-R |
CTGTCGTCCCCAAATCCT |
| NACA-F |
CAAAGGTCCCTCTACCATCTC |
| NACA-R |
CAACAGAAGAAACGGGCAT |
| POT1-F |
ATTTTAGGCAAGAAGTTCCACA |
| POT1-R |
TTGAAAGCGGGAGAATACC |
| TFRC-F |
TTCCTCAAGCCAAATAATGC |
| TFRC-R |
CAAAGGCATAGATAATGTCAGC |
| NIN-F1 |
GCTCTGGGATTGAAGGGAC |
| NIN-R1 |
CAGGCTGGAGTACAGTGGC |
Whole exome sequencing
Whole exome sequencing was performed on the
peripheral blood sample of the proband. Firstly, to construct a
library, whole exome DNA from the patient's whole blood was treated
using Agilent SureSelect Human All ExonV5 kits following the
manufacturer's instructions (Agilent Technologies Inc., Santa
Clara, CA, USA). Subsequent to the quality test, the qualified
library was sequenced as 100-bp paired-end reads on an Illumina
Hiseq 2000 platform (Illumina, San Diego, CA, USA) according to the
manufacturer's instructions.
Data analysis
For whole exome sequencing, clean data was obtained
after filtering the reads of low quality (reads with adapter
sequence, reads with proportion of N >10% and reads with low
quality base numbers of >5). Burrows-Wheeler transform methods
(16) were adopted to map these reads
in a human reference (UCSC hg19). Next, the Picard and Genome
Analysis Toolkit (GATK) methods (17,18) were
adopted for duplicate removal, local realignment and base quality
recalibration. Finally, the GATK Unified Genotyper (version 3.0;
Broad Institute, Cambridge, MA, USA) was used for single nucleotide
variation (SNV)/InDel annotation.
Variants were annotated using the ANNOVAR software
tool (http://www.openbioinformatics.org/annovar/).
Annotations for function (exonic, intronic and untranslated
region), reference genes, exonic function (synonymous,
non-synonymous, stop-gain, frameshift and unknown), amino acid
changes, 1000 Genomes Project data and dbSNP reference number were
performed.
The whole exome sequencing generated large volumes
of data, and several filtering criteria were applied to the data
set. Firstly, variants with a low quality score (depth <20 or
genotype quality <20) were filtered. Secondly, the variants with
a reported frequency of >0.01 were filtered. Thirdly, synonymous
changes were removed, taking only the protein-altering
variants.
Results
Description of the pedigree
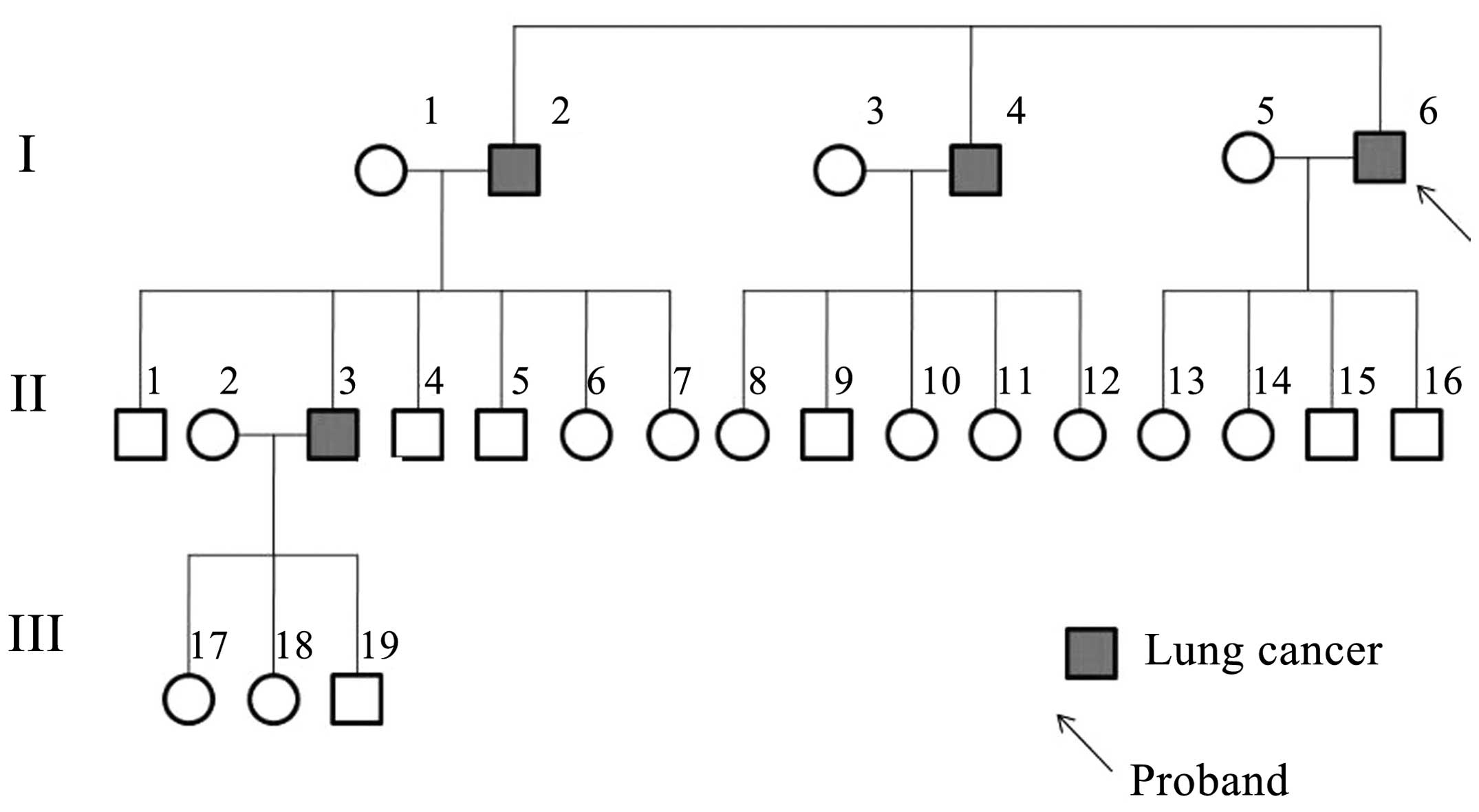
The lung cancer family were from Northwest China,
Shaanxi province, and four members of the family were diagnosed
with lung cancer (Fig. 1). The
proband was a 65-year-old male who presented with a tumor in the
inferior lobe of the right lung. Histological examination revealed
individual cell keratinization, as well as intercellular bridge and
squamous pearl formation, which resulted in a diagnosis of lung
squamous cell carcinoma, stage IIIa (T3N2M0), according to the
National Comprehensive Cancer Network TNM staging system (19). The patient was a heavy smoker, with
1.6 pack-year history of >10 years. Following lobectomy by
thoracotomy with lymph node dissection, the patient underwent
chemotherapy (120 mg/m2 cisplatin day 1, and 30
mg/m2 vinorelbine days 1 and 8 every 21 days for 4
cycles). A good response to chemotherapy was recorded. The proband
had suffered from no respiratory or malignant diseases prior to the
lung cancer diagnosis. Two elder brothers of the proband, who were
also smokers, had succumbed to lung squamous carcinoma metastasis
at the ages of 71 and 69 years old, respectively. One of the
proband's nephews had been diagnosed with lung squamous cancer at
the age of 51. All the lung cancer cases in the family were
histologically confirmed. Other members in the family had no
history of tumors or respiratory diseases. Peripheral blood samples
were collected from four family members, including three of the
cancer patients (I4, I6 and II3) and one healthy family member
(II1). II1 was the only surviving healthy control who was elder
than at least one of the lung cancer patients (Fig. 1).
Whole exome sequencing
Whole exome sequencing was conducted using the
peripheral blood sample of the proband, obtaining ~2.0 Gb of data
and achieving an average of 60x depth for each targeted base, with
98.3% of the exomic positions covered >10x. In all, 41,210 SNVs
and 3,299 InDels were identified. As described in the Materials and
methods section, 2,845 variations, including SNVs and InDels, were
removed due to a low quality score.
Sanger sequencing technology was applied to validate
the variants identified by high throughput sequencing. In all, 10
variants were selected randomly for testing. The sequence results
by Sanger sequencing confirmed the variants identified by whole
exome sequencing.
Candidate gene validation
To identify the causative variants, the no reported
alterations, or the alterations with low frequency (<=0.01, or
no reported frequency), and the alterations which were predicted to
alter the protein product (non-synonymous SNVs, splice-site
mutations and InDels) were selected. In all, 975 SNVs and 1,052
InDels in 767 genes satisfied the aforementioned criteria.
Next, the candidate alterations were further
restricted to 11 variants, which were located in genes with
definite functions and recorded in the COSMIC driver gene database
(http://cancer.sanger.ac.uk/cosmic;
Table II) (20).
 | Table II.Variants for validation. |
Table II.
Variants for validation.
| Gene | Ref. | Alt. | Amino acid
change | Mutation type | 1000G MAF | DP | GQ |
|---|
| ARID2 | A | G |
NM_152641:exon14:c.A1759G:p.S587G | Non-synonymous
SNV | 0.01 | 69 | 99 |
| GNAS | A | G |
NM_080425:exon1:c.A484G:p.M162V | Non-synonymous
SNV |
0.0014 | 116 | 99 |
| MLH1 | T | A |
NM_001167619:exon11:c.T428A:p.V143D | Non-synonymous
SNV | 0.01 | 83 | 99 |
| NACA | T | C |
NM_001113203:exon3:c.A1247G:p.K416R | Non-synonymous
SNV | 0.01 | 105 | 99 |
| NIN | G | A |
NM_016350:exon28:c.C3856T:p.R1286C | Non-synonymous
SNV |
0.0014 | 22 | 99 |
| POT1 | C | G |
NM_001042594:exon13:c.G835C:p.D279H | Non-synonymous
SNV |
0.0027 | 70 | 99 |
| TFRC | G | C |
NM_001128148:exon6:c.C634G:p.L212V | Non-synonymous
SNV | 0.01 | 273 | 99 |
| CLTCL1 | G | T |
NM_001835:exon8:c.C1342A:p.L448I | Non-synonymous
SNV | 0.01 | 61 | 99 |
| PDE4DIP | GT | G |
NM_001002811:exon2:c.1218delA:p.E406fs | Frameshift
deletion | – | 169 | 99 |
| CLTCL1 | A | AC |
NM_001835:exon23:c.3601_3602insG:p.V1201fs | Frameshift
insertion | – | 60 | 99 |
| MLL3 | G | GT |
NM_170606:exon14:c.2447_2448insA:
p.Y816_I817delinsX | Stop-gain SNV | – | 373 | 99 |
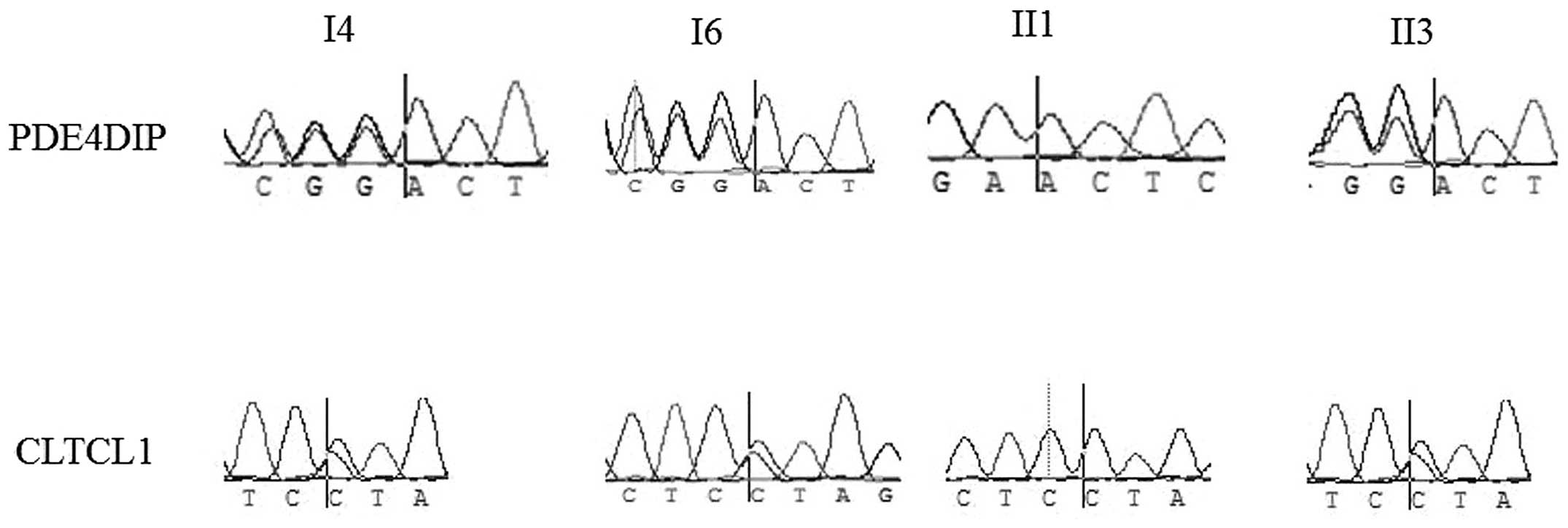
Subsequently, Sanger sequencing was conducted using
four peripheral blood samples from this family, from the three
patients and one non-cancer family member (II1), to validate the
variations in the aforementioned genes. As shown in Fig. 2, for the CLTCL1 L448I mutation, the
genotypes of the three affected individuals were heterozygous,
while that of the healthy man was homozygous wild-type. For the
PDE4DIPE406fs mutation, the three patients carried heterozygous
frameshift mutations, while the healthy individual carried the
homozygous wild-type. For variations in other genes, namely ARID2,
CLTCL1, GNAS, MLH1, NACA, NIN, POT1, TFRC, PDE4DIP and MLL3, the
variations did not segregate well.
Discussion
The present study recruited a Chinese family in
which four members had developed lung cancer, and is the first time
a study of Chinese familial lung cancer has been reported. Lung
cancer is one of the most common cancer types all over the world,
and a number of previous studies have identified a large quantity
of lung cancer driver genes, such as EGFR, Her2, AKT1, NRAS,
PIK3CA, BRAF, ALK-fusion and RET-fusion (5–8). Somatic
mutations of these genes have been recurrently identified in lung
cancer samples (5–8). However, unlike in sporadic lung cancer,
the molecular mechanisms behind inherited lung cancer remains
largely unknown.
Previous studies reported the germline mutations of
EGFR and HER2 as driver mutations in familial lung cancer patients
(13–15). In the present study, whole exome
sequencing identified 8 SNVs in the EGFR gene. However, 7 of these
were synonymous, and 1 was a common variant with a frequency of
>0.2. No variants that cause amino acid or splice changes were
identified in the HER2 gene. The study also analyzed mutations in
other inherited cancer-related genes, namely BRCA1/2, MSH2 and
MLH1, but found no non-synonymous mutations in these genes. Thus,
an unreported causative variation may drive lung cancer development
in this family.
In this study, whole exome sequencing using a
peripheral blood sample of one lung cancer member of the family
totally identified >2,000 unreported or low frequency
non-synonymous variants (975 SNVs and 1,052 InDels). Alterations
were validated in 10 definitive cancer driver genes, and two
plausible candidate genes, PDE4DIP and CLTCL1, were identified. For
the PDE4DIP gene, all the patients carried a frameshift variation,
while the non-cancer family member did not. The PDE4DIP protein
acts as an anchor phosphodiesterase, localized in the
Golgi/centrosome region of the cell. The protein is able to
interact with a phosphodiesterase superfamily protein member
(21,22). Functional mutations of PDE4DIP have
been found to be associated with several disorders. In the study by
DeWan et al, it was found that one mutation, I303L, in
PDE4DIP may be the causative variant in hereditary asthma (23). CLTCL1 is a member of the clathrin
family, which plays essential roles in intracellular traffic and
centrosomal stabilization (24).
Mutations or the abnormal expression of CLTCL1 have been reported
to be associated with breast cancer, meningioma and pulmonary valve
stenosis (25). Further functional
studies are required to verify whether the mutations in the
aforemenioned two genes cause functional change. Other variants in
the ARID2, GNAS, MLH1, NACA, NIN, POT1, TFRC and MLL3 genes did not
segregate perfectly with lung cancer. It is also possible that one
or several alterations that were not validated are the causative
mutations for this familial case.
Overall, the present study focused on a Chinese
family in which four members had developed lung cancer. Whole exome
sequencing was conducted using the peripheral blood sample of the
proband, and 11 variants were validated in more samples. Two
potentially functional variants were identified in two genes,
PDE4DIP and CLTCL1. Although the present study has certain
limitations, this study provides useful sources for the further
study of hereditary lung cancer.
References
|
1
|
World Health Organization GLOBOCAN 2012, .
Estimated cancer incidence, mortality and prevalence worldwide in
2012. http://globocan.iarc.fr/Default.aspxAccessed.
October 10–2014.
|
|
2
|
Suh JH: Current readings: Pathology,
prognosis, and lung cancer. Semin Thorac Cardiovasc Surg. 25:14–21.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Detterbeck FC, Postmus PE and Tanoue LT:
The stage classification of lung cancer: Diagnosis and management
of lung cancer, 3rd ed: American College of Chest Physicians
evidence-based clinical practice guidelines. Chest. 143
(Suppl):e191S–e210S. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
VanSchil PE, Sihoe AD and Travis WD:
Pathologic classification of adenocarcinoma of lung. J Surg Oncol.
108:320–326. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kris MG, Johnson BE, Kwiatkowski DJ, et
al: Identification of driver mutations in tumor specimens from
1,000 patients with lung adenocarcinoma: The NCI's lung cancer
mutation consortium (LCMC). J Clin Oncol. 29:CRA75062011.
|
|
6
|
Palmer JD, Zaorsky NG, Witek M and Lu B:
Molecular markers to predict clinical outcome and radiation induced
toxicity in lung cancer. J Thorac Dis. 6:387–398. 2014.PubMed/NCBI
|
|
7
|
Ulivi P, Zoli W, Capelli L, Chiadini E,
Calistri D and Amadori D: Target therapy in NSCLC patients:
Relevant clinical agents and tumour molecular characterisation. Mol
Clin Oncol. 1:575–581. 2013.PubMed/NCBI
|
|
8
|
Oxnard GR, Binder A and Jänne PA: New
targetable oncogenes in non-small-cell lung cancer. J Clin Oncol.
31:1097–1104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wooster R, Bignell G, Lancaster J, Swift
S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C and Micklem G:
Identification of the breast cancer susceptibility gene BRCA2.
Nature. 378:789–792. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Miki Y, Swensen J, ShattuckEidens D,
Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM,
Ding W, et al: A strong candidate for the breast and ovarian cancer
susceptibility gene BRCA1. Science. 266:66–71. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fishel R, Lescoe MK, Rao MR, Copeland NG,
Jenkins NA, Garber J, Kane M and Kolodner R: The human mutator gene
homolog MSH2 and its association with hereditary nonpolyposis colon
cancer. Cell. 75:1027–1038. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bronner CE, Baker SM, Morrison PT, Warren
G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A,
et al: Mutation in the DNA mismatch repair gene homologue hMLH1 is
associated with hereditary non-polyposis colon cancer. Nature.
368:258–261. 1994. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
van Noesel J, van der Ven WH, van Os TA,
Kunst PW, Weegenaar J, Reinten RJ, Kancha RK, Duyster J and van
Noesel CJ: Activating germline R776H mutation in the epidermal
growth factor receptor associated with lung cancer with squamous
differentiation. J Clin Oncol. 31:e161–e164. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ohtsuka K, Ohnishi H, Kurai D, Matsushima
S, Morishita Y, Shinonaga M, Goto H and Watanabe T: Familial lung
adenocarcinoma caused by the EGFR V843I germ-line mutation. J Clin
Oncol. 29:e191–e192. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamamoto H, Higasa K, Sakaguchi M, Shien
K, Soh J, Ichimura K, Furukawa M, Hashida S, Tsukuda K, Takigawa N,
et al: Novel germline mutation in the transmembrane domain of HER2
in familial lung adenocarcinomas. J Natl Cancer Inst.
106:djt3382014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li H and Durbin R: Fast and accurate
long-read alignment with Burrows-Wheeler transform. Bioinformatics.
26:589–595. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The genome analysis toolkit: A MapReduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
DePristo MA, Banks E, Poplin R, Garimella
KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA,
Hanna M, et al: A framework for variation discovery and genotyping
using next-generation DNA sequencing data. Nat Genet. 43:491–498.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ettinger DS, Akerley W, Bepler G, et al:
NCCN Non-Small Cell Lung Cancer Panel Members: Non-small cell lung
cancer. J Natl Compr Canc Netw. 8:740–801. 2010.PubMed/NCBI
|
|
20
|
Forbes SA, Bindal N, Bamford S, et al:
COSMIC: mining complete cancer genomes in the Catalogue of Somatic
Mutations in Cancer. Nucleic Acids Res. 39:945–950. 2011.
View Article : Google Scholar
|
|
21
|
Wong KA, Wilson J, Russo A, et al:
Intersectin (ITSN) family of scaffolds function as molecular hubs
in protein interaction networks. PLoS One. 7:e360232012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vinayagam A, Stelzl U, Foulle R, et al: A
directed protein interaction network for investigating
intracellular signal transduction. Sci Signal. 4:rs82011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
DeWan AT, Egan KB, Hellenbrand K,
Sorrentino K, Pizzoferrato N, Walsh KM and Bracken MB: Whole-exome
sequencing of a pedigree segregating asthma. BMC Med Genet.
13:952012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu SH, Towler MC, Chen E, Chen CY, Song
W, Apodaca G and Brodsky FM: A novel clathrin homolog that
co-distributes with cytoskeletal components functions in the
trans-Golgi network. EMBO J. 20:272–284. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sens-Abuázar C, Napolitano E, Ferreira E,
Osório CA, Krepischi AC, Ricca TI, Castro NP, da Cunha IW, Maciel
Mdo S, Rosenberg C and Brentani MM: Down-regulation of ANAPC13 and
CLTCL1: Early events in the progression of preinvasive ductal
carcinoma of the breast. Transl Oncol. 5:113–123. 2012. View Article : Google Scholar : PubMed/NCBI
|