Introduction
microRNAs (miRs) are known to be involved in the
development of hepatocellular carcinoma (HCC). They are non-coding
microregulators of ~22 nucleotides in length, which are involved in
the regulation of hepatocarcinogenesis (1). miRs may function as oncogenes or as
tumor suppressors. An example of an oncogenic miR, or oncomiR, is
miR-221. HCC cells were shown to overexpress miR-221, which, in
turn, enhances HCC cell proliferation, migration and invasion by
enhancing AKT signaling (2–4). By contrast, miR-122, a tumor suppressor,
is the most abundantly expressed miR in the healthy liver, while it
is downregulated in HCC (5). Inducing
the expression of miR-122 has been shown to induce apoptosis and
suppress proliferation in HepG2 and Hep3B cells, which is in part
due to direct targeting of Cyclin G1 (6,7).
miR-155 has been demonstrated to be involved in the
development of a number of types of malignancies, such as B cell
malignancies (8), breast cancer
(9) and colon cancer (10), in addition to hepatocellular carcinoma
(11), which was the focus of the
present study. It has been reported that miR-155 is an oncomiR that
is frequently upregulated in HCC (12). miR-155 was found to be involved in the
initiation of hepatocarcinogenesis, in which it provided a link
between inflammation and HCC. Zhang et al (13) reported that the inflammation-related
transcription factor, nuclear factor κB, induces upregulation of
miR-155, which in turn promotes tumorigenesis by activating Wnt
signaling. Similarly, loss of function studies demonstrated that
silencing miR-155 leads to G0/G1 cell cycle arrest and increased
cell death (13). This is in
accordance with other observations, where the upregulation of
miR-155 in HCC has been shown to correlate with the upregulation of
oncogenes, such as cyclin D1, c-MYC and matrix metalloproteinase-9
(14), and the downregulation of
tumor suppressors, such as phosphatase and tensin homolog and
CCAAT/enhancer binding protein β (12). Therefore, miR-155 is hypothesized to
be a potential target for the treatment of HCC.
Insulin-like growth factor-II (IGF-II) is a member
of a complex system, termed the IGF axis. The IGF axis consists of
two ligands, two receptors and six IGF-binding proteins (IGFBPs).
Upon binding of the ligands to the tyrosine kinase receptor, IGF
type-1 receptor (IGF-1R), activation of the RAF/MEK/ERK and the
PI3K/AKT/mTOR mitogenic signaling pathways occurs, which leads to
the induction of cell proliferation, differentiation and survival
(15). Uncontrolled IGF-II-IGF-1R
interaction, and consequent excessive intracellular mitogenic
signals, is a common feature of a number of types of cancer. This
is ordinarily restricted by the presence of IGFBPs, which sequester
IGF-II in the blood and thereby limit its interaction with IGF-1R
(16).
Three IGF axis members are known to be involved in
the development of HCC. IGF-II has been reported to be upregulated
in HCC tissues, and this increase in expression was correlated with
cancer cell proliferation and tumor neovascularization (17,18).
IGF-1R has also been demonstrated to be upregulated in HCC.
Rodriguez-Tarduchy et al (19)
reported that the primary tumorigenic effects of IGFs are regulated
by IGF-IR. By contrast, the expression of IGFBP-3 has been found to
be downregulated in HCC, and appears to exhibit tumor suppressor
effects, by neutralizing IGFs and inducing the expression of p53
(20).
Based on preliminary in silico miR-mRNA
binding predictions, the current study aimed to investigate the
effect of miR-155 on the three IGF axis family members, and on HCC
cellular functions.
Patients and methods
Patients
The present study included 23 patients with HCC, who
underwent liver transplant surgery in the Kasr El Einy Hospital
(Cairo University, Cairo, Egypt). Four samples of cirrhotic tissues
were taken from a subsection of these patients with focal HCC
lesions. Ten biopsies from healthy livers were obtained. According
to the agreement between the ethical review committee of the German
University in Cairo and the institutional review board of Cairo
University, ethical approval was obtained for the present study. In
addition, all participants provided written informed consent. The
institutional ethics committees approving this research comply with
the principles set forth in the international reports and
guidelines of the Helsinki Declaration and the International
Ethical Guidelines for Biomedical Research Involving Human
Subjects, issued by the Council for International Organizations of
Medical Sciences. The majority of the patients (66.6%) had >1
focal lesion, as indicated in the pathology report, and were
subjected to clinical assessment, as summarized in Table I.
 | Table I.Clinical assessment of 23 patients
with HCC. |
Table I.
Clinical assessment of 23 patients
with HCC.
| Parameter | Value |
|---|
| Age (years) |
49±13.5 |
| Aspartate
aminotransferase (U/l) |
100.5±65.8 |
| Alanine
aminotransferase (U/l) |
85.6±95.6 |
| Alkaline
Phosphatase (U/l) |
110.2±60.7 |
| Serum albumin
(g/dl) |
4.6±1.5 |
| Serum α fetoprotein
(ng/ml) |
155.7±22.3 |
Cell cultures
HuH-7 and HepG2 cells were maintained in Dulbecco's
modified Eagle's medium (Lonza, Basal, Switzerland), supplemented
with 4.5 g/l glucose, 4 mmol/l L-glutamine, 10% fetal bovine serum
and Mycozap (1:500, Lonza) at 37°C in a 5% CO2
atmosphere.
Transfection of miR
oligonucleotides
HuH-7 and HepG2 cell lines were transfected with
miScript™ miRNA mimics/inhibitors of miR-155, and scrambled miRs
(Qiagen, Venlo, The Netherlands). All transfection experiments were
conducted at room temperature using HiPerfect Transfection Reagent
(Qiagen), according to the manufacturer's instructions. Experiments
were performed in triplicate and repeated 3 times. Cells that were
exposed only to the transfection reagent were designated mock
cells; cells transfected with scrambled miRs were designated as
Scr-miR cells; cells transfected with miR-155 mimics were
designated miR-155 cells; and cells transfected with the miR-155
inhibitor were designated anti-miR-155 cells. Cells were lysed 48 h
posttransfection and total RNA was extracted for further
analysis.
mRNA and miR extraction from liver
biopsies and HCC cell lines
mRNA and miR were extracted from liver biopsies and
the HCC cell lines. Fresh liver samples (HCC, cirrhotic and healthy
tissues) were collected during surgery and were immediately
snap-frozen in liquid nitrogen. The specimens were manually
pulverized in liquid nitrogen and ~100 mg of the resulting tissue
powder was used for large and small RNA extraction, using mirVana
miRNA Isolation kit (Ambion Life Technologies, Austin, TX, USA),
according to the manufacturer's instructions. Cells were harvested
48 h after transfection, according to the HiPerfect Transfection
Reagent instructions. For transfection of HuH-7 cell, 150 ng
oligonucleotides was used and for HepG2 cells, 300 ng
oligonucleotides were used in 6-well plates. The RNA yield was
quantified using a V-530 UV-Vis Spectrophotometer (Jasco
International Co., Ltd., Tokyo, Japan), and RNA integrity was
tested by 18S rRNA bands detection on 1% agarose gel
electrophoresis. RNA samples with an Optical Density 260/280 of
>2 were excluded.
miR and mRNA quantification
Extracted miRs were reverse transcribed into single
stranded cDNA, using a TaqMan® MicroRNA Reverse Transcription kit
(Applied Biosystems Life Technologies, Foster City, CA, USA), and
specific primers for hsa-miR-155 and RNU6B. IGF-II mRNA was reverse
transcribed into cDNA, using the high-capacity cDNA reverse
transcription kit (Applied Biosystems Life Technologies), according
to the manufacturer's instructions. The relative expression of
miR-155 to that of RNU6B, as well as that of IGF-II, IGF-1R and
IGFBP-3 to that of β-microglobulin, was quantified with TaqMan
Real-Time Q-PCR (ABI Assay IDs: 002623, 001093, Hs01005963_m1,
Hs00609566_m1, Hs00365742_g1 and Hs00984230_m1 respectively), using
StepOne™ Systems (Applied Biosystems Life Technologies). The
polymerase chain reaction (PCR) for miR quantification included 1
µl TaqMan Small RNA Assay (20X) specific for miR-155 or RNU6B and
1.33 µl cDNA from the miR-155 or RNU6B RT reactions, respectively.
TaqMan Gene Expression Assay (1.25 µl) specific for each of the
three target genes, IGF-II, IGF-1R and IGFBP-3, and 5 µl cDNA from
the mRNA reverse transcription reaction were used for
quantification. For both reactions, TaqMan Universal PCR Master Mix
II (2X) was used for quantifications (Applied Biosystems Life
Technologies) and the cycling conditions were as follows: 2 min at
50°C and 10 min at 95°C followed by 40 cycles of 15 sec at 95°C and
60 sec at 60°C. The relative expression was calculated using the
2−∆∆Ct method. All PCR reactions, including controls,
were run in duplicate.
Cell proliferation assay (BrdU)
HuH-7 and HepG2 cells were seeded 24 h prior to
transfection in black 96-well plates, and transfected with 12.5 ng
and 25 ng oligonucleotides, respectively, according to the
HiPerfect instructions, with an initial constant cell count of
5×104 cells/well. At 48 h following oligonucleotide
transfection, cells were labeled with BrdU labeling reagent for 4 h
(final concentration, 100 uM), using the Cell Proliferation ELISA
(Roche Applied Science, Penzberg, Germany). Cells were then fixed
using FixDenate for 30 min and incubated with Anti-BrdU POD (final
concentration, 10 uM) for 90 min. Colorimetric measurements were
performed using Wallac 1420 Victor2™ Multilabel Counter
(PerkinElmer, Inc., Waltham, MA, USA). All cell proliferation
experiments were conducted in triplicate and repeated three
times.
Cell scratch wound healing assay
HuH-7 and HepG2 cells were cultured to 80–90%
confluence in 6-well plates. At 48 h post-transfection, 5
scratches/well were made in each plate with 200-µl pipette tip.
Detached cells were removed using serum-free medium. Medium
[Dulbecco's modified Eagle's medium supplemented with 4.5 g/l
glucose, 4 mmol/l L-glutamine, 1% fetal bovine serum and Mycozap
(1:500)] was then added and culture plates were incubated at 37°C
for 48 h. Migration was then documented and wound closure was
quantified with Image J software for Windows (version 1.48;
http://rsbweb.nih.gov/ij/download.html), by measuring
the surface area covered by the cells. All Scratch assays were
conducted in duplicate (two wells/test, representing 10
scratches/test) and repeated three times.
Colony-forming assay
HuH-7 cells were seeded at an initial count of 1,000
cells/well and left to adhere overnight. Cells were then
transfected with miR-155 mimics, inhibitors or small interfering
RNA to IGF-II (siIGF-II). At 24 h post-transfection, cells were
detached by trypsinization, and embedded on a gel composed of a
soft 0.36% (w/v) agarose bottom layer and a hard 0.76% cell-free
media-enriched agarose top layer. Cells were incubated at 37°C and
colonized for 2 weeks. Colonies were observed under a light
microscope (Axio Observer A1; Zeiss, Oberkochen, Germany) and the
number of cells in each well was counted. All colony-forming assays
were conducted in duplicate (two wells/test) and repeated three
times.
Bioinformatics
Bioinformatics algorithms; microrna.org (www.microrna.org), miRDB (www.mirdb.org/miRDB/), DIANA Lab (www.diana.cslab.ece.ntua.gr/) and Target Scan
(www.targetscan.org/) were used to
predict 3′-UTR downstream targets for miR-155. 5′-UTR regions were
predicted using bibiserv software (Bielefeld University
Bioinformatics Server; http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html).
Statistical analysis
All data are presented as the mean ± standard error
of the mean. Statistical significance was analyzed by performing
one-way analysis of variance or paired Student's t-tests using
GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-155 screening in non-metastatic
liver cancer tissues and HCC cell lines
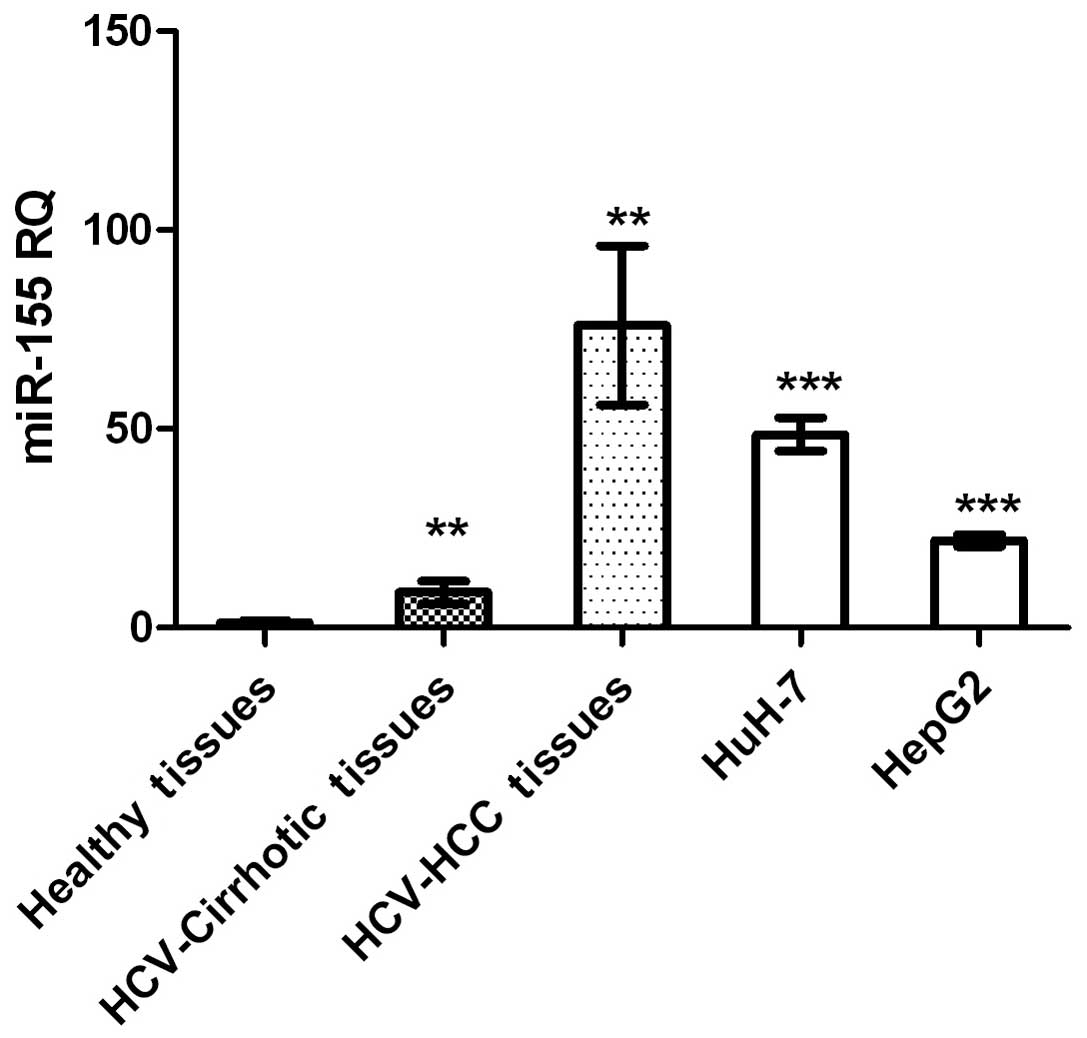
miR-155 expression was significantly higher in HCC
and cirrhotic tissues, and in HCC cell lines, compared with that in
healthy tissues (P=0.0015, P=0.0036 and P<0.0001, respectively;
Fig. 1).
Bioinformatics
miR-155 accession numbers and mature sequences were
retrieved using the miRBase database (http://www.mirbase.org/). Using different in silico
algorithms for predicting binding to the downstream targets at the
3′-UTR and 5′-UTR positions, miR-155 was shown to target IGF-II
IGF-1R and IGFBP-3 mRNA. Microrna.org
and miRDeep2 computational algorithms, demonstrated that miR-155
targeted the 3′-UTR of the IGF-II ligand and IGF-1R, although not
that of IGFBP-3 (Table II). By
contrast, miR-155 was predicted to target the 5′-UTR of IGF-II,
IGF-1R and IGFBP-3, according to bibiserv software (Table II).
 | Table II.Binding sites of miR-155 to IGF-II,
IGF-1R and IGFBP-3 UTRs. |
Table II.
Binding sites of miR-155 to IGF-II,
IGF-1R and IGFBP-3 UTRs.
| A, 5′-UTR target
regions |
|
|---|
|
|---|
| IGF family
member | miR-155 binding
position |
|---|
| IGF-II | Pos: 658 |
| IGF-1R | Pos: 127 |
| IGFBP-3 | Pos: 114 |
|
| B, 3′-UTR target
regions |
|
|
| IGF family
member | miR-155 binding
position |
|
|---|
| IGF-II | Pos: 3838 |
| IGF-1R | Pos: 4205 |
| IGFBP-3 | N/A |
Transfection efficiency of miR-155
oligonucleotides
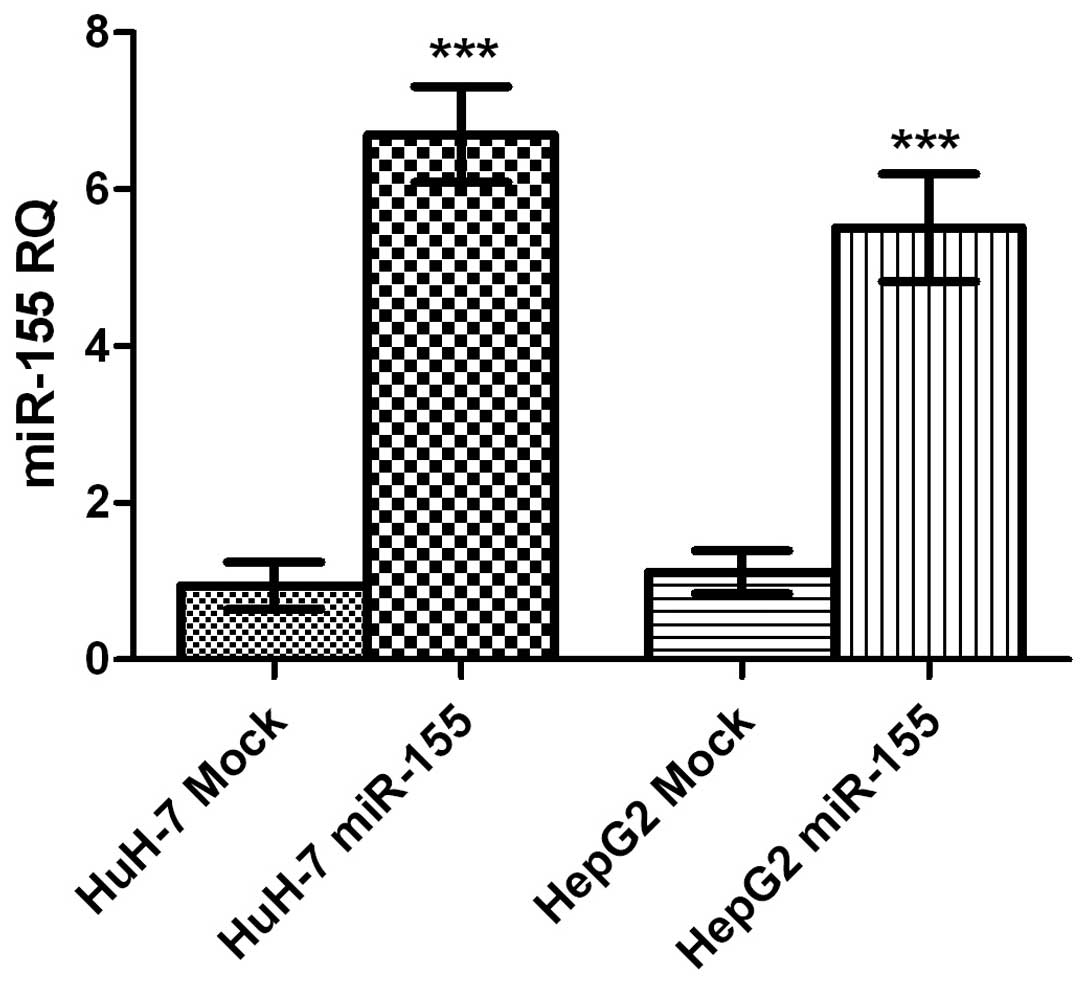
HuH-7 and HepG2 cells transfected with miR-155
mimics, exhibited an upregulation in miR-155 expression of 20-fold
and 17-fold compared with mock HuH-7 and HepG2 cells, respectively
(both P<0.0001; Fig. 2).
IGF-II, IGF-1R and IGFBP-3 mRNA
expression following induction and inhibition of miR-155
expression
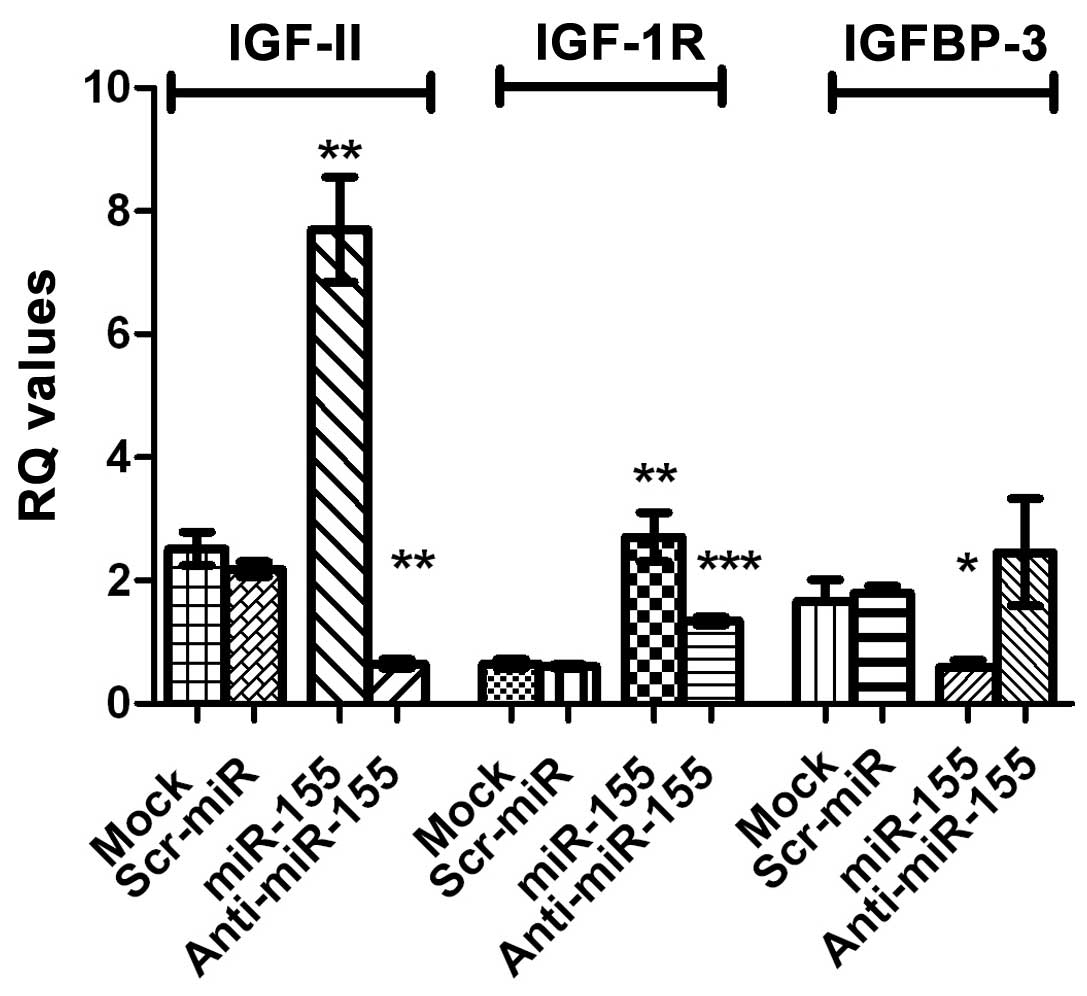
Transfection of HuH-7 with miR-155 mimics resulted
in a significant increase in the expression of IGF-II mRNA
(P=0.0046) as well as that of IGF-1R mRNA (P=0.0078), compared with
control cells. By contrast, a significant downregulation of IGFBP-3
mRNA expression, compared with that of mock cells, was observed
(P=0.0458). Inhibition of miR-155, significantly repressed IGF-II
and IGF-1R mRNA expression in HuH-7 cells (P=0.0008 and P=0.0006,
respectively). By contrast, IGFBP-3 mRNA expression was increased
in cells transfected with miR-155, compared with miR-155
mimic-transfected cells, as shown in Fig.
3. The same statistically significant effects were observed in
HepG2 cells; for example, a significant increase in IGF-II mRNA
expression was observed in HepG2 cells tranfected with miR-155
mimics (P=0.029; data not shown).
Effect of miR-155 inhibition on
cellular functions
Cellular proliferation
Transfection with the miR-155 inhibitor,
significantly decreased HuH-7 and HepG2 cell proliferation
(P=0.0005 and P=0.0009, respectively), compared with that of
negative control and mock cells (Fig.
4A). By contrast, transfection with miR-155 mimics
significantly enhanced the proliferation HuH-7 and HepG2 cells
(P=0.0017 and P=0.0292, respectively Fig.
4A). It is worth noting that cells transfected with siIGF-II
were used for confirmation and showed a negative effect on cell
proliferation in Huh7 and HepG2 cells (P=0.0005 and P=0.0042,
respectively; Fig. 4A), similar to
the effect of miR-155 inhibitors.
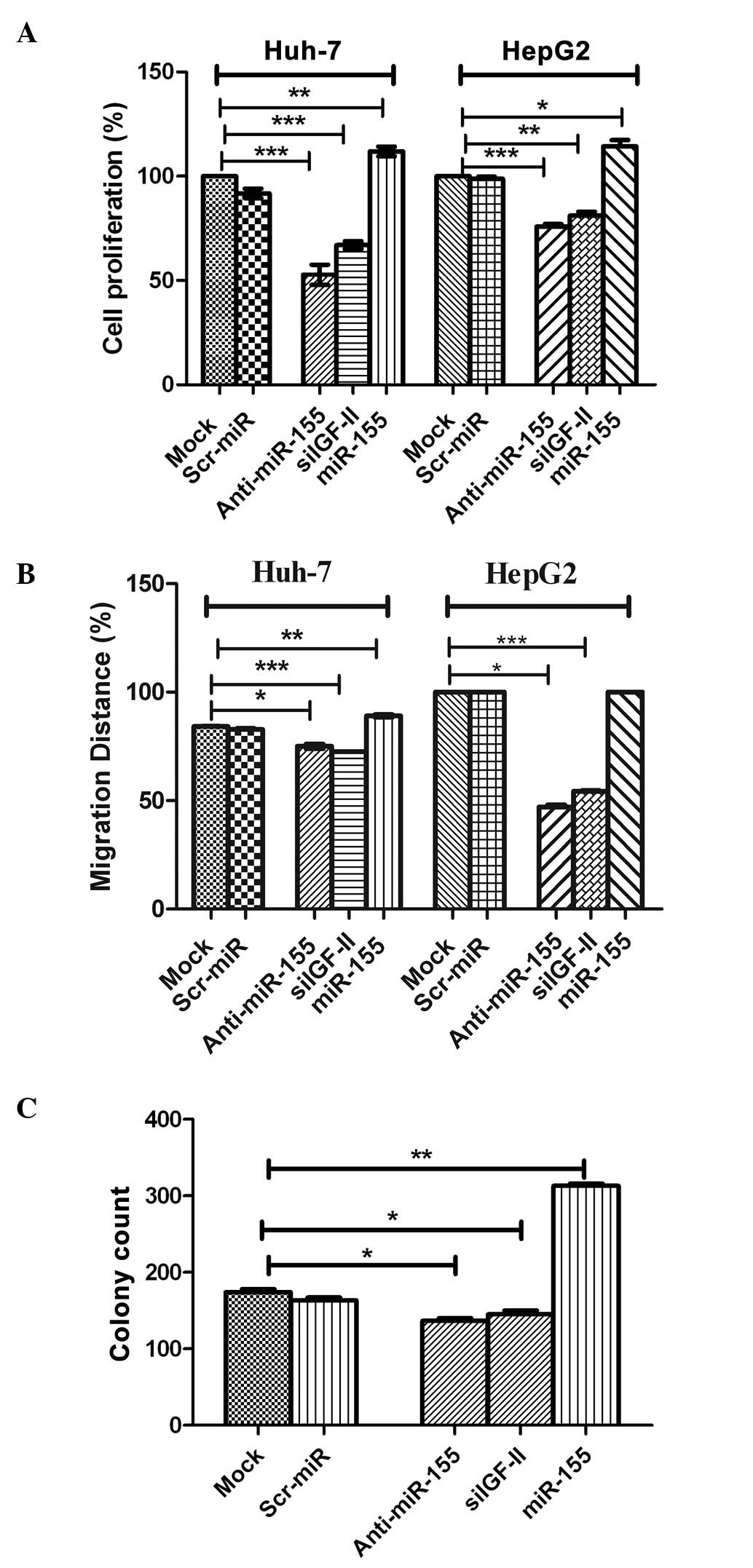 | Figure 4.Effect of miR-155 inhibition on
cellular functions. (A) Cellular proliferation. Inhibition of
miR-155 significantly decreased HuH-7 and HepG2 cell proliferation
compared with negative control and mock cells (P=0.0005 and
P=0.0009, respectively). This was in contrast to induced miR-155
expression using miR-155 mimics, which significantly enhanced the
proliferation of HuH-7 and HepG2 cells (P=0.0017 and P=0.0292,
respectively). (B) Cellular migration. Inhibition of miR-155
abrogated the effect of miR-155 on the promotion of migration in
HuH-7 and HepG2 cells compared with the controls, with coverage of
48 and 76% (P=0.0002 and P=0.0001, respectively) of the original
scratch, respectively. By contrast, transfection of miR-155 mimics
resulted in the promotion of tumor cell migration in HuH-7 and
HepG2 cells, with coverage of 89 and 100% of the original scratch,
compared with mock and Scr-miR, which had an original scratch
coverage of 84.2 and 82.7%, and 100 and 100%, in these cell lines,
respectively. (C) Cellular clonogenicity. Inhibition of miR-155
suppressed its tumorigenic effect by significantly decreasing the
colony-forming ability of HuH-7 cells compared with that of with
untransfected mock cells (P=0.0195). Induction of miR-155
overexpression induced high clonogenicity in HuH-7 cells compared
with mock cells (P=0.0013). miR, microRNA; Scr-miR, scrambled miR;
siIGF, small interfering RNA specific to insulin-like growth factor
II. *P<0.05; **P<0.01; ***P<0.001. |
Cellular migration
Images for the 2D scratch-migration assay were
captured at 5× magnification (21).
Migration inhibition by miR-155 antagomirs (inhibitors) abrogated
the migratory effect of miR-155 in HuH-7 and HepG2 cells compared
with the controls, with coverage of 48 and 76% of the original
scratch, respectively (Fig. 4B). By
contrast, transfection of miR-155 mimics resulted in the promotion
of tumor cell migration in HuH-7 and HepG2 cells, with coverage of
89 and 100% of the original scratch, respectively, compared with
mock and Scr-miR, which exhibited original scratch coverage of 84.2
and 82.7%, and 100 and 100%, in the HuH-7 and HepG2 cell lines,
respectively (Fig. 4B). siIGF-II also
significantly decreased the migration of Huh7 and HepG2 cells
compared with mock untransfected cells, with coverage of 72.5 and
54.0% (P=0.0002 and P=0.0001, respectively) of the original
scratch, respectively (Fig. 4B).
Cellular clonogenicity
Inhibition of miR-155 suppressed its tumorigenic
effect by significantly decreasing the colony-forming capability of
HuH-7 cells, compared with that of untransfected mock cells
(P=0.0195; Fig. 4C). This was in
contrast to the induction of miR-155 overexpression, which induced
high clonogenicity in HuH-7 cells, compared with that of mock cells
(P=0.0013; Fig. 4C). Furthermore,
siIGF-II significantly suppressed colony formation in Huh7 cells
compared with mock untransfected cells (P=0.0455; Fig. 4C).
Discussion
One of the pathways that may be involved in the
pathogenesis of human HCC, is the IGF axis. Aberrant expression of
members of the IGF axis is a hallmark of the initiation,
progression and metastasis of HCC (21–25). In
addition, miR-155 was recently demonstrated to be involved in HCC
proliferation and migration (11,26).
Therefore, the present study aimed to investigate whether an IGF
axis-miR-155 interaction exists in HCC. In silico analysis
demonstrated that miR-155 had putative target sites in the 3′-UTR
regions of two IGF axis members, involved in HCC, namely IGF-II and
IGF-1R (Table II). The aim of the
current study was to examine the interaction between miR-155 and
certain IGF axis members, in order to improve the understanding of
the mechanisms underlying the development of HCC.
Expression profiling of miR-155 demonstrated higher
expression of this molecule in liver biopsies from patients with
non-metastatic HCC and cirrhosis, compared with those from healthy
controls (Fig. 1). Similarly, when
measured in HuH-7 and HepG2 cell lines, miR-155 expression was
significantly upregulated compared with healthy controls (Fig. 1). These findings are in accordance
with those from other studies, which reported the elevation of
miR-155 expression in non-neoplastic tissues compared with that in
normal liver tissues (27), and in
biopsies from HCV-induced and HBV-induced HCC, compared with
healthy tissues (11,13). Since the increase in miR-155
expression occurs at an early stage of hepatocarcinogenesis, even
prior to the development of cirrhosis (28), it was hypothesized to act as a
oncomir, bridging inflammation with carcinogenesis (13,29).
Functional analysis, performed in HuH-7 and HepG2 cell lines,
demonstrated that miR-155 promoted cellular proliferation (Fig. 4A), migration (Fig. 4B) and anchorage-independent growth
(Fig. 4C). This data is in accordance
with previous studies, which demonstrated that miR-155 promotes
invasiveness of HCC cells and tumor recurrence, and predicts poor
survival in patients with HCC following liver transplantation
(14,26). Transfection of HuH-7 cells with
inhibitors of miR-155, resulted in a significant reduction in the
proliferation, migration and anchorage-independent growth of these
cells, compared with mock cells (Fig.
4A–C). This data is in accordance with that from a previous
study, which reported reduced cell growth and enhanced cell death
in xenograft tumors, following knockdown of miR-155 expression in
J7 and Mahlavu hepatoma cells (30).
In addition, a separate study showed that miR-155 overexpression
leads to the promotion of cell proliferation and tumorigenesis, and
the inhibition of hepatocyte apoptosis, through the activation of
Wnt signaling, by enhancing β-catenin nuclear accumulation with a
simultaneous increase in cyclin D1 and c-myc expression (13).
This indicates that miR-155 may act through
alternative mechanisms, to link inflammation with tumorigenesis.
Preliminary in silico analysis demonstrated that miR-155 has
potential target sites in the 3′-UTR regions of IGF-II and IGF-1R,
although not that of IGFBP-3 (Table
II). Despite this prediction, IGF-II and IGF-1R mRNA expression
was significantly upregulated following ectopic induction of
miR-155 expression in HuH-7 and HepG2 cells, compared with that in
the respective untransfected mock cells (Fig. 3). This overexpression of the
downstream targets was restored to normal levels following
transfection of HuH-7 cells with specific antagomirs or miR-155
(Fig. 3). Ordinarily, miRNAs act by
base-pairing with 3′-UTR of their target mRNAs, thereby hindering
their translation into proteins or enhancing their degradation
(31,32). However, the anticipated downregulation
by miR-155 of its predicted targets did not occur in the present
study. Further analysis using bioinformatic tools, demonstrated
that miR-155 has potential target sites in the 5′-UTR regions of
IGF-II and IGF-1R (Table II). The
present findings were in accordance with those of another study,
which proposed a new mechanism of action of miRNAs (33). The authors reported that 5′-UTR
binding may lead to the activation of gene expression, through
stabilization of the mRNA rather than the induction of its
degradation, which may, in part, explain the observed induction of
IGF-II and IGF-1R mRNAs by miR-155. This was not the first study to
report the enhancement of target mRNA expression by miRNAs as a
result of binding to the 5′-UTR region. Other studies have
demonstrated that miR-122 and miR-346 may bind to the 5′-UTR of HCV
RNA and Receptor-Interacting Protein 140, respectively, resulting
in the induction of translation and thereby the upregulation of the
expression of these proteins (34–37).
By contrast, the tumor suppressor, IGFBP-3, was
predicted to bear a putative miR-155 binding site in its 5′-UTR,
although not in its 3′-UTR (Table
II). However, it was observed that IGFBP-3 mRNA expression was
significantly downregulated in HuH-7 cells transfected with miR-155
mimics, compared with the untransfected mock cells (Fig. 3). The translational repression by
miRNA-mediated 5′-UTR binding has been reported for miR-2 in
Drosophila (38). The authors
reported 6 miRNA binding sites in the 5′-UTR and in the open
reading frame, where miR-2 binding inhibits translation initiation
in a cap-dependent manner, induces mRNA deadenylation and decrease
80S complex formation, resulting in inhibition of the translation
initiation step.
The present results suggest a possible mechanism
underlying the involvement of miR-155 in the development of HCC.
Following transfection of HuH-7 cells with miR-155 mimics, there
was a concomitant increase in the mRNA expression of IGF-II and
IGF-1R, which are mitogenic, and a decrease in the mRNA expression
of IGFBP-3 mRNA, which is a tumor suppressor. This variability in
the expression of the IGF axis family members, potentiates the
tumorigenic effect of miR-155 by activating the downstream
signaling cascade, eventually leading to tumor formation,
progression and metastasis.
In conclusion, the present study demonstrated that
miR-155 expression is higher in cirrhotic and non-metastatic HCC
tissues, compared with that in healthy controls (Fig. 1), which is in accordance with the
results of previous studies, implicating its upregulation in each
stage of tumor formation beginning with inflammation, passing
through cirrhosis, and culminating with HBV- and HCV-induced HCC
(11,27,28). This
is in contrast to other metastamiRs, such as miR-17-5p, which was
found to be significantly downregulated in non-metastatic HCC,
while its expression was upregulated only in metastatic HCC
(39,40). The current data indicate that miR-155
is an oncogenic miR that is involved early in the development of
HCC. To the best of our knowledge, the present study demonstrated,
for the first time, a novel mechanism through which miR-155
regulates multiple oncogenic IGF pathway components in HCC. The
current results provide a rationale for the potential use of
miR-155 in therapeutic approaches as a ‘one for all’ target,
through which multiple oncogenic targets may be inhibited and tumor
suppressors induced.
Acknowledgements
The authors would like to thank the patients and
healthy volunteers for their participation. This study was funded
by the Science and Technology Development Fund.
References
|
1
|
Sidhu K, Kapoor NR, Pandey V and Kumar V:
The ‘macro’ world of microRNAs in hepatocellular carcinoma. Front
Oncol. 5:682015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pineau P, Volinia S, McJunkin K, Marchio
A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM and
Dejean A: MiR-221 overexpression contributes to liver
tumorigenesis. Proc Natl Acad Sci USA. 107:264–269. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yuan Q, Loya K, Rani B, Möbus S,
Balakrishnan A, Lamle J, Cathomen T, Vogel A, Manns MP, Ott M, et
al: MicroRNA-221 overexpression accelerates hepatocyte
proliferation during liver regeneration. Hepatology. 57:299–310.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jensen MR, Factor VM, Fantozzi A, Helin K,
Huh CG and Thorgeirsson SS: Reduced hepatic tumor incidence in
cyclin G1-deficient mice. Hepatology. 37:862–870. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Girard M, Jacquemin E, Munnich A, Lyonnet
S and Henrion-Caude A: miR-122, a paradigm for the role of
microRNAs in the liver. J Hepatol. 48:648–656. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gramantieri L, Ferracin M, Fornari F,
Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E,
Grazi GL, et al: Cyclin G1 is a target of miR-122a, a microRNA
frequently down-regulated in human hepatocellular carcinoma. Cancer
Res. 67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Datta J, Kutay H, Nasser MW, Nuovo GJ,
Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, et
al: Methylation mediated silencing of MicroRNA-1 gene and its role
in hepatocellular carcinogenesis. Cancer Res. 68:5049–5058. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang M, Tan LP, Dijkstra MK, van Lom K,
Robertus JL, Harms G, Blokzijl T, Kooistra K, van T'veer MB, Rosati
S, et al: miRNA analysis in B-cell chronic lymphocytic leukaemia:
Proliferation centres characterized by low miR-150 and high
BIC/miR-155 expression. J Pathol. 215:13–20. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu
H, Liu MF and Wang ED: MicroRNA-155 functions as an OncomiR in
breast cancer by targeting the suppressor of cytokine signaling 1
gene. Cancer Res. 70:3119–3127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bakirtzi K, Hatziapostolou M,
Karagiannides I, Polytarchou C, Jaeger S, Iliopoulos D and
Pothoulakis C: Neurotensin signaling activates microRNAs-21 and
−155 and Akt, promotes tumor growth in mice and is increased in
human colon tumors. Gastroenterology. 1411749–1761. (e1)2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xie Q, Chen X, Lu F, Zhang T, Hao M, Wang
Y, Zhao J, McCrae MA and Zhuang H: Aberrant expression of microRNA
155 may accelerate cell proliferation by targeting sex-determining
region Y box 6 in hepatocellular carcinoma. Cancer. 118:2431–2442.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang B, Majumder S, Nuovo G, Kutay H,
Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K and Jacob ST:
Role of microRNA-155 at early stages of hepatocarcinogenesis
induced by choline-deficient and amino acid-defined diet in C57BL/6
mice. Hepatology. 50:1152–1161. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Wei W, Cheng N, Wang K, Li B,
Jiang X and Sun S: Hepatitis C virus-induced up-regulation of
microRNA-155 promotes hepatocarcinogenesis by activating Wnt
signaling. Hepatology. 56:1631–1640. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yan XL, Jia YL, Chen L, Zeng Q, Zhou JN,
Fu CJ, Chen HX, Yuan HF, Li ZW, Shi L, et al: Hepatocellular
carcinoma-associated mesenchymal stem cells promote hepatocarcinoma
progression: Role of the S100A4-miR155-SOCS1-MMP9 axis. Hepatology.
57:2274–2286. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pollak M: Insulin and insulin-like growth
factor signalling in neoplasia. Nat Rev Cancer. 8:915–928. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fürstenberger G and Senn HJ: Insulin-like
growth factors and cancer. Lancet Oncol. 3:298–302. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bae MH, Lee MJ, Bae SK, Lee OH, Lee YM,
Park BC and Kim KW: Insulin-like growth factor II (IGF-II) secreted
from HepG2 human hepatocellular carcinoma cells shows angiogenic
activity. Cancer Lett. 128:41–46. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lahm H, Gittner K, Krebs O, Sprague L,
Deml E, Oesterle D, Hoeflich A, Wanke R and Wolf E:
Diethylnitrosamine induces long-lasting re-expression of
insulin-like growth factor II during early stages of liver
carcinogenesis in mice. Growth Horm IGF Res. 12:69–79. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rodriguez-Tarduchy G, Collins MK, García I
and López-Rivas A: Insulin-like growth factor I inhibits apoptosis
in IL-3-dependent hemopoietic cells. J Immunol. 149:535–540.
1992.PubMed/NCBI
|
|
20
|
Rajah R, Valentinis B and Cohen P:
Insulin-like growth factor (IGF)-binding protein-3 induces
apoptosis and mediates the effects of transforming growth
factor-beta1 on programmed cell death through a p53- and
IGF-independent mechanism. J Biol Chem. 272:12181–12188. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nussbaum T, Samarin J, Ehemann V,
Bissinger M, Ryschich E, Khamidjanov A, Yu X, Gretz N, Schirmacher
P and Breuhahn K: Autocrine insulin-like growth factor-II
stimulation of tumor cell migration is a progression step in human
hepatocarcinogenesis. Hepatology. 48:146–156. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gong Y, Cui L and Minuk GY: The expression
of insulin-like growth factor binding proteins in human
hepatocellular carcinoma. Mol Cell Biochem. 207:101–104. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Aleem E, Nehrbass D, Klimek F, Mayer D and
Bannasch P: Upregulation of the insulin receptor and type I
insulin-like growth factor receptor are early events in
hepatocarcinogenesis. Toxicol Pathol. 39:524–543. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Khandwala HM, McCutcheon IE, Flyvbjerg A
and Friend KE: The effects of insulin-like growth factors on
tumorigenesis and neoplastic growth. Endocr Rev. 21:215–244. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
LeRoith D and Roberts CT Jr: The
insulin-like growth factor system and cancer. Cancer Lett.
195:127–137. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Han ZB, Chen HY, Fan JW, Wu JY, Tang HM
and Peng ZH: Up-regulation of microRNA-155 promotes cancer cell
invasion and predicts poor survival of hepatocellular carcinoma
following liver transplantation. J Cancer Res Clin Oncol.
138:153–161. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yoon SO, Chun SM, Han EH, Choi J, Jang SJ,
Koh SA, Hwang S and Yu E: Deregulated expression of microRNA-221
with the potential for prognostic biomarkers in surgically resected
hepatocellular carcinoma. Hum Pathol. 42:1391–1400. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Eis PS, Tam W, Sun L, Chadburn A, Li Z,
Gomez MF, Lund E and Dahlberg JE: Accumulation of miR-155 and BIC
RNA in human B cell lymphomas. Proc Natl Acad Sci USA.
102:3627–3632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tili E, Michaille JJ, Wernicke D, Alder H,
Costinean S, Volinia S and Croce CM: Mutator activity induced by
microRNA-155 (miR-155) links inflammation and cancer. Proc Natl
Acad Sci USA. 108:4908–4913. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang YH, Lin KH, Chen HC, Chang ML, Hsu
CW, Lai MW, Chen TC, Lee WC, Tseng YH and Yeh CT: Identification of
postoperative prognostic microRNA predictors in hepatocellular
carcinoma. PLoS One. 7:e371882012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mattick JS and Makunin IV: Small
regulatory RNAs in mammals. Hum Mol Genet. 14:R121–R132. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Henke JI, Goergen D, Zheng J, Song Y,
Schüttler CG, Fehr C, Jünemann C and Niepmann M: MicroRNA-122
stimulates translation of hepatitis C virus RNA. EMBO J.
27:3300–3310. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Villanueva RA, Jangra RK, Yi M, Pyles R,
Bourne N and Lemon SM: MiR-122 does not modulate the elongation
phase of hepatitis C virus RNA synthesis in isolated replicase
complexes. Antiviral Res. 88:119–123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Norman KL and Sarnow P: Modulation of
hepatitis C virus RNA abundance and the isoprenoid biosynthesis
pathway by microRNA miR-122 involves distinct mechanisms. J Virol.
84:666–670. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jopling CL: Regulation of hepatitis C
virus by microRNA-122. Biochem Soc Trans. 36:1220–1223. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tsai NP, Lin YL and Wei LN: MicroRNA
mir-346 targets the 5′-untranslated region of receptor-interacting
protein 140 (RIP140) mRNA and up-regulates its protein expression.
Biochem J. 424:411–418. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Moretti F, Thermann R and Hentze MW:
Mechanism of translational regulation by miR-2 from sites in the 5′
untranslated region or the open reading frame. RNA. 16:2493–2502.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang F, Yin Y, Wang F, Wang Y, Zhang L,
Tang Y and Sun S: miR-17-5p Promotes migration of human
hepatocellular carcinoma cells through the p38 mitogen-activated
protein kinase-heat shock protein 27 pathway. Hepatology.
51:1614–1623. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
El Tayebi HM, Omar K, Hegy S, El Maghrabi
M, El Brolosy M, Hosny KA, Esmat G and Abdelaziz AI: Repression of
miR-17-5p with elevated expression of E2F-1 and c-MYC in
non-metastatic hepatocellular carcinoma and enhancement of cell
growth upon reversing this expression pattern. Biochem Biophys Res
Commun. 434:421–427. 2013. View Article : Google Scholar : PubMed/NCBI
|