Introduction
Esophageal cancer is one of the seven leading causes
of cancer-related mortality and is highly malignant (1). China has the highest incidence of
esophageal cancer worldwide, and more specifically esophageal
squamous cell carcinoma, with the mortality rate associated with
this cancer ranking fourth among malignant tumors (2). Atypical early symptoms, middle-to-late
stage diagnosis, low treatment remission rates and high local
recurrence rates all contribute to the poor prognosis of patients
with esophageal cancer. The development and incidence of esophageal
cancer involves a multi-factor, multi-step and multi-stage process.
The necessary strategies to improve the prognosis and survival
rates in patients with esophageal cancer require early discovery,
diagnosis and treatment, which rely on studying and exploring the
factors that influence the prognosis of esophageal cancer.
The P53 gene displays the highest correlation with
human types of cancer thus far. The past decade has witnessed three
shifts in the understanding of the association between P53 and
cancer, starting from P53 as a protein antigen to P53 as a
cancer-associated gene, and finally, to P53 as a tumor-suppressor
gene (3). This last advancement arose
from the identification of an important dominant-negative mutated
P53 gene product acting as an oncogene alleviating the normal tumor
suppressor function of wild-type P53 (3). The human Cox-2 gene is located on
chromosome 1q25.2-q25.3 and participates in the occurrence and
development of tumors by promoting cell proliferation, restraining
cell apoptosis, promoting angiogenesis and suppressing immune
functions (4). The aims of the
present study were to assess the P53 and Cox-2 expression levels in
esophageal cancer and to analyze the correlation between P53 and
Cox-2 co-expression and the prognosis of esophageal cancer.
Materials and methods
Clinical data
Tumor samples from 195 patients (150 men and 45
women, aged 34–83 years, with a median age of 62 years) diagnosed
with esophageal cancer and who underwent radical surgery at
Changzhou First People's Hospital (Changzhou, China) between May
2010 and December 2011 were studied. The present study was approved
by the Institutional Review Board of Soochow University (Changzhou,
China), according to the Declaration of Helsinki. Data regarding
age, demographics, tumor location, staging, pathology, adjuvant
radiotherapy and survival outcomes were obtained with the written
informed consent of each patient. All specimens were associated
with a definite pathological immunohistochemical report and
detailed follow-up and prognosis data. Of these 195 cases, 194 were
identified as squamous carcinoma and 1 case as adenosquamous
carcinoma. According to the seventh edition of the international
TNM staging criteria of esophageal cancer (5): 11 cases were in stage IA; 11, in stage
IB; 33, in stage IIA; 58 in stage IIB; 41, in stage IIIA; 15, in
stage IIIB; and 26 in stage IIIC.
Immunohistochemical analysis
The archived formaldehyde-fixed paraffin-embedded
esophageal cancer specimens were serially cut into 4-µm slices and
stained using the two-step Envision Immunochemistry kit (Dako,
Glostrup, Denmark). Validated breast cancer specimen sections were
used as a positive control, and phosphate-buffered saline was used
instead of primary antibodies as a negative control. Monoclonal
anti-P53 antibody was obtained from Fuzhou Maixin Biotechnology
Development Co., Ltd. (Fuzhou, China), and monoclonal anti-Cox-2
antibody was obtained from Beijing Zhongshan Jinqiao Biotechnology
Co. Ltd. (Beijing, China).
Evaluation standards for results
All staining results were analyzed by two
double-blinded pathological evaluations. Tan nuclear staining
indicated positive P53 expression, and tan cytoplasmic staining
indicated positive Cox-2 expression. Five randomly selected fields
were analyzed for a total of 500 scored cells using a Leica DM2500
microscope (Leica Camera AG, Wetzlar, Germany). For unstained
cells, a score of 0 was specified. For stained cells, 1–19% of
cells indicated weak staining intensity (1 point), 20–49% indicated
moderate staining intensity (2 points), and ≥50%, appearing as dark
brown staining, indicated strong staining intensity (3 points). The
scores were then divided into two groups: Scores of 0 and 1 as the
negative expression group (-), and scores of ≥2 points as the
positive expression group (+).
Follow-up
The 195 patients were followed up over a minimal
period of 2 years until December 31, 2013, and the median follow-up
time was 30 months (range, 2–43 months). No cases were lost
resulting in a follow-up rate of 100.00%.
Statistical analysis
The statistical analysis was performed using the
SPSS statistical software for Windows version 17.0 (SPSS, Inc.,
Chicago, IL, USA). Survival curves are presented as Kaplan-Meier
curves, and significance was classified by the log-rank test. The
Cox regression model was used for multivariate prognostic analysis,
and a binary logistic regression model was used for the correlation
analyses to analyze the influencing clinical factors.
Results
Mortality rate due to recurrence or
metastasis
On December 31, 2013, 144 patients had survived and
51 patients had succumbed to tumor recurrence or metastasis. Of
those 51 mortality cases, 15 patients exhibited anastomotic
recurrence; 13, regional lymph node recurrence; 7, liver
metastasis; 8, lung metastasis; 2, bone metastasis; 3, pleural
metastasis; and 3, multi-organ metastasis.
Correlation analyses between P53 and
Cox-2 expression, P53/Cox-2 co-expression and clinical factors
Positive P53 expression, assessed by tan granular
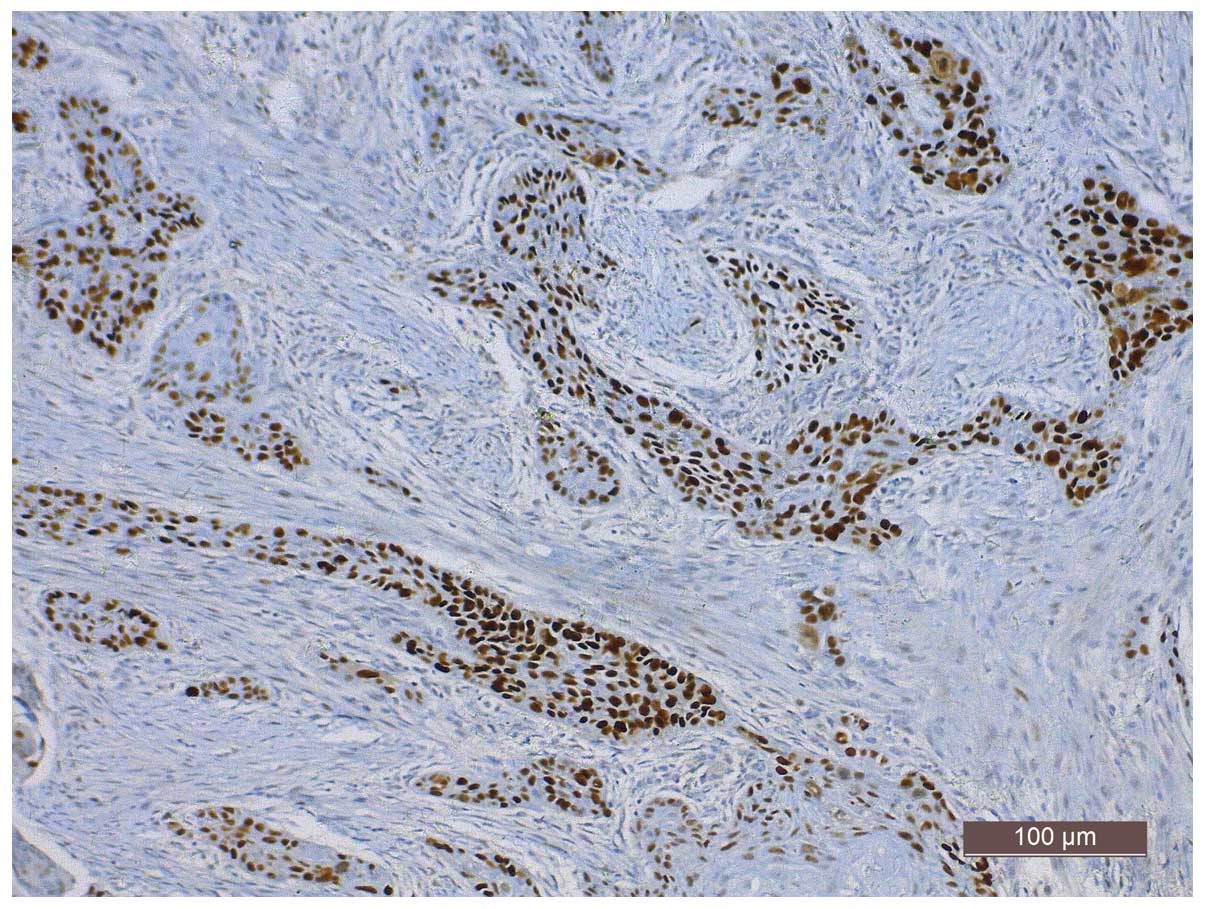
staining in tumor cell nuclei, was observed in 60.5% (118/195) of
the specimens (Fig. 1). Positive
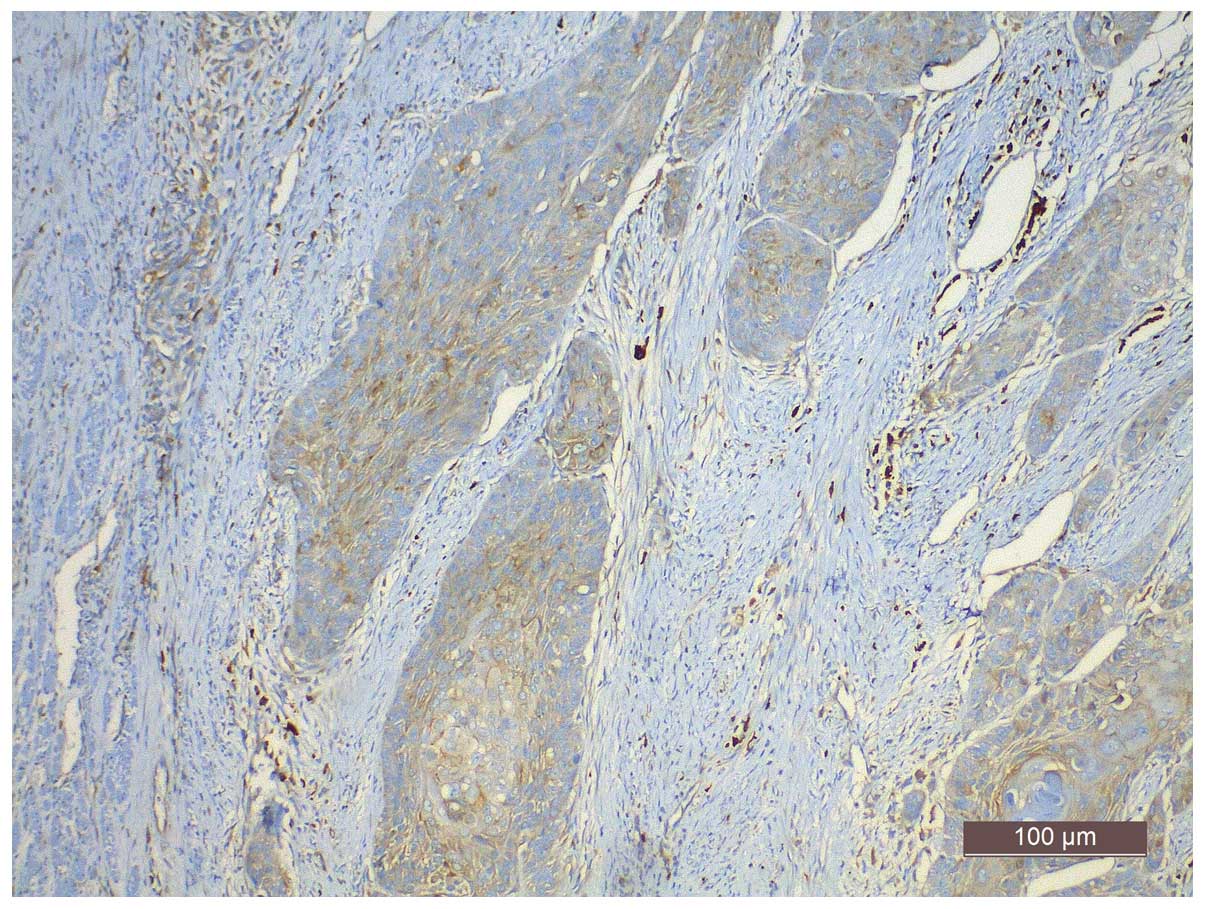
Cox-2 expression, assessed by cytoplasmic yellow staining, was
observed in 69.7% (136/195) of the specimens (Fig. 2). In 43.1% (84/195) of the specimens,
the co-expression of P53 and Cox-2 was observed, while 17.4%
(34/195) of the specimens expressed P53 only and 26.7% (52/195)
expressed Cox-2 only. A total of 12.8% (25/195) of the specimens
were negative for P53 and Cox-2. P53 expression and P53/Cox-2
co-expression were associated with the age of the patient (P=0.028)
and tumor differentiation status (P=0.015; Table I).
 | Table I.The associations between P53 and Cox-2
expression and P53/Cox-2 co-expression and the assessed clinical
factors. |
Table I.
The associations between P53 and Cox-2
expression and P53/Cox-2 co-expression and the assessed clinical
factors.
|
| P53 expression |
| Cox-2 expression |
| P53 and Cox-2
co-expression |
|
|---|
|
|
|
|
|
|
|
|
|---|
| Clinical factors | Total | High n=118 | Low n=77 | P-value | High n=136 | Low n=59 | P-value | P53(+) Cox-2(+)
n=84 | Other groups
n=111 | P-value |
|---|
| Age (years) |
|
|
| 0.028a |
|
| 0.641 |
|
| 0.056 |
|
<60 |
68 | 34 | 34 |
|
46 | 22 |
| 23 | 45 |
|
| ≥60 | 127 | 84 | 43 |
|
90 | 37 |
| 61 | 66 |
|
| Gender |
|
|
| 0.538 |
|
| 0.378 |
|
| 0.635 |
|
Female |
45 | 29 | 16 |
|
29 | 16 |
| 18 | 27 |
|
| Male | 150 | 89 | 61 |
| 107 | 43 |
| 66 | 84 |
|
| Differentiation |
|
|
| 0.015a |
|
| 0.200 |
|
| 0.020a |
| High |
16 | 13 | 3 |
|
10 | 6 |
|
8 | 8 |
|
|
Moderate |
108 | 56 | 52 |
|
71 | 37 |
| 37 | 71 |
|
|
Poor |
71 | 49 | 22 |
|
55 | 16 |
| 39 | 32 |
|
| Position |
|
|
| 0.078 |
|
| 0.285 |
|
| 0.560 |
|
Upper |
7 |
7 | 0 |
|
3 | 4 |
|
3 | 4 |
|
|
Middle |
155 | 90 | 65 |
| 110 | 45 |
| 64 | 91 |
|
|
Lower |
33 | 21 | 12 |
| 23 | 10 |
| 17 | 16 |
|
| TNM stage |
|
|
| 0.499 |
|
| 0.309 |
|
| 0.662 |
| I |
22 | 12 | 10 |
| 14 | 8 |
|
8 | 14 |
| II |
91 | 59 | 32 |
| 60 | 31 |
| 38 | 53 |
|
III |
82 | 47 | 35 |
| 62 | 20 |
| 38 | 44 |
Correlation analyses between P53 and
Cox-2 expression and P53/Cox-2 co-expression, and overall survival
(OS) or disease-free survival (DFS)
Single factor log-rank analysis by Kaplan-Meier
survival analysis were used to assess the association between P53
and Cox-2 expression as well as P53/Cox-2 co-expression and DFS or
OS following radical surgery in patients with esophageal cancer.
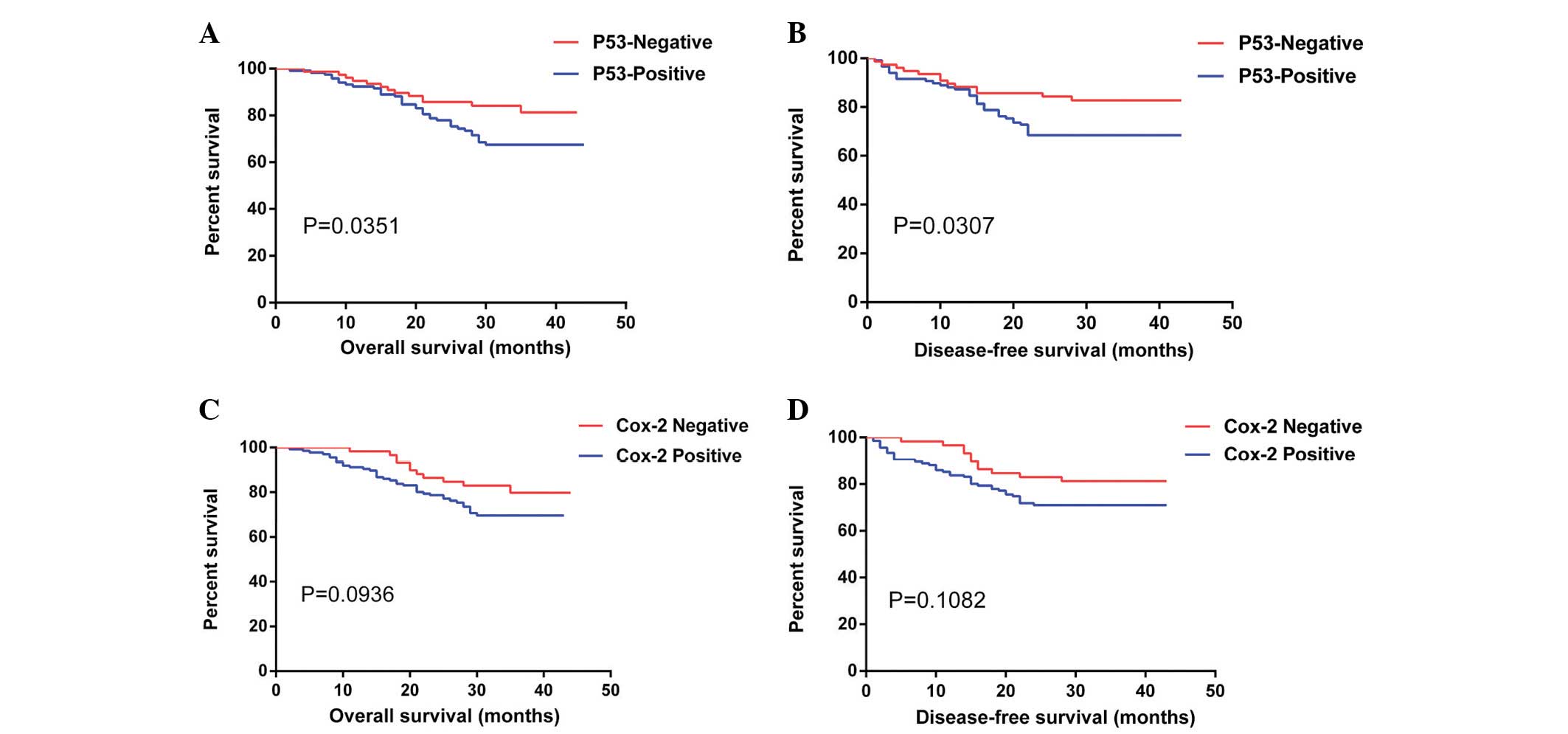
Differences between the OS (χ2=4.440, P=0.0351) and DFS
(χ2=4.672, P=0.0307) curves according to P53 expression
were observed, with a two-year OS of 78.0% in the P53-positive
group compared with 85.7% in the P53-negative group (Fig. 3). The DFS of the P53-positive group
was 68.4% compared with 82.8% for the P53-negative group. No
statistically significant differences (P>0.05) were observed for
Cox-2 expression in the OS and DFS curves. DFS
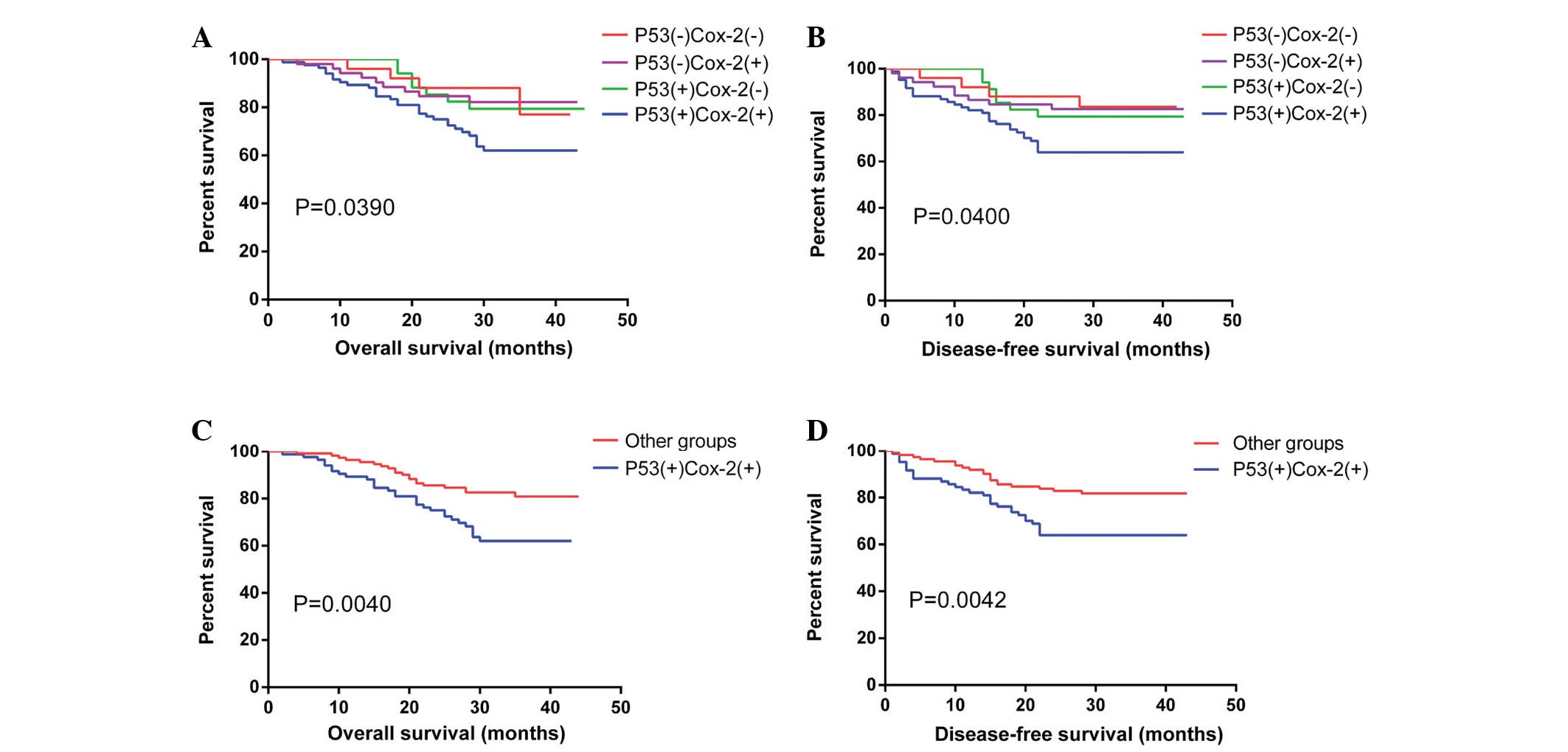
(χ2=8.277, P=0.0040), and OS (χ2=8.203,
P=0.0042) curves were also affected by the P53/Cox-2 co-expression
status, with a two-year OS of 75.0% for the double-positive group
compared with 85.6% for the other groups, and a DFS of 63.9% in
double-positive patients compared with 82.8% in the other groups
(Fig. 4).
Relevance of clinical pathological
factors with prognosis
Eight risk factors (gender, age, tumor location, TNM
stage, tumor differentiation degree, P53 and Cox-2 expression and
P53/Cox-2 co-expression) were included in a multifactor analysis
using the Cox multivariate regression model with a forced entry
method. The results showed that TNM staging [hazard ratio
(HR)=3.379, P<0.001], P53 expression (HR=2.102, P=0.023) and
P53/Cox-2 co-expression (HR=2.212, P=0.009) were all independent
factors affecting the OS curves of patients with esophageal cancer.
The same independent prognostic factors also influenced the DFS
curves (TNM staging, HR=3.497, P<0.001; P53 expression,
HR=2.138, P=0.020; P53/Cox-2 co-expression, HR=2.221, P=0.008)
(Table II). The same eight risk
factors were also analyzed by the binary logistic regression model
with a forced entry method. The results showed that the tumor
differentiation degree [odds ratio (OR)=1.964, P=0.023], TNM
staging (OR=3.206, P<0.001), P53 expression (OR=2.510, P=0.012)
and P53/Cox-2 co-expression (OR=2.204, P=0.021) were associated
with the local recurrence or distant metastasis of esophageal
cancer (Table III).
 | Table II.Cox multivariate analysis: The
associations between clinical factors and esophageal cancer
survival rates. |
Table II.
Cox multivariate analysis: The
associations between clinical factors and esophageal cancer
survival rates.
|
| Overall
survival | Disease-free
survival |
|---|
|
|
|
|
|---|
| Characteristic | HR | 95% CI | P-Value | HR | 95% CI | P-value |
|---|
| Age (≥60 vs. <60
years) | 1.129 | 0.613–2.077 | 0.697 | 1.157 | 0.628–2.130 | 0.641 |
| Gender (female vs.
male) | 0.863 | 0.423–1.763 | 0.686 | 0.908 | 0.445–1.855 | 0.792 |
| Position (upper vs.
middle vs. lower) | 1.329 | 0.690–2.560 | 0.396 | 1.319 | 0.686–2.538 | 0.406 |
| Differentiation
(high vs. moderate vs. poor) | 1.254 | 0.796–1.974 | 0.329 | 1.252 | 0.795–1.971 | 0.332 |
| TNM stage (I vs. II
vs. III) | 3.379 | 1.919–5.952 |
<0.001a | 3.497 | 1.979–6.181 |
<0.001a |
| P53 expression (low
vs. high) | 2.102 | 1.108–3.991 | 0.023a | 2.138 | 1.127–4.056 | 0.020a |
| Cox-2 expression
(low vs. high) | 1.473 | 0.742–2.923 | 0.268 | 1.453 | 0.734–2.875 | 0.283 |
| P53(+) Cox-2(+) vs.
other groups | 2.212 | 1.219–4.012 | 0.009a | 2.221 | 1.228–4.017 | 0.008a |
 | Table III.Binary logistic regression analysis:
The associations between clinical factors and recurrence or
metastasis in esophageal carcinoma. |
Table III.
Binary logistic regression analysis:
The associations between clinical factors and recurrence or
metastasis in esophageal carcinoma.
|
| Recurrence or
metastasis |
|
|---|
|
|
|
|
|---|
| Characteristic | OR | 95% CI | P-value |
|---|
| Age (≥60 vs.
<60) | 0.862 | 0.425–1.747 | 0.680 |
| Gender (female vs.
male) | 1.432 | 0.609–3.371 | 0.411 |
| Position (upper vs.
middle vs. lower) | 1.456 | 0.670–3.164 | 0.343 |
| Differentiation
(high vs. moderate vs. poor) | 1.964 | 1.099–3.508 | 0.023a |
| TNM (I vs. II vs.
III) | 3.206 | 1.763–5.830 |
<0.001a |
| P53 expression (low
vs. high) | 2.510 | 1.228–5.131 | 0.012a |
| Cox-2 expression
(low vs. high) | 1.583 | 0.740–3.383 | 0.236 |
| P53(+) Cox-2(+) vs.
other groups | 2.204 | 1.124–4.322 | 0.021a |
Discussion
P53 is a known tumor-suppressor gene that
participates in the occurrence and development of esophageal
cancer. The P53 gene is located on human chromosome 17p13 and is
composed of 10 exons and 11 introns, encoding a protein 393 amino
acids in length. P53 gene products can be divided into wild-type
(wtp53) and mutant (mtp53). Upon DNA damage, increased P53 protein
expression regulates target genes involved in preventing cells in
the G1 phase from entering the S phase, which favors DNA
repair. If the DNA is seriously damaged, P53 will trigger apoptosis
to remove the cells with the overly damaged DNA. Tumor growth
requires angiogenesis, and Kang et al (6) demonstrated that the P53 gene functions
by inhibiting tumor angiogenesis via the adjustment of platelet
response protein 1 (TSP-1) levels, which is the main angiogenesis
inhibiting factor. However, mtp53 acts as a proto-oncogene by
promoting the occurrence and development of tumor cells. Huang
et al (7) showed that the P53
expression level in normal tissue is only one-eighth of that in
tumor tissues; furthermore, since the P53 protein has a short
half-life, it can hardly be detected in normal cells. However, when
cells become damaged or mutated by various factors, P53 expression
increases significantly. Mtp53, instead of inhibiting tumor cell
proliferation, promotes cell proliferation and eventually alters
the cellular phenotype in a malignant manner (8).
Previous studies have demonstrated that the P53 gene
mutation is associated with poor prognosis in various types of
cancer, including colon, breast, lung, gastric and esophageal
cancer (9,10). Overexpression of P53 in esophageal
tumor cells increases their potential to invade tissue and blood
vessels, and promotes the local recurrence and metastasis of
esophageal cancer, leading the progression towards late
pathological staging and poor prognosis (11). In the present study, it was revealed
that P53 expression was associated with age and tumor
differentiation degree (P<0.05). In patients ≥60 years old, P53
expression was found in 66.1% (84/127) of the cases, and in
patients with poorly differentiated cancer, P53 expression was
observed in 69.0% (49/71) of the cases. Han et al (12) showed that P53 expression was
positively correlated with tumor stage and lymph node metastasis.
Ye et al (13) noted that P53
expression was not associated with the gender or age of the
patient, but was associated with tumor differentiation degree and
lymph node metastasis. Finally, Chino et al (14) showed that P53 expression was not
associated with tumor infiltration depth, lymph node metastasis or
venous or lymphatic invasion. Such differences in findings between
studies may be caused by the different stages and sources of
samples, different P53 antibodies or variations in the experimental
methods. Jin et al (15) used
an immunohistochemical method to detect the expression level of P53
in 80 specimens of esophageal carcinoma and different diseased
tissues in situ, which implied that positive P53 expression
was associated with the occurrence and stage of esophageal squamous
cell carcinoma and could be used to identify high-risk individuals
in a precancerous population. In the present study, single factor
Kaplan-Meier analysis showed a difference in OS curves according to
P53 expression (χ2=4.440, P=0.0351), with a two-year OS
of 85.7% in the P53-negative group compared with 78.0% in the
P53-positive group. Similarly, P53 expression also influenced the
DFS curves (χ2=4.672, P=0.0307), with a two-year DFS of
82.8% in the P53-negative group compared with 68.4% in the
P53-positive group. In addition, a Cox multivariate regression
analysis identified P53 expression as an independent factor
affecting patient survival rate, and a binary logistic regression
analysis showed that P53 expression was associated with local
recurrence or distant metastasis following esophagectomy.
Cox-2 plays a role in the development of esophageal
cancer. Prostaglandin-endoperoxide synthase (PTGS), also known as
cyclooxygenase, is a monotopic membrane protein which acts as a
rate-limiting enzyme for the conversion of arachidonic acid into
prostaglandins. The PTGS family comprises Cox-1 and Cox-2, which
regulate different cellular functions despite their homology
(16). Cox-1 is expressed in the
majority of normal tissues, whereas the Cox-2 enzyme is induced
rapidly in response to pathological states, such as inflammation
and tumor formation (17,18). A previous study has also shown that
Cox-1 has an induced type and Cox-2 has a structured type, and that
a variant named Cox-3 (an isomer of Cox-1) also possibly exists
(19). The human Cox-2 gene, located
on chromosome 1q25.2-q25.3, is composed of 9 introns and 10 exons
encoding a protein of 604 amino acid residues. In normal tissue,
Cox-2 expression is low or absent. Cox-2 expression is induced by
various cellular factors, including proinflammatory responses, and
is involved in tumor development, invasion and metastasis (20). According to Misra et al
(21), Cox-2 participates in the
occurrence and development of esophageal cancer in multiple ways,
including by inhibiting the apoptosis or promoting the
proliferation of tumor cells and accelerating invasion and
metastasis; however, the specific mechanism remains unclear.
Okumura et al (22) noted the
important role of Cox-2 in the synthesis of prostaglandin and its
role in mediating angiogenesis, tumor growth, invasion and
metastasis. Kashiwagi et al (23) revealed that Cox-2 may increase
vascular endothelial growth factor-C expression by generating
prostaglandin, thus promoting the generation of lymphatic vessels
in tumor tissues and favoring metastasis possibly through the lymph
nodes. Zhou et al (24)
measured Cox-2 expression and lymphatic vessel density (MLD) in
esophageal cancer tissues by an immunohistochemical method and
observed that MLD increased together with the increase in Cox-2
expression. Consequently, the authors proposed that Cox-2 could be
contributing to the formation of lymphatic vessels in esophageal
cancer, thereby promoting metastasis. The present study revealed no
correlation between Cox-2 expression and clinical factors in
esophageal cancer. Cox-2 expression did not affect the DFS and OS
curves of the patients and was not identified as a significant
independent factor affecting survival rate, recurrence or
metastasis of esophageal cancer. However, the OS and DFS curves
were clustered according to Cox-2 expression, and the prognosis for
the patients with negative Cox-2-expressing tumors was improved
compared with patients with positive Cox-2-expressing tumors. Prins
et al (25) noted that Cox-2
expression was associated with prognosis in esophageal
adenocarcinoma and could be used as a risk stratification parameter
in esophageal adenocarcinoma. However, in China, the more prevalent
subtype of esophageal cancer is esophageal squamous cell carcinoma,
and consequently, all cases included in the present study are of
esophageal squamous carcinoma. The tumor subtype may account for
the difference between the two studies.
P53/Cox-2 co-expression may have prognostic value in
esophageal cancer. P53 and Cox-2 are expressed at higher levels in
esophageal cancerous tissues compared with normal tissues.
Mutations in the P53 gene are induced by various factors and lead
to a loss of P53 tumor cell growth-inhibiting functions. Mutated
P53 promotes tumor cell proliferation, inhibits apoptosis and
promotes the occurrence and development of esophageal cancer.
Although the specific mechanism remains unclear, Cox-2 acts as a
cancer-promoting gene and plays a role in mediating angiogenesis,
tumor growth, invasion and metastasis. This resulted in the
hypothesis that there may be a synergistic association between
Cox-2 and P53 in the occurrence and development of esophageal
cancer. Benoit et al (26)
reported that the P53 tumor suppressor gene could recruit nuclear
factor (NF)-κB to transcriptionally activate Cox-2 expression and
activity. Song et al (27)
noted that blocking Cox-2 expression using small interfering RNA
reinforced P53 transcriptional activity. Cheng et al
(28) reported that specific Cox-2
inhibitors could completely reverse the inhibition of apoptosis
induced by P53 and hepatitis virus X, suggesting that HBx could
block P53-induced apoptosis through the Cox-2/prostaglandin E
(2) signaling pathway. Choi et
al (29) showed that Cox-2
expression reduced the expression of P53 and led to the
inactivation of the P53 gene, thus promoting tumor development. Ma
et al (30) observed that in
precancerous lesions, tumor development is promoted via a cell
survival mechanism by the interaction between Cox-2 and wild-type
P53. However, in the late stage of tumor development, cells could
resist apoptosis by relying on Cox-2 alone, without wild-type P53.
This outcome may be due to the independence of the Cox-2 activation
mechanism on P53 and NF-κB activity, or the occurrence of other
cellular modifications to avoid apoptosis. Another mechanism may
exist in lesions under inflammatory stress, where growth promoting
signaling cascades (including Wnt/β-catenin, KRAS or c-Myb)
activate the promoter and upregulate Cox-2 expression levels.
Therefore, P53 and NF-κB action may not be the factors that
activate Cox-2 expression. In addition, mutated P53 proteins may
coexist with Cox-2 in the same cells and could synergize to inhibit
cell apoptosis, thereby enhancing the malignant behavior of tumors
and resulting in a significantly poorer prognosis. In the present
study, P53 expression was observed in 60.5% (118/195), Cox-2
expression in 69.7% (136/195) and co-expression of P53 and Cox-2 in
43.1% (84/195) of the cases, with the expression of the two
proteins being positively correlated. Using single factor
Kaplan-Meier analysis, differences in survival rate between the
P53/Cox-2 double-positive group compared with the other groups were
identified. Furthermore, the two-year OS and DFS in the P53/Cox-2
double-positive group were significantly reduced compared with
those in the other groups (75.0% vs. 85.6% and 63.9% vs. 81.8%,
respectively) and have smaller P-values when compared with the
group expressing P53 alone. Cox multivariate regression model
analysis identified P53/Cox-2 co-expression as an independent
factor influencing DFS and OS in esophageal carcinoma due to a
larger HR and a smaller P-value compared with P53 expression alone.
Analysis using a binary logistic regression model revealed that
P53/Cox-2 co-expression also influenced the recurrence and
metastasis of esophageal cancer, further implying that this may be
used as a risk stratification parameter for the prognosis of
esophageal cancer.
In conclusion, in the present study P53 and Cox-2
were markedly expressed in esophageal cancer tissues. P53 and Cox-2
co-expression was associated with increased malignant behavior of
tumors and predicted a poor prognosis. Therefore, P53/Cox-2
co-expression may be used as a potential risk stratification
parameter in esophageal cancer and may also be a promising
therapeutic target. The pitfall of the present study lies in the
short follow-up period following surgery, therefore, further
complementary studies will aid in the thorough elucidation of the
mechanisms behind P53 and Cox-2 interactions.
Acknowledgements
The authors would like to thank Mr. Bin Xu (Soochow
University) for statistical guidance.
References
|
1
|
Huang JX, Chen WC, Lin M, Zhang YL, Li FY,
Song ZX, Xiao W, Chen P, Qian RY, Salminen E, et al:
Clinicopathological significance of cyclooxygenase-2 and cell
cycle-regulatory proteins expression in patients with esophageal
squamous cell carcinoma. Dis Esophagus. 25:121–129. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Meng XY, Zhu ST, Zong Y, Wang YJ, Li P and
Zhang ST: Promoter hypermethylation of cyclooxygenase-2 gene in
esophageal squamous cell carcinoma. Dis Esophagus. 24:444–449.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Surget S, Khoury MP and Bourdon JC:
Uncovering the role of p53 splice variants in human malignancy: A
clinical perspective. Onco Targets Ther. 7:57–68. 2014.
|
|
4
|
Chandrasekharan NV and Simmons DL: The
cyclooxygenases. Genome Biol. 5:2412004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen LQ: Understanding and appraisal of
the new TNM classification for esophageal cancer in the AJCC cancer
staging manual (7th edition). Zhonghua Zhong Liu Za Zhi.
32:237–240. 2010.(In Chinese). PubMed/NCBI
|
|
6
|
Kang SY, Halvorsen OJ, Gravdal K,
Bhattacharya N, Lee JM, Liu NW, Johnston BT, Johnston AB, Haukaas
SA, Aamodt K, et al: Prosaposin inhibits tumor metastasis via
paracrine and endocrine stimulation of stromal p53 and Tsp-1. Proc
Natl Acad Sci USA. 106:12115–12120. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang H, Wang LF, Tian HM, Liu Y, Li M, Qu
P, Wang WR and Zhang W: Expression of retinoic acid receptor-beta
mRNA and p16, p53, Ki67 proteins in esophageal carcinoma and its
precursor lesions. Zhonghua Zhong Liu Za Zhi. 27:152–155. 2005.(In
Chinese). PubMed/NCBI
|
|
8
|
Do PM, Varanasi L, Fan SQ, Li CY, Kubacka
I, Newman V, Chauhan K, Daniels SR, Boccetta M, Garrett MR, et al:
Mutant p53 cooperates with ETS2 to promote etoposide resistance.
Genes Dev. 26:830–845. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee MH and Lozano G: Regulation of the
p53-MDM2 pathway by 14-3-3 and other proteins. Semin Cancer Biol.
16:225–234. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kirsch DG and Kastan MB: Tumor-suppressor
p53: Implications for tumor development and prognosis. J Clin
Oncol. 16:3158–3168. 1998.PubMed/NCBI
|
|
11
|
Fagundes RB, Mello CR, Tollens P, Pütten
ACK, Wagner MB, Moreira LF and Barros SG: P53 protein in esophageal
mucosa of individuals at high risk of squamous cell carcinoma of
the esophagus. Dis Esophagus. 14:185–190. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han U, Can OI, Han S, Kayhan B and Onal
BU: Expressions of p53, VEGF C, p21: Could they be used in
preoperative evaluation of lymph node metastasis of esophageal
squamous cell carcinoma? Dis Esophagus. 20:379–385. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ye B, Wang X, Yang Z, Sun Z, Zhang R, Hu
Y, Lu Y and Du J: P53 and p73 expression in esophageal carcinoma
correlate with clinicopathology of tumors. Hepatogastroenterology.
59:2192–2195. 2012.PubMed/NCBI
|
|
14
|
Chino O, Kijima H, Shimada H, Nishi T,
Tanaka H, Kise Y, Kenmochi T, Himeno S, Machimura T, Tanaka M, et
al: Accumulation of p53 in esophageal squamous cell carcinoma. Int
J Mol Med. 8:359–363. 2001.PubMed/NCBI
|
|
15
|
Jin Y, Zhang W and Liu B: Abnormal
expression of p53, Ki67 and iNOS in human esophageal carcinoma in
situ and pre-malignant lesions. Zhonghua Zhong Liu Za Zhi.
23:129–131. 2001.(In Chinese). PubMed/NCBI
|
|
16
|
Feletou M, Huang Y and Vanhoutte PM:
Endothelium-mediated control of vascular tone: COX-1 and COX-2
products. Br J Pharmacol. 164:894–912. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
O'Banion MK: Cyclooxygenase-2: Molecular
biology, pharmacology, and neurobiology. Crit Rev Neurobiol.
13:45–82. 1999.PubMed/NCBI
|
|
18
|
Williams CS and DuBois RN: Prostaglandin
endoperoxide synthase: Why two isoforms? Am J Physiol.
270:G393–G400. 1996.PubMed/NCBI
|
|
19
|
Willoughby DA, Moore AR and Colville-Nash
PR: COX-1, COX-2 and COX-3 and the future treatment of chronic
inflammatory disease. Lancet. 355:646–648. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xin X, Majumder M, Girish GV, Mohindra V,
Maruyama T and Lala PK: Targeting COX-2 and EP4 to control tumor
growth, angiogenesis, lymphangiogenesis and metastasis to the lungs
and lymph nodes in a breast cancer model. Lab Invest. 92:1115–1128.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Misra S and Sharma K: COX-2 signaling and
cancer: New players in old arena. Curr Drug Targets. 15:347–359.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Okumura H, Uchikado Y, Setoyama T,
Matsumoto M, Owaki T, Ishigami S and Natsugoe S: Biomarkers for
predicting the response of esophageal squamous cell carcinoma to
neoadjuvant chemoradiation therapy. Surg Today. 44:421–428. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kashiwagi S, Hosono K, Suzuki T, Takeda A,
Uchinuma E and Majima M: Role of COX-2 in lymphangiogenesis and
restoration of lymphatic flow in secondary lymphedema. Lab Invest.
91:1314–1325. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou BT, Yang W, Xu XH, Ai YG, Li XL and
Huang YJ: Correlation of cyclooxygenase 2 expression with
microlymphatic density and its clinical significance. Zhonghua Wei
Chang Wai Ke Za Zhi. 13:699–702. 2010.(In Chinese). PubMed/NCBI
|
|
25
|
Prins MJ, Verhage RJ, ten Kate FJ and van
Hillegersberg R: Cyclooxygenase isoenzyme-2 and vascular
endothelial growth factor are associated with poor prognosis in
esophageal adenocarcinoma. J Gastrointest Surg. 16:956–966. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Benoit V, de Moraes E, Dar NA, Taranchon
E, Bours V, Hautefeuille A, Tanière P, Chariot A, Scoazec JY, de
Moura Gallo CV, et al: Transcriptional activation of
cyclooxygenase-2 by tumor suppressor p53 requires nuclear
factor-kappaB. Oncogene. 25:5708–5718. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Song J, Wei Y, Chen Q and Xing D:
Cyclooxygenase 2-mediated apoptotic and inflammatory responses in
photodynamic therapy treated breast adenocarcinoma cells and
xenografts. J Photochem Photobiol B. 134:27–36. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cheng AS, Yu J, Lai PB, Chan HL and Sung
JJ: Cox-2 mediates hepatitis B virus X protein abrogation of
p53-induced apoptosis. Biochem Biophys Res Commun. 374:175–180.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Choi EM, Kim SR, Lee EJ and Han JA:
Cyclooxygenase-2 functionally inactivates p53 through a physical
interaction with p53. Biochim Biophys Acta. 1793:1354–1365. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ma XL, XS G and Sun HJ: Advance in
research on relations between cyclooxygenase-2 and P53 during
inflammatory stress and carcinogenesis. Int J Genet. 32:372–376.
2009.(In Chinese).
|