Introduction
Ovarian cancer is one of the most common malignant
tumors of the female reproductive system, and is the third most
frequently occurring cancer in women, following cervical and
endometrial cancer (1). Ovarian
cancer possesses the poorest prognosis of all gynecological tumors,
and is typically undetectable at onset (2). Thus, the majority of patients are
diagnosed at advanced stages, resulting in a high rate of
mortality. Therefore, ovarian cancer is a significant threat to
female health (3). Due to the fact
that patients with ovarian cancer exhibit no symptoms during the
early stages, as well as a lack of effective screening measures,
>80% patients with ovarian cancer are diagnosed at an advanced
stage (4). These patients possess a
five-year survival rate of just 20–30%; however, the five-year
survival rate of patients with ovarian cancer diagnosed at an early
stage is >90% (5). Therefore,
early diagnosis is significant for the prognosis of patients with
ovarian cancer. At present, certain tumor markers, including cancer
antigen 125 (CA125) and human epididymis protein 4 (HE-4), aid in
the early diagnosis of ovarian cancer (6,7).
Furthermore, these markers are significant for monitoring the
development and progression of ovarian cancer. However, there are
various associated disadvantages, including, high false-positive or
-negative rates and low specificity (7). In order to improve the accuracy of
diagnosis for ovarian cancer, the use of cooperative detection
methods may be required.
Since Leon et al (8) identified cell-free DNA (cf-DNA) in the
serum of cancer patients in 1977, cf-DNA has presented a number of
advantages in the early stages of tumor diagnosis (9,10),
particularly in solid tumors (11).
Previous studies of cf-DNA have included qualitative and
quantitative investigation. Qualitative analysis primarily detects
tumor-specific gene alterations (12,13),
including gene mutations, promoter hypermethylation of tumor
suppressor genes (14) and
microsatellite alterations (15).
Quantitative analyses primarily involve radioimmunity methods
(16), polymerase chain reaction
(PCR) amplification techniques (17),
DNA nick translation labeling techniques (18) and DNA DipStick kits (19). These methods are associated with a
number of issues, including radioactivity, environmental pollution
problems, complex instructions and DNA loss during the process
itself, which result in reduced sensitivity.
Branched DNA (bDNA), in contrast to PCR (which
amplifies a portion of the gene sequence), is a signal
amplification technology that detects the presence of specific
nucleic acids by measuring the signal generated by specific
hybridization of numerous branched, labeled DNA probes on an
immobilized target nucleic acid (20).
The present study utilized a bDNA technique in order
to quantitatively detect the levels of cf-DNA in patients
exhibiting ovarian cancer, and analyzed the association between
cf-DNA levels and clinicopathological characteristics, in order to
investigate the value of cf-DNA in the diagnosis of ovarian cancer
by quantitative measures.
Patients and methods
Patients
Samples were obtained from 36 patients exhibiting
ovarian cancer from the Maternity and Child Healthcare Hospital of
Nantong and the Tumor Hospital of Nantong (Nantong, China) between
October 2011 and October 2012. The average age of patients
exhibiting ovarian cancer was 58.6 years (range, 40–85 years) and
all diagnoses were confirmed via pathological examination. Tumor
staging was performed according to the International Federation of
Gynecology and Obstetrics (FIGO) criteria (2010) (21). In addition, samples from 29 healthy
women served as the control group, and the average age of these
patients was 57.7 years (range, 42–75 years). A total of 22 cases
of benign ovarian tumor were also investigated, and the average age
of these patients was 51.6 years (range, 37–72 years). All
diagnoses were confirmed by surgical and pathological examination.
The present study was conducted in accordance with the Declaration
of Helsinki, and was approved by the Ethics Committee of the
Maternity and Child Healthcare Hospital of Nantong (Nantong,
China). Written informed consent was obtained from all
participants.
Specimen collection
Venous blood (5 ml) was collected from patients with
ovarian cancer and benign tumors, and healthy women, on an empty
stomach in the morning. The blood specimens were collected in
tubes, and subsequently centrifuged at 2,800 × g and 4°C for 15
min. The serum was transferred into Eppendorf tubes (Eppendorf,
Hamburg, Germany) and stored at −80°C.
cf-DNA detection using a bDNA
technique
Specimens were diluted with distilled water
(dilution, 1:20), heated at 95°C for 5 min and subsequently placed
rapidly into cold water. A preparation solution (90 µl) containing
33.0 µl lysate, 55.3 µl Tris-EDTA buffer solution, 0.1 µl
proteinase K, 1.0 µl confining liquid, 0.3 µl capture extender
probe, 0.3 µl label extender probe and 10 µl DNA samples, including
samples, double controls and three standards, was added into
96-well plates, and subsequently blocked using tin foil papers. All
aforementioned reagents were provided by Fuzhou Maixin
Biotechnology Development Co., Ltd., (Fuzhou, China). Plates were
immediately placed in the HB-1000 hybridization oven (Nanjing
Xincheng Health Biotech Co., Ltd., Nanjing, China) and hybridized
at 55°C for 16–21 h. Subsequently, each well was washed three times
with 300 µl scrubbing solution (Fuzhou Maixin Biotechnology
Development Co., Ltd.), followed by addition of 100 µl labeling
probe (dilution, 1:1,000) and incubation at 55°C for 60 min. This
process was repeated once. Subsequently, 100 µl working solution
(Fuzhou Maixin Biotechnology Development Co., Ltd.) was added to
each well following washing, and plates were incubated at room
temperature for 5–10 min. Finally, 96-well plates were placed in
the LMax microtiter plate luminometer (MaxLine Inc., CA, USA);
absorbance values (A) were acquired, and the mean absorbance value
was calculated. cf-DNA standard curves were constructed based on
standard concentrations and A values. cf-DNA levels were calculated
using the standard curves.
CA125 and HE-4 detection
CA125 and HE-4 levels were detected using the Roche
E170 chemiluminescence detector (Roche Diagnostics, Indianapolis,
IN, USA), and CA125 and HE-4 kits (Shanghai Sangon Biological
Engineering Co., Ltd., Shanghai, China), according to the
manufacturer's instructions.
Statistical analysis
Statistical analysis was performed using SPSS
version 18.0 (SPSS Inc., Chicago, IL, USA). Measured data are
expressed as the mean ± standard deviation. Serum cf-DNA levels are
presented as a skew distribution: Median (25th percentile-75th
percentile) [M (P25-P75)], following a test
of normality. The Mann-Whitney U test was utilized for comparisons
between two groups and the Spearman's rank order correlation was
utilized to analyze double variable correlations. Receiver
operating characteristic (ROC) curves and the area under the ROC
curves (AUC) were utilized to evaluate the sensitivity and
specificity of ovarian cancer detection. P<0.05 was considered
to indicate a statistically significant difference.
Results
Serum levels of cf-DNA are
significantly increased in ovarian cancer
The serum levels of cf-DNA were 197.176
(94.757–303.367), 199.943 (78.730–396.208) and 811.354
(450.714–1307.185) µg/l in the healthy control, benign tumor and
ovarian cancer groups, respectively (Table I).
 | Table I.Comparison of the serum levels of
cf-DNA. |
Table I.
Comparison of the serum levels of
cf-DNA.
| Clinicopathological
parameter | n | cf-DNA level, µg/l [M
(P25-P75)] | P-value |
|---|
| Group |
|
|
|
|
Control | 19 | 197.176
(102.600–303.367) |
|
| Benign
tumor | 22 | 199.943
(78.730–396.208) |
|
| Ovarian
cancer | 36 |
881.181
(624.110–1409.170) |
<0.001a |
| FIGO stage |
|
|
|
| I–II | 11 | 593.401
(349.743–737.295) |
<0.001b |
| III | 16 | 1104.975
(829.756–1805.888) |
0.610c |
| IV | 9 |
954.370
(411.354–1507.910) |
|
According to the Mann-Whitney U test, the serum
levels of cf-DNA were significantly increased in the ovarian cancer
group compared with those of the healthy control and benign tumor
groups (P<0.01). There was no significant difference between the
benign tumor group and control group (P=0.917). Furthermore,
according to FIGO staging of ovarian cancer, the serum cf-DNA
levels of stage I–II ovarian cancer were significantly different
from those observed in stage III and IV (P<0.01), although there
was no significant difference between stage III and stage IV
(P=0.610).
No association is identifiable between
the serum levels of cf-DNA and clinicopathological features
No statistically significant differences were
observed between cf-DNA levels and age, differentiation, tumor
classification and complications among the ovarian cancer, healthy
control or benign tumor groups (Table
II).
 | Table II.Comparison of cf-DNA levels among
various clinicopathological parameters in patients with ovarian
cancer. |
Table II.
Comparison of cf-DNA levels among
various clinicopathological parameters in patients with ovarian
cancer.
| Clinicopathological
parameter | n | cf-DNA level, µg/l [M
(P25-P75)] | P-value |
|---|
| Age, years |
|
| 0.928 |
|
<60 | 19 | 646.757
(326.429–1034.856) |
|
| ≥60 | 17 | 953.347
(477.989–1527.393) |
|
| Degree of
differentiation |
|
| 0.785 |
| High | 10 | 800.951
(429.309–1411.844) |
|
|
Moderate | 15 | 865.411
(343.617–1243.754) |
|
| Low | 11 | 729.056
(452.638–1461.874) |
|
| Complications |
|
| 0.689 |
| Yes | 15 | 908.910
(593.401–1507.910) |
|
| No | 21 | 971.621
(642.051–1526.560) |
|
| Tumor type |
|
| 0.824 |
|
Endometrioid adenocarcinoma of
ovary | 14 | 727.152
(129.725–1216.922) |
|
| Ovarian
serous adenocarcinoma | 22 | 811.354
(467.924–1471.326) |
|
cf-DNA levels increase following
surgery and, subsequently, gradually decrease
Postoperative cf-DNA levels were significantly
increased in patients exhibiting ovarian cancer, these levels then
gradually decreased. This may potentially be due to surgical trauma
causing cells to enter the bloodstream and generate significant
quantities of cf-DNA. Following metabolism of this increased
cf-DNA, cf-DNA levels are able to decrease (Table III).
 | Table III.Alterations in cf-DNA level prior to
and following surgery. |
Table III.
Alterations in cf-DNA level prior to
and following surgery.
| Time-point | cf-DNA level, µg/l [M
(P25-P75)] | P-value |
|---|
| Pre-operation | 881.181
(624.110–1409.170) |
|
| Postoperative day
1 | 1531.718
(929.120–3271.500) | 0.001a |
| Postoperative day
3 | 1386.810
(914.410–2595.596) | 0.444b |
| Postoperative day
7 | 914.410
(93.401–1507.910) | 0.002c |
There is no correlation between the
serum levels of cf-DNA and CA125 and HE-4
Spearman correlation analysis demonstarted that the
correlation coefficient between cf-DNA and CA125 was 0.060
(P=0.729), and the correlation coefficient between cf-DNA and HE-4
was 0.043 (P=0.802). This indicated that there was no significant
difference in the level of correlation between these factors and
cf-DNA.
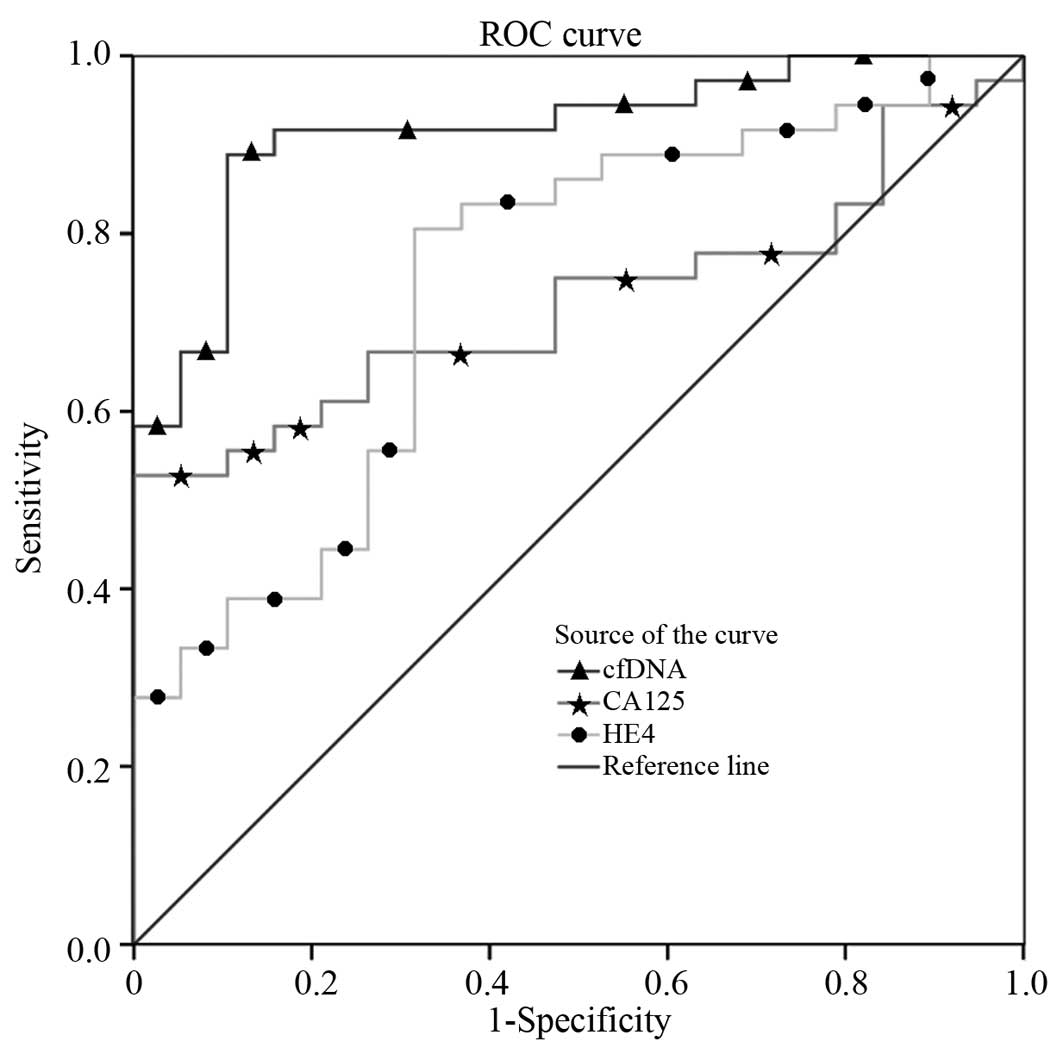
According to the ROC curve (Fig. 1), the cut-off values for cf-DNA, CA125
and HE-4 were 478.435 µg/l, 33.745 U/ml and 137.627 pmol/l,
respectively. Furthermore, the AUC, sensitivity and specificity of
cf-DNA in the ovarian cancer group were 0.917, 88.9% and 89.5%,
respectively, which were higher than those of CA125 (0.724, 75% and
52.6%) and HE-4 (0.743, 80.6% and 68.4%) (Table IV). cf-DNA levels combined with CA125
and HE-4 detection may improve the sensitivity and specificity of
ovarian cancer detection. When two or three of these biomarkers
were identified as positive during combined detection, the
sensitivity and specificity were 91.67% (33/36) and 84.21% (16/19)
respectively.
 | Table IV.Efficacy comparison between cf-DNA,
CA125 and HE-4 levels. |
Table IV.
Efficacy comparison between cf-DNA,
CA125 and HE-4 levels.
| Index | AUC | Baseline | Sensitivity, % | Specificity, % | 95% CI | P-value |
|---|
| cf-DNA | 0.917 | 478.435 | 88.9 | 89.5 | 0.842–0.991 | <0.001 |
| CA125 | 0.724 |
33.745 | 75.0 | 52.6 | 0.592–0.855 |
0.007 |
| HE-4 | 0.743 | 137.627 | 80.6 | 68.4 | 0.605–0.881 |
0.003 |
Discussion
Due to developments in tumor cellular and molecular
biology, there has been increased interest in serum cf-DNA as a
potential biomarker in oncology (10,22).
cf-DNA alteration, as a tumor indicator, has been observed in the
serum of patients with certain tumors, and cf-DNA alteration has
been associated with the expression of certain genes in tumor
tissue (23). As described in the
Introduction, existing qualitative and quantitative methods exhibit
a number of limitations (12–19). By contrast, bDNA techniques may be
regarded as an improvement compared with other monitoring methods
associated with existing tumor markers, due to their convenience
and lack of invasiveness (24). The
present study used bDNA techniques to detect the levels of cf-DNA
in patients exhibiting ovarian cancer. The associations between
cf-DNA levels and clinicopathological characteristics were
analyzed, and the value of cf-DNA in the diagnosis of ovarian
cancer was investigated.
Generally, cf-DNA is maintained at low and constant
levels in normal human serum, and is significantly increased in
patients exhibiting certain tumors. A previous study demonstrated
that serum cf-DNA levels are directly associated with the presence,
development, recovery from and recurrence of malignant tumors
(25). In the present study, healthy
volunteers, patients with benign ovarian tumors and patients
exhibiting ovarian cancer were compared, and the results indicated
that the serum cf-DNA levels were significantly increased in the
ovarian cancer group compared with those of the control group
(P<0.01). However, there was no significant difference between
those of the benign tumor and healthy control groups (P>0.05).
Furthermore, according to FIGO staging, the serum levels of cf-DNA
were significantly increased in patients with stage III and IV
ovarian cancer compared with stage I–II patients (P<0.01).
cf-DNA levels were also significantly increased following surgery
(P<0.01), and then decreased gradually; there was no significant
difference observed between pre-operative and postoperative day 7
cf-DNA levels (P>0.05). The results of the present study
suggested that the serum levels of cf-DNA increased gradually with
the development of disease, and monitoring the serum levels of
cf-DNA may aid in the analysis of patient status, prognosis and
efficacy observations. Serum CA125 and HE-4 are common markers of
ovarian cancer and possess diagnostic value; however, their
sensitivity and specificity have been observed to be lower than
that of the cf-DNA assay. According to Spearman's related analysis,
cf-DNA levels did not correlate with CA125 and HE-4 levels, which
indicated that cf-DNA was an independent prognostic biomarker due
to alternative pharmacokinetics (26).
ROC curves of cf-DNA revealed that the sensitivity
and specificity of serum cf-DNA levels were 88.9 and 89.5%,
respectively, while the baseline was 478.435 µg/l as diagnosed for
ovarian cancer, which were higher than those of CA125 and HE-4.
Furthermore, the AUC of the ROC curves of cf-DNA, CA125 and HE-4
were 0.917, 0.724 and 0.743, respectively; therefore, cf-DNA
combined with CA125 and HE-4 detection may improve sensitivity up
to 91.67%.
In conclusion, the serum levels of cf-DNA were
significantly increased in patients exhibiting ovarian cancer,
compared with those of patients with benign ovarian tumors and
healthy controls. Furthermore, cf-DNA levels were observed to
increase as ovarian cancer progressed to later disease stages.
Thus, quantitative detection of cf-DNA may possess value for the
diagnosis of ovarian cancer, and bDNA techniques demonstrated
higher sensitivity and specificity for detecting serum cf-DNA
levels compared with existing methods.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81000775/H2006), the
Social Development Foundation of Nantong City (grant no. S2010021),
the Technology Innovation and Demonstration Program of Nantong City
(grant no. S2011025) and the Medical Innovation Team and Leader
Program of Jiangsu Province (grant no. LJj201133).
References
|
1
|
Bhurgri Y, Shaheen Y, Kayani N, Nazir K,
Ahmed R, Usman A, Bashir I, Setna F, Bhurgri A, Hasan SH and Zaidi
SM: Incidence, trends and morphology of ovarian cancer in Karachi
(1995–2002). Asian Pac J Cancer Prev. 12:1567–1571. 2011.PubMed/NCBI
|
|
2
|
Tinelli A, Vergara D, Martignago R, Leo G,
Pisanò M and Malvasi A: An outlook on ovarian cancer and borderline
ovarian tumors: Focus on genomic and proteomic findings. Curr
Genomics. 10:240–249. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sharifian A, Pourhoseingholi MA,
Norouzinia M and Vahedi M: Ovarian cancer in Iranian women, a trend
analysis of mortality and incidence. Asian Pac J Cancer Prev.
15:10787–10790. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ohman AW, Hasan N and Dinulescu DM:
Advances in tumor screening, imaging, and avatar technologies for
high-grade serous ovarian cancer. Front Oncol. 4:3222014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kajiyama H, Shibata K, Mizuno M, Umezu T,
Suzuki S, Nawa A, Kawai M, Nagasaka T and Kikkawa F: Long-term
survival of young women receiving fertility-sparing surgery for
ovarian cancer in comparison with those undergoing radical surgery.
Br J Cancer. 105:1288–1294. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhen S, Bian LH, Chang LL and Gao X:
Comparison of serum human epididymis protein 4 and carbohydrate
antigen 125 as markers in ovarian cancer: A meta-analysis. Mol Clin
Oncol. 2:559–566. 2014.PubMed/NCBI
|
|
7
|
Karlan BY, Thorpe J, Watabayashi K,
Drescher CW, Palomares M, Daly MB, Paley P, Hillard P, Andersen MR,
Anderson G, et al: Use of CA125 and HE4 serum markers to predict
ovarian cancer in elevated-risk women. Cancer Epidemiol Biomarkers
Prev. 23:1383–1393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Leon SA, Shapiro B, Sklaroff DM and Yaros
MJ: Free DNA in the serum of cancer patients and the effect of
therapy. Cancer Res. 37:646–650. 1977.PubMed/NCBI
|
|
9
|
Salani R, Davidson B, Fiegl M, Marth C,
Müller-Holzner E, Gastl G, Huang HY, Hsiao JC, Lin HS, Wang TL, et
al: Measurement of cyclin E genomic copy number and strand length
in cell-free DNA distinguish malignant versus benign effusions.
Clin Cancer Res. 13:5805–5809. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fox BP and Kandpal RP: DNA-based assay for
EPHB6 expression in breast carcinoma cells as a potential
diagnostic test for detecting tumor cells in circulation. Cancer
Genomics Proteomics. 7:9–16. 2010.PubMed/NCBI
|
|
11
|
Esposito A, Bardelli A, Criscitiello C,
Colombo N, Gelao L, Fumagalli L, Minchella I, Locatelli M,
Goldhirsch A and Curigliano G: Monitoring tumor-derived cell-free
DNA in patients with solid tumors: Clinical perspectives and
research opportunities. Cancer Treat Rev. 40:648–655. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Q, Hu G, Yang Q, Dong R, Xie X, Ma
D, Shen K and Kong B: A multiplex methylation-specific PCR assay
for the detection of early-stage ovarian cancer using cell-free
serum DNA. Gynecol Oncol. 130:132–139. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Manderson EN, Presneau N, Provencher D,
Mes-Masson AM and Tonin PN: Comparative analysis of loss of
heterozygosity of specific chromosome 3, 13, 17, and X loci and
TP53 mutations in human epithelial ovarian cancer. Mol Carcinog.
34:78–90. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tian F, Yip SP, Kwong DL, Lin Z, Yang Z
and Wu VW: Promoter hypermethylation of tumor suppressor genes in
serum as potential biomarker for the diagnosis of nasopharyngeal
carcinoma. Cancer Epidemiol. 37:708–713. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carpagnano GE, Costantino E, Palladino GP,
Lacedonia D, Martinelli D, Orlando S and Foschino-Barbaro MP:
Microsatellite alterations and cell-free DNA analysis: Could they
increase the cytology sensitivity in the diagnosis of malignant
pleural effusion? Rejuvenation Res. 15:265–273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Portincasa P, Conti G and Chezzi C: Radio
immune Western blotting: A possible solution to indeterminate
conventional Western blotting results for the serodiagnosis of HIV
infections. New Microbiol. 19:85–90. 1996.PubMed/NCBI
|
|
17
|
Wong IH: Qualitative and quantitative
polymerase chain reaction-based methods for DNA methylation
analyses. Methods Mol Biol. 336:33–43. 2006.PubMed/NCBI
|
|
18
|
Maehara Y, Anai H, Kusumoto T, Sakaguchi Y
and Sugimachi K: Nick translation detection in situ of cellular DNA
strand break induced by radiation. Am J Pathol. 134:7–10.
1989.PubMed/NCBI
|
|
19
|
Kalogianni DP, Litos IK, Christopoulos TK
and Ioannou PC: Dipstick-type biosensor for visual detection of DNA
with oligonucleotide-decorated colored polystyrene microspheres as
reporters. Biosens Bioelectron. 24:1811–1815. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cao Y, Ho DD, Todd J, Kokka R, Urdea M,
Lifson JD, Piatak M Jr, Chen S, Hahn BH, Saag MS, et al: Clinical
evaluation of branched DNA signal amplification for quantifying HIV
type 1 in human plasma. AIDS Res Hum Retroviruses. 11:353–361.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liutkauskiene S, Janciauskiene R,
Jureniene K, Grizas S, Malonyte R and Juozaityte E: Retrospective
analysis of the impact of platinum dose reduction and chemotherapy
delays on the outcomes of stage III ovarian cancer patients. BMC
Cancer. 15:1052015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kohler C, Radpour R, Barekati Z,
Asadollahi R, Bitzer J, Wight E, Bürki N, Diesch C, Holzgreve W and
Zhong XY: Levels of plasma circulating cell free nuclear and
mitochondrial DNA as potential biomarkers for breast tumors. Mol
Cancer. 8:1052009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zane M, Agostini M, Enzo MV, Casal Ide E,
Del Bianco P, Torresan F, Merante Boschin I, Pennelli G, Saccani A,
Rubello D, et al: Circulating cell-free DNA, SLC5A8 and SLC26A4
hypermethylation, BRAF (V600E): A non-invasive tool panel for early
detection of thyroid cancer. Biomed Pharmacother. 67:723–730. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rass U: Resolving branched DNA
intermediates with structure-specific nucleases during replication
in eukaryotes. Chromosoma. 122:499–515. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hao TB, Shi W, Shen XJ, Qi J, Wu XH, Wu Y,
Tang YY and Ju SQ: Circulating cell-free DNA in serum as a
biomarker for diagnosis and prognostic prediction of colorectal
cancer. Br J Cancer. 111:1482–1489. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Beiter T, Fragasso A, Hudemann J, Niess AM
and Simon P: Short-term treadmill running as a model for studying
cell-free DNA kinetics in vivo. Clin Chem. 57:633–666. 2011.
View Article : Google Scholar : PubMed/NCBI
|