Introduction
Gastric carcinoma is one of the most common
gastrointestinal malignant tumors worldwide. It develops through a
multifactorial, multistage process resulting from an imbalance
between oncogenes and tumor suppressor genes. The global five-year
survival rate for gastric carcinoma is <10% (1) and cancer cell metastasis is the major
reason for gastric carcinoma lethality (2). Metastasis and proliferation rely on a
series of interactions between tumor cells and the extracellular
matrix, as well as other cells (3).
In addition, tumor cell surface adhesion molecules may have a
significant role in this process (2,3).
Cluster of differentiation 44 (CD44) is an integral
cell membrane glycoprotein with a molecular weight of 85–250 kD.
CD44 was initially identified as a lymphocyte homing receptor on
circulating lymphocytes, and exhibits homing, adhesion and
migration functions (4). The CD44
protein exists in various isoforms [variants 1–10 (v1-10)]
generated from the primary transcript by the alternate splicing of
10 exons. The generation of CD44 splice variants is hypothesized to
be closely associated with gastric carcinoma tumorigenesis and
differentiation (5,6). For example, it has been reported that
the formation and metastasis of gastric carcinoma cells promoted by
CD44 is associated with the CD44v adherence function (7). Cancer cells expressing CD44v are able to
utilize a camouflage function against lymphocytes to escape
identification and destruction by the human immune system, thus,
enabling easier metastasis (8). Among
these CD44 variants, CD44v8-10 expression is considered to be a
prognostic indicator of gastric carcinoma (9). A previous study of gastric cancer
evaluated the expression of standard CD44 (CD44s) (10), while other studies have investigated
the expression of the CD44v8-10 isoform or CD44s and CD44v
(11–15). The study utilized antibodies against
CD44s, an isoform lacking all variant exons that is widely
expressed in lymphoid and non-lymphoid tissues.
In the present study, reverse
transcription-polymerase chain reaction (RT-PCR) combined with
nested PCR technology (nested RT-PCR) was used to detect CD44 mRNA
expression in 153 dual biopsy samples obtained during gastroscopy
(one sample was obtained from the lesion and one from the
corresponding adjacent non-cancerous tissue). Pathologically
diagnosed gastric carcinoma and atypical hyperplasia tissues were
subsequently subjected to immunohistochemical analysis, to
investigate the value of nested RT-PCR for the early diagnosis and
prognosis of patients with gastric cancer or precancerous
lesions.
Materials and methods
Reagents and antibodies
The RNA isolation kit and RT-PCR kit were purchased
from Promega Corp. (Madison, WI, USA). The monoclonal mouse
anti-human CD44v antibody (cat. no. SM1126F) was obtained from
Acris Antibodies GmbH (Herford, Germany) and the
streptavidin-peroxidase immunohistochemistry kit was obtained from
Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). The DNA quick
purification kit was from Biodev-Tech Scientific and Technical Co.,
Ltd (Beijing, China). All other chemicals were purchased from DAAN
Gene Co., Ltd of Sun Yat-Sen University (Guangzhou, China), unless
otherwise stated.
Sample preparations and pathological
diagnosis
The present study included 153 patients (108 males
and 45 females; mean age, 65.4±16.7 years) who underwent a
gastroscopic examination in the Beijing Chao Yang Hospital
(Beijing, China) between March 2009 and April 2012. During the
procedure, 6–8 tumor tissue biopsies and 3–4 corresponding adjacent
non-cancerous tissue biopsies were obtained from each patient. The
samples were immediately frozen and stored at −80°C prior to use.
Pathological examination diagnosed 128 cases of gastric cancer,
including 56 cases of well-differentiated adenocarcinoma, 40 cases
of poorly differentiated adenocarcinoma and 32 cases of mucinous
carcinoma (16). Furthermore, 25
patients exhibited precancerous lesions, including 19 cases of
atypical hyperplasia and 6 cases of intestinal metaplasia (Table I). The present study was approved by
the Ethics Committee of Beijing Chao Yang Hospital Affiliated to
Capital Medical University, and written informed consent was
obtained from all patients.
 | Table I.Pathological diagnoses of 153 patients
with gastric cancer (n=128) and precancerous lesions (n=25). |
Table I.
Pathological diagnoses of 153 patients
with gastric cancer (n=128) and precancerous lesions (n=25).
| Pathological
diagnosis | Patients, n |
|---|
| Gastric cancer |
|
|
Well-differentiated
adenocarcinoma | 56 |
| Poorly
differentiated adenocarcinoma | 40 |
| Mucinous
carcinoma | 32 |
| Precancerous
lesion |
|
| Atypical
hyperplasia | 19 |
|
Intestinal hyperplasia | 6 |
Nested RT-PCR
All primers were synthesized by Beijing SBS Genetech
Co., Ltd (Beijing, China), as described by Matsumura and
Tarin (17). The primer sequences
were as follows: Forward, 5′-GACACATATTGCTTCAATGCTTCAGC-3′ and
reverse, 5′-GATGCCAAGATGATCAGCCATTCTGGAAT-3′ for full-length CD44s
(482-bp PCR product); forward, 5′-CCTGAAGAAGATTGTACATACGTCACAGAC-3′
and reverse, 5′-TGTCCTTATAGGACCAGA-3′ for nested PCR (417-bp PCR
product); and forward, 5′-ATGGACTCCAGTCATAGTACAACG-3′ and reverse,
5′-TAAGGAACGATTGACATTAGAG-3′ for CD44V8-10 (396-bp PCR product)
(18). Total RNA was extracted using
the acid-guanidinium-phenol-chloroform method (19), according to the manufacturer's
instructions. Subsequently, RT-PCR was conducted in accordance with
the manufacturer's instructions. Briefly, 20 µl RT reaction mixture
[containing 1 µg total RNA, 100 ng Oligo dT (15mer), 50 mmol/l
Tris-HCl (pH 8.3), 3.75 mmol/l KCl, 3 mmol/l MgCl2, 10
mmol/l DTT, 500 µmol/l of each dNTP, 25 U M-MLV reverse
transcriptase and 20 U RNasin] was added to 20 µl PCR Master Mix.
The mixture was initially denatured at 94°C for 5 min, and then
subjected to 30 cycles of 94°C for 1 min, 55°C for 1 min and 72°C
for 1 min. β-actin was used as the internal control, and nested PCR
was performed immediately following RT-PCR using the RT-PCR product
as the template and the same primers as described previously.
Sequencing analysis of PCR
products
The PCR products were separated on 1.8% agarose gels
and stained with ethidium bromide (10 µg/ml). Notable bands were
excised from the gel, recovered and purified using the Wizard® SV
Gel and PCR Clean-Up System (Promega Corp.), according to the kit
manual. The purified PCR products were sequenced using the
5′-AGACCAAGACACATTCAA-3′ primer and an ABI 373A DNA sequencer
(Applied Biosystems Life Technologies, Foster City, CA, USA).
Immunostaining assay
Seventy-six gastric cancer samples and 12 atypical
hyperplasia samples that exhibited a strong PCR band (CD44v; 600
bp), were selected to undergo immunohistochemical analysis.
Paraffin-embedded sections (4-µm) were deparaffinized, rehydrated
and blocked for endogenous peroxidase activity. The sections were
incubated with primary CD44v monoclonal antibodies at a dilution of
1:200 overnight at 4°C. Following washing three times in
Tris-buffered saline, the sections were incubated with horseradish
peroxidase-labeled immunoglobulin G for 2 h at room temperature.
Immunoreactive complexes were detected using ImmunoCruz™ mouse ABC
Staining System (Santa Cruz Biotechnology, Inc.) prior to
counterstaining the slides with hematoxylin. Cells stained brown in
the membrane or cytoplasm were considered positive. The
immunostaining was graded based on the percentage of positive cells
observed, as follows: Negative (−), <10%; weak positive (+),
10–25%; positive (++), 25–50%; strong positive (+++), >50%
(20).
Statistical analysis
All data were analyzed using the SPSS statistical
package for Windows (version 10.0; SPSS, Inc., Chicago, IL, USA).
The χ2 test was used to determine statistically
significant differences among the groups. P<0.05 was considered
to indicate a statistically significant difference.
Results
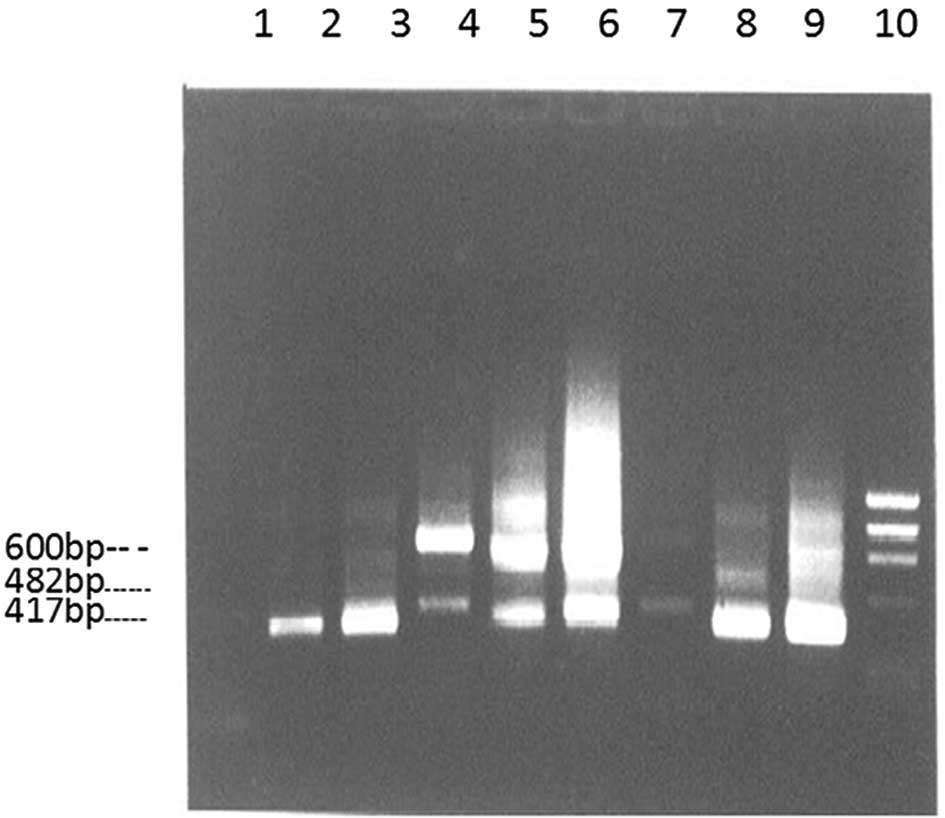
As indicated in Fig.
1, the nested RT-PCR products obtained from the biopsy samples
(tumor and non-cancerous) of 153 patients all exhibited 482 bp CD44
bands. The CD44v band (>600 bp) was observed in 132/153 total
tumor samples (86.3%), including 114/128 gastric cancer samples
(89.1%), 16/19 atypical hyperplasia samples (84.2%) and 2/6
intestinal metaplasia samples (33.3%). Furthermore, the CD44v band
was only observed in 18/153 non-cancerous tissue samples
(11.8%).
CD44v expression was significantly higher in gastric
cancer tissues and precancerous lesions compared with that in
non-cancerous tissues (P<0.05; Table
II). In addition, there was a significant difference in
CD44v8-10 expression between gastric cancer tissue and adjacent
non-cancerous tissues (P<0.05; Table
III). Among the 25 patients with precancerous lesions,
CD44v8-10 expression was positive in 8/19 atypical hyperplasia
cases and 1/6 intestinal metaplasia cases. Furthermore, no
significant difference was identified in CD44v8-10 expression rate
between the various pathological types of gastric cancer
(P>0.05; Table IV).
 | Table II.CD44v expression in gastric tumor,
precancerous lesion and non-cancerous tissue samples. |
Table II.
CD44v expression in gastric tumor,
precancerous lesion and non-cancerous tissue samples.
|
|
| CD44v positive |
|---|
|
|
|
|
|---|
| Pathological
diagnosis | Total cases, n | n | Rate, % |
|---|
| Gastric cancer | 128 | 114 | 89.1 |
| Precancerous
lesion |
|
|
|
| Atypical
hyperplasia | 19 | 16 | 84.2 |
|
Intestinal metaplasia | 6 | 2 | 33.3 |
| Adjacent
non-cancerous tissues | 153 | 18 | 11.8 |
 | Table III.CD44v8-10 expression in gastric tumor
and adjacent non-cancerous tissue samples. |
Table III.
CD44v8-10 expression in gastric tumor
and adjacent non-cancerous tissue samples.
|
|
| CD44v8-10
positive |
|---|
|
|
|
|
|---|
| Pathological
diagnosis | Total cases, n | n | Rate, % |
|---|
| Gastric cancer | 128 | 97 | 75.8 |
| Adjacent
non-cancerous tissues | 128 | 14 | 10.9 |
 | Table IV.CD44v and CD44v8-10 expression in
various pathological types of gastric cancer. |
Table IV.
CD44v and CD44v8-10 expression in
various pathological types of gastric cancer.
|
|
| CD44v positive | CD44v8-10
positive |
|
|---|
|
|
|
|
|
|
|---|
| Gastric cancer
type | Total cases, n | n | Rate, % | n | Rate, % | Cumulative rate,
% |
|---|
| Well-differentiated
adenocarcinoma | 56 | 50 | 89.3 | 42 | 75.0 | 84.0 |
| Poorly differentiated
adenocarcinoma | 40 | 36 | 90.0 | 32 | 80.0 | 88.0 |
| Mucinous
carcinoma | 32 | 28 | 87.5 | 23 | 71.9 | 82.1 |
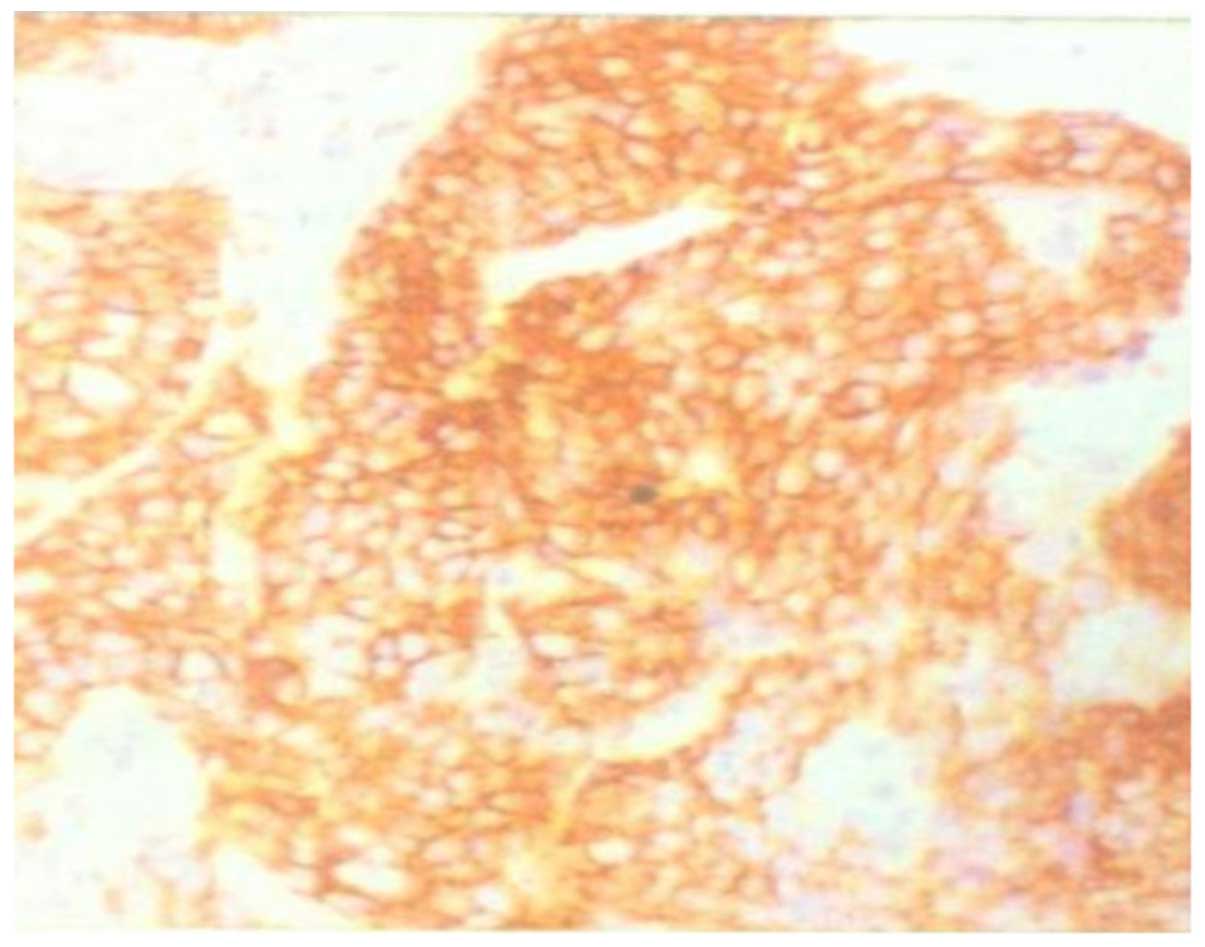
To verify the nested RT-PCR results, 76 gastric
cancer samples and 12 atypical hyperplasia samples were examined by
immunohistochemical staining (data not shown). The sections were
CD44v-positive (++) in 59/76 (77.6%) cases of gastric cancer and
5/12 cases of atypical hyperplasia (Fig.
2). To compare the immunostaining and nested RT-PCR data, the
CD44v and CD44v8-10 PCR products were confirmed by sequencing
analysis.
Discussion
Previous studies identified that CD44 messenger
(m)RNA expression markedly varied between healthy and carcinomatous
tissues, particularly with respect to the number, size and signal
intensity of amplified PCR bands. In all tumor tissues analyzed,
the amplified bands exhibited strong signals, high molecular
weights and a relatively uniform distribution (21–23).
Therefore, the abnormal differential expression of CD44 may be used
to distinguish between carcinomatous gastric tissues and healthy
gastric mucosa. Furthermore, abnormal expression of CD44 mRNA was
determined to be relatively high irrespective of whether the
gastric carcinoma tissues were highly or poorly differentiated
(24,25). A number of studies have proposed that
the role of CD44v in enhancing the infiltration and metastasis of
tumor cells may be associated with its ability to alter the
structure and function of cell surface adhesion molecules (10,26). Miwa
et al (27) revealed that
gastric carcinoma tissues exhibited higher CD44v expression
compared with that of adjacent healthy tissues, with 76.6% (23/30
cases) positive expression. In addition, a Chinese study detected
the expression of CD44v in gastric carcinoma tissue samples using
RT-PCR technology and determined that CD44v expression was as high
as 80.0% (16/20) (28). In the
present study, RT-PCR was performed in combination with nested PCR,
whereby the initial RT-PCR product was used as the template for
nested PCR, to detect CD44v expression. A positive expression rate
of 86.3% (132/153 cases) was obtained, marginally higher than the
aforementioned expression rates.
CD44v expression appears to be significantly higher
in metastasized gastric cancer compared with that of the primary
tumor. According to currently available literature, positive
CD44v8-10 expression may be an indication of metastasis and poor
clinical prognosis (17).
Furthermore, the survival rate was significantly lower in
gastrointestinal malignant cancer patients with positive CD44v8-10
expression compared with patients not exhibiting CD44v8-10
expression (9,29). Therefore, CD44v8-10 expression may be
considered an important indicator of gastric carcinoma
metastasis.
In the present study, RT-PCR combined with nested
PCR revealed that the positive rate of CD44v expression in all the
investigated lesions was 86.3% (132/153 cases). Of these cases, 128
were gastric carcinoma specimens, with 114 cases (89.1%) exhibiting
positive CD44v expression. In addition, 25 cases were diagnosed as
precancerous lesions (including 19 cases of atypical hyperplasia
and 6 cases of intestinal metaplasia), with positive CD44v rates of
84.2% (16/19 cases) and 33.3% (2/6 cases), respectively. Among the
153 healthy adjacent gastric mucosa specimens, 18 cases (11.8%)
exhibited positive CD44v expression. Furthermore, CD44v8-10
expression was observed in 97 cases of gastric cancer, resulting in
rates of 85.1% (97/114) of the CD44v positive gastric carcinoma
cases and 75.8% (97/128) of the total gastric carcinoma cases.
These values are higher than those reported previously, indicating
that the combination of RT-PCR and nested PCR may improve the
detection sensitivity of CD44v compared with current techniques,
including RT-PCR alone. Using RT-PCR and nested PCR in combination
increases the number of rounds of amplification performed and thus,
we hypothesize that this achieves a higher level of sensitivity. In
addition, it was identified that CD44v expression was not
associated with pathological disease type.
Previous studies have typically employed RT-PCR
combined with Southern blot hybridization or immunohistochemical
techniques to detect CD44v and CD44v8-10 (30,31).
However, in the present study, nested RT-PCR was combined with
immunohistochemical analysis of 76 cases of gastric carcinoma and
12 cases of atypical hyperplasia. The positive rate of CD44v
reported using nested RT-PCR in the present study was significantly
higher than the positive rates detected in previous studies
(9,10,25,30),
however, the possibility of false positives was not considered.
Furthermore, the use of RT-PCR with nested PCR as opposed to
Southern blot hybridization resulted in experimental results that
were confirmed by DNA sequencing. In particular, the detection rate
and sensitivity of CD44v was markedly improved. Additionally, the
nested RT-PCR method is simple and quick, does not require
radioisotopes, and uses samples that are easily obtained from
endoscopic examination. Therefore, nested RT-PCR represents a
promising technique for the widespread evaluation of gastropathies,
for the detection of gastric carcinoma.
References
|
1
|
Orditura M, Galizia G, Sforza V, et al:
Treatment of gastric cancer. World J Gastroenterol. 20:1635–1649.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wielenga VJ, Heider KH, Offerhaus GJ, et
al: Expression of CD44 variant proteins in human colorectal cancer
is related to tumor progression. Cancer Res. 53:4754–4756.
1993.PubMed/NCBI
|
|
3
|
Hsu KH, Tsai HW, Shan YS and Lin PW:
Significance of CD44 expression in gastrointestinal stromal tumors
in relation to disease progression and survival. World J Surg.
31:1438–1444. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Quiding-Järbrink M, Ahlstedt I, Lindholm
C, Johansson EL and Lönroth H: Homing commitment of lymphocytes
activated in the human gastric and intestinal mucosa. Gut.
49:519–525. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
East JA and Hart IR: CD44 and its role in
tumour progression and metastasis. Eur J Cancer. 29A:1921–1922.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Herrlich P, Zöller M, Pals ST and Ponta H:
CD44 splice variants: Metastases meet lymphocytes. Immunol Today.
14:395–399. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xin Y, Grace A, Gallagher MM, Curran BT,
Leader MB and Kay EW: CD44V6 in gastric carcinoma: A marker of
tumor progression. Appl Immunohistochem Mol Morphol. 9:138–142.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Finn L, Dougherty G, Finley G, Meisler A,
Becich M and Cooper DL: Alternative splicing of CD44 pre-mRNA in
human colorectal tumors. Biochem Biophys Res Commun. 200:1015–1022.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamaguchi A, Saito M, Gio T, et al:
Expression of CD44 variant exons 8–10 in gastric cancer. Jpn J
Cancer Res. 86:1166–1171. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ghaffarzadehgan K, Jafarzadeh M, Raziee
HR, et al: Expression of cell adhesion molecule CD44 in gastric
adenocarcinoma and its prognostic importance. World J
Gastroenterol. 14:6376–6381. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamaguchi A, Goi T, Yu J, et al:
Expression of CD44v6 in advanced gastric cancer and its
relationship to hematogenous metastasis and long term prognosis. J
Surg Oncol. 79:230–235. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sasaki JI, Tanabe KK, Takahashi K, et al:
Expression of CD44 splicing isoforms in lung cancers: Dominant
expression of CD44v8-10 in non-small cell lung carcinomas. Int J
Oncol. 12:525–533. 1998.PubMed/NCBI
|
|
13
|
Ahn MJ, Noh YH, Yoon HJ, et al: Detection
of malignant cells in pleural fluid or ascites by CD44v8-10/CD44v10
competitive RT-PCR. Korean J Int Med. 16:30–35. 2001. View Article : Google Scholar
|
|
14
|
Miyake H, Eto H, Arakawa S, Kamidono S and
Hara I: Over expression of CD44V8-10 in urinary exfoliated cells as
an independent prognostic predictor in patients with urothelial
cancer. J Urol. 167:1282–1287. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lau WM, Teng E, Chong HS, et al: CD44v8-10
is a cancer-specific marker for gastric cancer stem cells. Cancer
Res. 74:2630–2641. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu B, El Hajj N, Sittler S, Lammert N,
Barnes R and Meloni-Ehrig A: Gastric cancer: Classification,
histology and application of molecular pathology. J Gastrointest
Oncol. 3:251–261. 2012.PubMed/NCBI
|
|
17
|
Matsumura Y and Tarin D: Significance of
CD44 gene products for cancer diagnosis and disease evaluation.
Lancet. 340:1053–1058. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takeuchi K, Yamaguchi A, Urano T, Goi T,
Nakagawara G and Shiku H: Expression of CD44 variant exons 8–10 in
colorectal cancer and its relationship to metastasis. Jpn J Cancer
Res. 86:292–297. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chomczynski P and Sacchi N: Single-step
method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction. Anal Biochem.
162:156–159. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mathew J, Hines JE, Obafunwa JO, Burr AW,
Toole K and Burt AD: CD44 is expressed in hepatocellular carcinomas
showing vascular invasion. J Pathol. 179:74–79. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Haynes BF, Telen MJ, Hale LP and Denning
SM: CD44 - a molecule involved in leukocyte adherence and T-cell
activation. Immunol Today. 10:423–428. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mackay CR, Terpe HJ, Stauder R, Marston
WL, Stark H and Günthert U: Expression and modulation of CD44
variant isoforms in humans. J Cell Biol. 124:71–82. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shiozaki H, Oka H, Inoue M, Tamura S and
Monden M: E-cadherin mediated adhesion system in cancer cells.
Cancer. 77(Suppl): S1605–S1613. 1996. View Article : Google Scholar
|
|
24
|
Wang T, Ong CW, Shi J, et al: Sequential
expression of putative stem cell markers in gastric carcinogenesis.
Br J Cancer. 105:658–665. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yokozaki H, Ito R, Nakayama H, Kuniyasu H,
Taniyama K and Tahara E: Expression of CD44 abnormal transcripts in
human gastric carcinomas. Cancer Lett. 83:229–234. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takaishi S, Okumura T, Tu S, et al:
Identification of gastric cancer stem cells using the cell surface
marker CD44. Stem Cells. 27:1006–1020. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Miwa T, Watanabe A, Yamada Y, et al:
Progression in gastric carcinoma relative to the ratio of CD44
epithelial variant transcript to CD44 hematopoietic variant
transcript. Cancer. 77:25–29. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen GY and Wang DR: The expression and
clinical significance of CD44v in human gastric cancers. World J
Gastroenterol. 6:125–127. 2000.PubMed/NCBI
|
|
29
|
Mayer B, Jauch KW, Günthert U, et al:
De-novo expression of CD44 and survival in gastric cancer. Lancet.
342:1019–1022. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Miyake H, Okamoto I, Hara I, et al: Highly
specific and sensitive detection of malignancy in urine samples
from patients with urothelial cancer by CD44v8-10/CD44v10
competitive RT-PCR. Int J Cancer. 79:560–564. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Okamoto I, Morisaki T, Sasaki J, et al:
Molecular detection of cancer cells by competitive reverse
transcription-polymerase chain reaction analysis of specific CD44
variant RNAs. J Natl Cancer Inst. 90:307–315. 1998. View Article : Google Scholar : PubMed/NCBI
|