Introduction
Primary liver cancer is the fifth most common cause
of cancer-associated mortality worldwide (1), and up to 90% of primary liver cancers
are hepatocellular carcinomas (HCC) (2). Risk factors for hepatocarcinogenesis
include chronic hepatitis B or C infection, alcohol consumption and
aflatoxin B1 contamination (2). HCC
development, similarly to that of other types of cancer, is a
multistep process, involving numerous genetic and epigenetic
alterations (3). It has been reported
that promoter methylation of certain tumor suppressor genes occurs
frequently during HCC development and progression (4,5).
The GATA gene family comprises a group of zinc
finger transcription factors that are able to bind to the GATA
motif (WGATAR), which is present in the promoters of certain genes
(6). GATA1, GATA2 and GATA3 are
functionally involved in cellular lineage determination (7), while GATA4, GATA5 and GATA6 regulate the
development of endoderm-derived organs, including the heart and gut
(8). During development, GATA4 and
GATA5 induce differentiation of embryonic stem cells into specific,
mature gut cells (9). In adults,
GATA4 and GATA5 regulate epithelial cell differentiation (10,11).
However, altered expression of GATA4 and GATA5 proteins is
associated with tumorigenesis in certain organs and tissues,
including gastric and colon cancer; thus, GATA4 and GATA5 may act
as putative tumor suppressor genes (11,12). A
previous study revealed that GATA4 and GATA5 expression was lost in
ovarian and gastric cancer, and demonstrated that chromosomal
regions of GATA4 (8p23.1-p22) and GATA5 (20q13.2-q13.3) loci were
frequently deleted in various types of human cancer (13). Additional studies identified that loss
of GATA4 and GATA5 expression during human neoplastic progression,
including that of pancreatic, non-small cell lung, esophageal and
renal cancer, was due to promoter methylation (14–16). Loss
of GATA4 and GATA5 expression may also alter the typical expression
patterns of numerous downstream gene networks with antineoplastic
properties (11). For example, GATA
transcription factors are able to integrate Wnt signaling during
heart muscle formation (17,18) and alteration of the Wnt/β-catenin
pathway has a critical role in the development of various types of
human cancer, including HCC (19).
Thus, in the present study, the expression levels
and promoter hypermethylation of GATA4 and GATA5 were evaluated in
human HCC tissue specimens and cell lines. The effects of GATA4 and
GATA5 expression restoration in HCC cells in vitro were
subsequently characterized.
Materials and methods
Cell lines and culture
Human liver cancer cell lines (HepG2, SMZ7721,
HBxF344, PLC/PRF/5, SK-Hepl, 97H and 7402) and an
SV-40-immortalized Lo-2 liver cell line were obtained from the Cell
Bank of the Chinese Academy of Sciences (Shanghai, China). Cells
were maintained in RPMI-1640 medium (Grand Island Biological
Company, Grand Island, NY, USA) or Dulbecco's modified Eagle medium
(DMEM; Invitrogen Life Technologies, Carlsbad, CA, USA),
supplemented with 10% fetal bovine serum (FBS; Grand Island
Biological Company) in a humidified incubator, with 5%
CO2 and 95% air, at 37°C. Cells were passaged with
trypsin (Sigma-Aldrich, St. Louis, MO, USA) into T-75 flasks
(Sarstedt, Nümbrecht, Germany) using a 1:3 split when 80%
confluence was achieved (~1×106 cells/flask).
For treatment with 5-aza-2′-deoxycytidine (5-AZA;
Sigma-Aldrich), cells were separated and cultured at a low density
(30% confluence) overnight and subsequently treated with 5-AZA (1
µmol/l) for 96 h. Growth medium was replaced every 24 h, and at the
conclusion of treatment, DNA and RNA was isolated as described
below.
HCC tissue specimens
A total of 38 surgically resected and pathologically
confirmed HCC specimens (8 females and 30 males; aged 30–79 years)
were obtained from the Chinese People's Liberation Army General
Hospital (Beijing, China) between January 2009 and June 2011 and
stored in liquid nitrogen prior to use. The present study was
approved by the Institutional Review Board of the Chinese People's
Liberation Army General Hospital. Written informed consent was
obtained from each patient or their guardian. Tumor and paired
adjacent tissue samples (≥1 cm away from tumor lesion) were
collected. Tumor staging was determined according to the American
Joint Committee on Cancer Cancer Staging Manual, 2010 (7th Edition)
(20). Normal hepatic tissue was
obtained from the edge of resected hemangiomas of the liver.
DNA extraction
HCC cell lines were cultured and treated with 5-AZA
as described and subsequently digested with 0.5% trypsin-EDTA
(Sangon Biotech Co., Ltd). Following digestion, DNA was extracted
using the proteinase-K method. Genomic DNA from normal and HCC
liver tissues was also extracted using the proteinase-K method
(21). DNA was dissolved in Tris-EDTA
buffer and stored at −20°C until use.
RNA isolation and semi-quantitative reverse
transcription-polymerase chain reaction (RT-PCR). Total cellular
RNA was isolated from the cells using TRIzol reagent (Invitrogen
Life Technologies) according to the manufacturer's instructions.
RNA quality and quantity was assessed using agarose gel
electrophoresis (1%) and spectrophotometric analysis using a
260–280 ratio. RNA was stored at −80°C until use. First strand
complementary DNA (cDNA) was synthesized with oligo-(dT) primers
using a PrimeScript™ 1st Strand cDNA Synthesis kit obtained from
Invitrogen Life Technologies. RNA (2 µg) was subjected to first
strand cDNA synthesis, and subsequently 1 µl cDNA from the RT
reaction was subjected to PCR amplification in a total reaction
volume of 25 µl. PCR amplification was performed using primer sets
derived from the published GATA4 and GATA5 gene sequences (22). PCR conditions were set to 94°C for 1
min, 64°C for 1 min and 72°C for 90 sec for 3 cycles and 94°C for 1
min, 61°C for 1 min and 72°C for 90 sec for additional 3 cycles,
and 94°C for 1 min, 58°C for 1 min, and 72°C for 90 sec for final 3
cycles and then 94°C for 1 min, 55°C for 1 min, and 72°C for 90 sec
for 23 cycles and final 72°C extension for 7 min. GATA4 primers
used were as follows: Sense, 5′-CTGGCCTGTCATCTCACTACG-3′ and
antisense, 5′-GGTCCGTGCAGGAATTTGAGG-3′. GATA5 primers used were as
follows: Sense, 5′-TCGCCAGCACTGACAGCTCAG-3′ and antisense,
5′-TGGTCTGTTCCAGGCTGTTCC-3′. A total of 32 PCR cycles (based on
preliminary investigation) were performed for semi-quantitative
measurement of GATA4 and GATA5 gene expression. GAPDH was amplified
for 25 cycles as an internal control, to ensure equal loading, as
well as cDNA quality and quantity. GAPDH primers were as follows:
Sense, 5′-GACCACAGTCCATGCCATCAC-3′ and antisense,
5′-GTCCACCACCCTGTTGCTGTA-3′. PCR products were electrophoresed in
1.5% agarose gels containing ethidium bromide and were evaluated
using UV light.
Methylation-specific PCR (MSP)
Genomic DNA from HCC tissues and cell lines was
modified using bisulfite modification as described previously
(23). MSP primers were designed
according to the genomic sequences surrounding the putative
transcription start sites of the GATA4 and GATA5 genes. Primer
sequences were synthesized by Sangon Biotech Co.,Ltd. (Shanghai,
China) to allow MSP to detect bisulfite-induced changes affecting
unmethylated and methylated alleles. MSP primers were synthesised
by Shanghai Sangon Biotech Co., Ltd; MSP primers for GATA4 were as
follows: Methylated sense, 5′-GTATAGTTTCGTAGTTTGCGTTTA GC-3′ and
antisense, 5′-AACTCGCGACTCGAATCCCCG-3′; unmethylated sense,
5′-TTTGTATAGTTTTGTAGTTTGTGTTTAGT-3′ and antisense,
5′-CCCAACTCACAACTCAAATCCCCA-3′. MSP primers for GATA5 were as
follows: Methylated sense, 5′-AGTTCGTTTTTAGGTTAGTTTTCGGC-3′ and
antisense, 5′-CCAATACAACTAAACGAACGAACCG-3′; unmethylated sense,
5′-TGGAGTTTGTTTTTAGGTTAGTTTTTGGT-3′ and antisense,
5′-CAAACCAATACAACTAAACAAACAAACCA-3′. Each MSP reaction incorporated
~100 ng bisulfite-treated DNA, 25 pM each primer, 100 pM
deoxynucleotides, 2.5 µl 10X PCR buffer and 1 unit JumpStart Red
Taq Polymerase (Sigma-Aldrich) in a final reaction volume of
25 µl. PCR cycling conditions were as follows: 95°C for 5 min,
followed by 35 cycles at 95°C for 30 sec, 60°C for 30 sec and 72°C
for 30 sec. MSP products were analyzed using 2% agarose gel
electrophoresis. Subsequently, MSP products were subcloned into a
pGEM®-T vector (Promega Corp., Madison, WI, USA), and transformed
into Escherichia coli DH5α cells (Invitrogen Life Technologies).
Candidate plasmid clones were sequenced by Sangon Biotech Co.,
Ltd.
Expression vector and gene
transfection
Full-length GATA4 and GATA5 cDNA were purchased from
Orignene Technologies, Inc. (Rockville, MD, USA) and subcloned into
pCMV6-XL4 (OriGene Technologies, Inc.) in Escherichia coli DH5α
cells (Invitrogen Life Technologies). Candidate plasmid clones were
sequenced by Sangon Biotech Co., Ltd. Following amplification and
sequence confirmation, these vectors were named pCMV-GATA4 and
pCMV-GATA5. For gene transfection, HepG2 cells were seeded in DMEM
containing 10% FBS overnight. On the following day, cells were
transfected with pCMV-GATA4 or pCMV-GATA5 using Lipofectamine® 2000
(Invitrogen Life Technologies) according to the manufacturer's
instructions. Twenty-four hours later, the cells were separated
into fresh dishes, at a 1:3 ratio, and treated with 400 µg/ml
gentamycin sulfate (G418; Invitrogen Life Technologies). Cell
culture media was replaced every three days. Following three weeks
of incubation, the cells formed stable gene-transfected clones and
were used for the following experiments.
Protein extraction and immunoblot
assay
Total protein was prepared from the vector control
(pCMV-con vector only) and pCMV-GATA4 (pc-GATA4) or pCMV-GATA5
(pc-GATA5)-transfected HepG2 cells, with ice-cold lysis buffer
(Beyotime Institute of Biotechnology, Haimen, China), according to
the manufacturer's instructions. Following centrifugation at 12,000
× g for 15 min at 4°C, supernatants were collected and protein
concentration was assessed using a bicinchoninic acid protein assay
kit (Beyotime Institute of Biotechnology). Equal quantities of
protein were separated using 10% SDS-PAGE and transferred onto
polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA,
USA). Western blot analysis was performed using mouse monoclonal
IgG2a anti-GATA4 (sc-25310, 1:400, Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) and rabbit monoclonal anti-GATA5 (G8669, 1:600,
Sigma-Aldrich), while anti-β-actin antibody served as the loading
control (Wuhan Boster Biological Technology, Ltd., Wuhan, China):
the membranes were incubated overnight at 4°C. Then the membranes
were incubated with a horseradish peroxidase conjugated-goat
anti-rabbit or mouse IgG (Shanghai Westang Bio Tech Co., Ltd,
China). Chemiluminescence substrate solution (Pierce Biotechnology,
Inc., Rockford, IL, USA) was applied to the membranes and X-ray
film (Eastman Kodak Co., Rochester, NY, USA) was used to visualize
protein bands.
Cell viability MTS assay
Cells transfected with a vector control, and
pc-GATA4 or pc-GATA5, were seeded at a density of 500 cells/well in
96-well plates, and grown for 6 days prior to commencement of the
cell proliferation assay. Cell proliferation was determined using
cell counting kit-8 solution (Beyotime Institute of Biotechnology),
according to the manufacturer's instructions. The optical density
was measured at 492 nm using an Multiskan TM FC ELISA plate reader
(ThermoFisher Scientific, Inc., Waltham, MA, USA). Experiments were
performed in triplicate and repeated three times.
Flow cytometric apoptosis assay
HCC cells were transfected with a vector-control and
pc-GATA4 or pc-GATA5 for 72 h; subsequently, attached and floating
cells were harvested and fixed using 70% ethanol for at least 48 h.
Cells were resuspended in 50 µg/ml propidium iodide
(Sigma-Aldrich)and 100 µg/ml RNase (Takara Biotechnology Inc.) for
30 min, and analyzed using flow cytometry (FACSCalibur™; BD
Biosciences, Franklin Lakes, NJ, USA).
Colony formation assay
HepG2 cells were plated in 6-well culture plates,
grown for 24 h, and subsequently transfected with a vector control
and pc-GATA4 or pc-GATA5 using FuGENE® 6 transfection reagent
(Roche Diagnostics, Indianapolis, IN, USA) according to the
manufacturer's instructions. Following 24 h of incubation,
transfected cells were diluted and reseeded into fresh dishes at
500 cells/well in 6-well culture plates, and were subsequently
grown and selected in 400 mg/ml G418. Following a further 14 days
of incubation, cells were fixed using 75% ethanol for 30 min and
stained with 0.2% crystal violet (Beijing Zhongshan Biotechnology
Co., Ltd, Beijing, China) for visualization and counting. The
experiment was performed in triplicate and repeated once.
Luciferase reporter assay
HepG2 cells were plated at a density of
5×104 cells/well in 24-well culture plates and grown for
24 h. Subsequently, the TOP/FOP flash system reporter construct
(Promega Corporation, Madison, WI, USA) pGL3-OT (0.4 µg) and
pc-GATA4 or pc-GATA5 (0.4 µg) were transfected into the HepG2 cells
using FuGENE® 6 transfection reagent according to the
manufacturer's instructions. The pGL3-OT plasmid is a
TCF/LEF-responsive reporter that contains three consensus TCF
binding sites fused to the firefly luciferase gene. The pRL-TK
vector served as the system internal control. Following 48 h of
incubation, relative luciferase activity was measured using a
GloMax® luminometer (Promega Corporation), and normalized for
background Renilla luciferase activity using the Dual
Luciferase Reporter Assay system (Promega Corporation), according
to the manufacturer's instructions. For each experiment, the
luciferase assay was performed three times.
Statistical analysis
Data are expressed as the mean ± standard error
where appropriate. All statistical analyses were performed using
SPSS 13.0 for Windows (SPSS, Inc., Chicago, IL, USA). P-values for
dichotomous variables were two-tailed and based on the Pearson
χ2 test or the Pearson χ2 test with
continuity correction. Continuous variables were analyzed using the
Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
GATA4 and GATA5 expression is lost due
to gene promoter methylation in certain HCC cell lines
The HepG2, SMZ7721 and 7402 cell lines expressed
undetectable levels of GATA4 mRNA, while PLC/PRF/5 cells expressed
reduced levels of GATA4 mRNA. By contrast, 97H and Lo-2 cell lines
expressed increased levels of GATA4 mRNA. Similarly, GATA5 mRNA was
not detectable in PLC/PRF/5, HBfX344, Sk-hepl and SMZ7721 cell
lines, whereas the HepG2, 97H, 7402 and Lo-2 cell lines expressed
high levels of GATA5 mRNA (Fig. 1A).
In order to investigate the underlying mechanisms responsible for
the reduced expression of GATA4 and GATA5 in the aforementioned HCC
cell lines, these cells were treated with a DNA-demethylating agent
(5-AZA) and the expression of GATA4 and GATA5 was subsequently
assessed. The results of the present study revealed that GATA4
expression was induced in HepG2 and 7402 cells, and GATA5
expression was significantly enhanced in SMZ7721 and PLC/PRF/5
cells following treatment with 5-AZA (Fig. 1A; P<0.05).
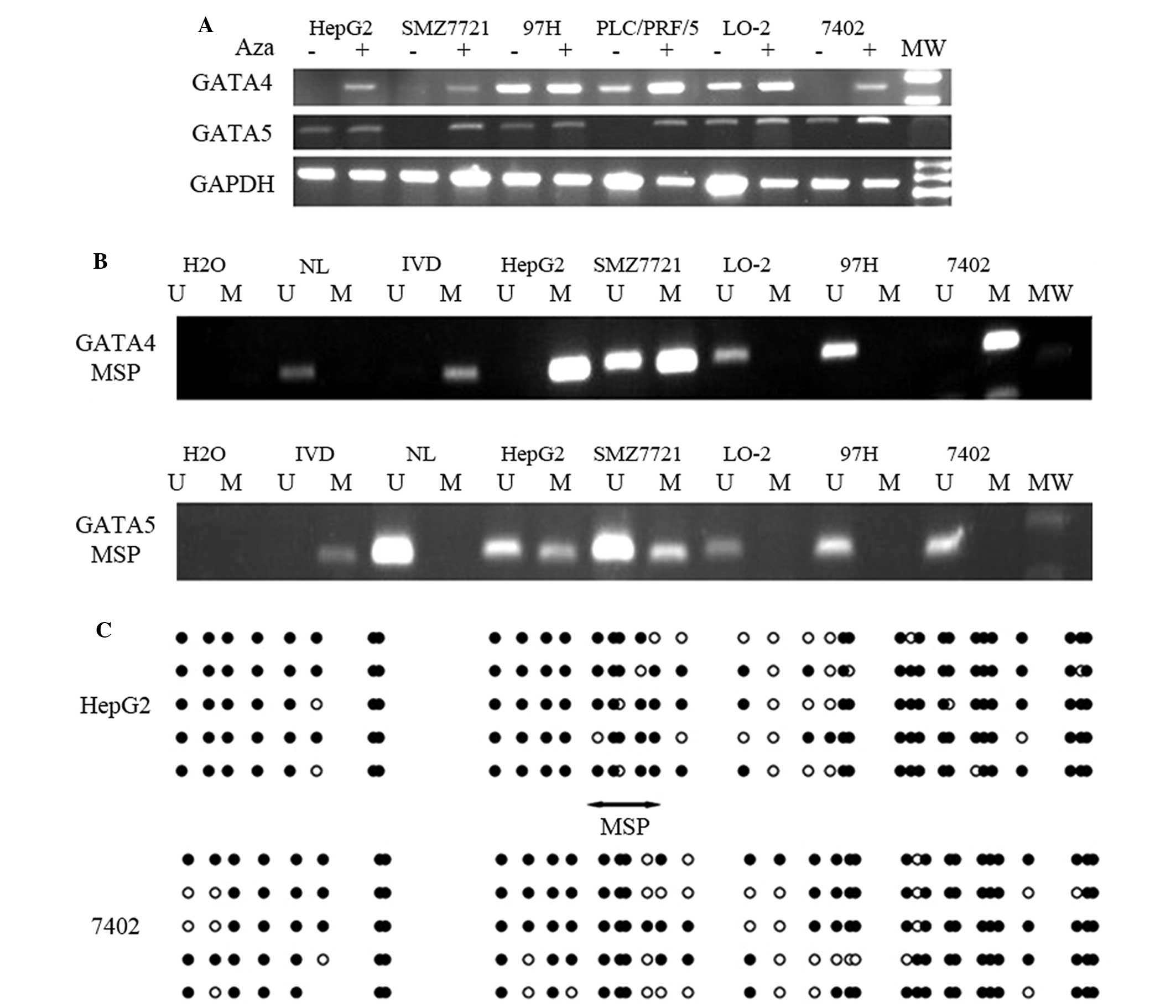 | Figure 1.GATA4 and GATA5 expression is
downregulated in HCC cell lines due to DNA methylation. (A) HCC
cell lines and an immortal liver cell line (Lo-2) were cultured and
treated with or without 10 µmol/l 5-aza-2′-deoxycytidine for up to
96 h. Reverse transcription-PCR analysis of GATA4 and GATA5 mRNA
expression was performed. GAPDH mRNA served as an internal control.
(B) MSP analysis of GATA4 and GATA5 promoter methylation in HCC
cell lines. In vitro methylated DNA and DNA from normal
human peripheral lymphocytes were used as methylated and
unmethylated controls, respectively. (C) Sequence analysis of the
GATA4 gene promoters following bisulfite modification in HCC cells.
Black circles indicate methylated cytosine and white circles
indicate unmethylated cytosine in the dinucleotide CpG. The region
of the CpG island studied by MSP (doubleheaded arrow) spanned 142
bp. Bisulfite sequencing focused on a 385 bp (+47 to +432 bp) CpG
island downstream of the GATA4 transcription start site. H2O, water
control; U, unmethylated alleles; M, methylated alleles; HCC,
hepatocellular carcinoma; IVD, in vitro methylated DNA; NL,
normal human peripheral lymphocytes; PCR, polymerase chain
reaction; MW, molecular weight; MSP, methylation-specific
polymerase chain reaction. |
Furthermore, MSP analysis was performed to determine
gene promoter methylation. The GATA4 promoter was entirely
methylated in 7402 cells, as evidenced by undetectable GATA4 mRNA
expression levels. Partial GATA4 promoter methylation was observed
in HepG2 and SMZ7721 cells; HepG2 and SMZ7721 cells possessed
methylated and unmethylated alleles (Fig.
1B). GATA4 expression was inversely associated with the levels
of gene promoter methylation in the remaining HCC cell lines
(Fig. 1A and B; Table I). Similarly, the GATA5 promoter was
entirely methylated in PLC/PRF/5 cells, which exhibited
undetectable levels of GATA5 mRNA. The GATA5 promoter was partially
methylated in HepG2 and SMZ7721 cells, which possessed methylated
and unmethylated alleles (Fig. 1B;
Table I). In the present study, MSP
products were cloned and sequenced and it was revealed that GATA4
promoter methylation in HepG2 and 7402 cell lines was present in
the CpG islands of the gene promoter region (Fig. 1C).
 | Table I.Expression and gene promoter region
methylation of GATA4 and GATA5 in HCC cell lines. |
Table I.
Expression and gene promoter region
methylation of GATA4 and GATA5 in HCC cell lines.
| A, GATA4 |
|---|
|
|---|
| HCC cell line | RT-PCR | +5-AZA | MSP |
|---|
| HepG2 | − | − + | M |
| SMZ7721 | − | − + | U/M |
| HBfX344 | + | + | U |
| PRC/PLC/5 | + | + | U |
| 97H | + | + | U |
| 7402 | − | + | M |
| Sk-hepl | + | + | U |
| Lo-2 | + | + | U |
|
| B, GATA5 |
|
| HCC cell line | RT-PCR | +5-AZA | MSP |
|
| HepG2 | − + | − + | U/M |
| SMZ7721 | − | − + | U/M |
| HBfX344 | − | − | U |
| PRC/PLC/5 | − | + | M |
| 97H | + | + | U |
| 7402 | + | + | U |
| Sk-hepl | − | + | M |
| Lo-2 | + | + | U |
Methylation of the GATA5 promoter is
associated with increased age of patients with HCC
To confirm that GATA4 and GATA5 expression was lost
due to gene promoter methylation in HCC cells ex vivo, HCC tissue
samples were collected and GATA4 and GATA5 promoter methylation was
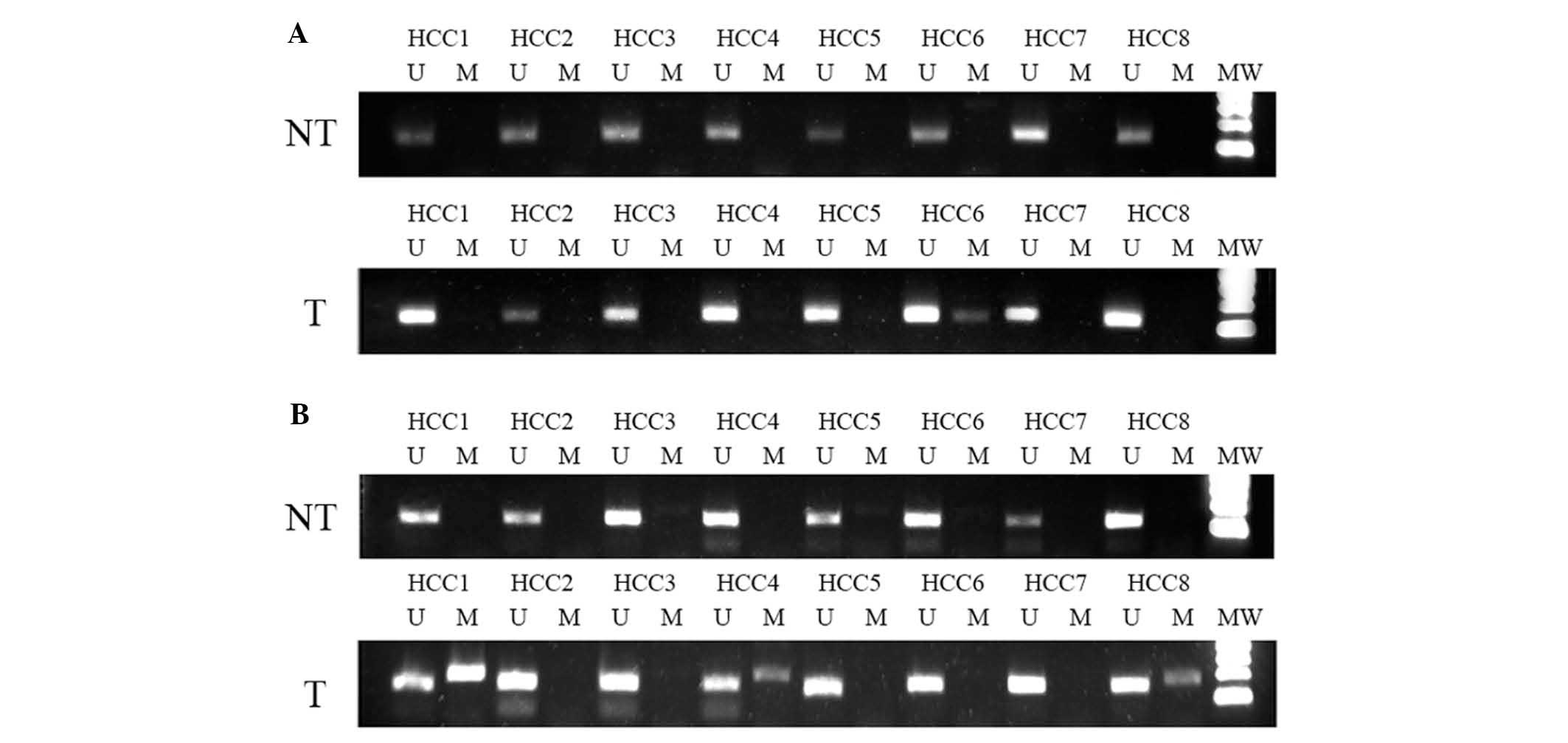
analyzed. Specifically, MSP was performed on 38 samples of human
HCC and surrounding non-tumor tissue obtained from patients with
HCC. MSP GATA4 experiments were only successful for 28 HCC and 26
non-tumor tissue specimens, whereas MSP experiments for GATA5 were
succesful for all 38 HCC and surrounding non-tumor tissue samples;
the results revealed GATA4 and GATA5 promoter methylation in 2/28
(7.14%) and 13/38 (34.20%) HCC tissues and 3/26 (11.54%) and 0/38
(0%) surrounding non-tumor tissues, respectively (Fig. 2A and B). For GATA4 methylation, there
was no statistically significant difference observed between HCC
tissues and the surrounding tissue (P=0.578). For GATA5,
methylation was only observed in tumor tissue.
Methylation of the GATA5 promoter was subsequently
observed to be correlated with certain clinicopathological
features. Notably, methylation of the GATA5 promoter in HCC tissues
was associated with increased age, but there were no associations
with other clinicopathological features, including gender, tumor
size, tumor stage, α-fetoprotein, cirrhosis, hepatitis B surface
antigen or tumor differentiation (Table
II).
 | Table II.Association of GATA5 promoter
methylation with clinicopathological factors in patients with
hepatocellular carcinoma. |
Table II.
Association of GATA5 promoter
methylation with clinicopathological factors in patients with
hepatocellular carcinoma.
| Clinicopathological
factor | Methylated, n
(n=13) | Unmethylated, n
(n=25) | P-value |
|---|
| Age, years |
|
| 0.01 |
|
<50 | 1 | 12 |
|
|
≥50 | 12 | 12 |
|
| Gender |
|
| 0.28 |
|
Male | 9 | 21 |
|
|
Female | 4 | 4 |
|
| α-fetoprotein,
µg/l |
|
| 0.13 |
|
<20 | 8 | 9 |
|
|
≥20 | 5 | 16 |
|
| Liver
cirrhosis |
|
| 0.57 |
|
Absent | 4 | 10 |
|
|
Present | 9 | 15 |
|
| Hepatitis B surface
antigen |
|
| 0.37 |
|
Negative | 3 | 3 |
|
|
Positive | 10 | 22 |
|
| Tumor size, cm |
|
| 0.29 |
| ≤5 | 10 | 15 |
|
|
>5 | 3 | 10 |
|
| Tumor
differentiation |
|
| 0.13 |
|
Well | 0 | 4 |
|
|
Moderate | 8 | 17 |
|
|
Poor | 5 | 4 |
|
| UICC
TNMa stage |
|
| 0.20 |
|
I–II | 8 | 10 |
|
|
III | 5 | 15 |
|
GATA4 and GATA5 expression reduces
cell viability, inhibits colony formation and induces apoptosis of
HCC cells
Since GATA4 and GATA5 expression was reduced in HCC
tissues and cell lines, the present study hypothesized that
restoration of GATA4 and GATA5 expression may suppress HCC.
Therefore expression vectors were constructed for GATA4 and GATA5,
which were then transfected into HepG2 cells. The results of the
present study revealed that overexpression of GATA4 or GATA5 in
HepG2 cells (Fig. 3A) significantly
reduced cell viability compared with those transfected with the
control vector, as indicated by MTS assay (Fig. 3B; 30.14 and 39.31% inhibition
following 6 days of incubation, respectively; P<0.05). In
addition, flow cytometric analysis indicated that the cell cycle
was arrested. In particular, the fraction of HepG2 cells in the
G2-M phase following GATA4 and GATA5 transfection was 8.83 and
13.31%, respectively, compared with 15.14% in pc-con-transfected
HepG2 cells (n=3; P<0.05; Fig.
3C). Furthermore, restoration of GATA4 and GATA5 gene
expression inhibited the colony formation ability of HepG2 cells
(Fig. 3D; P<0.05) compared with
that of control cells. Restoration of GATA4 and GATA5 gene
expression induced HepG2 cells to undergo apoptosis (20.46 and
24.79%, respectively, vs. 11.28%; P<0.05; Fig. 3E).
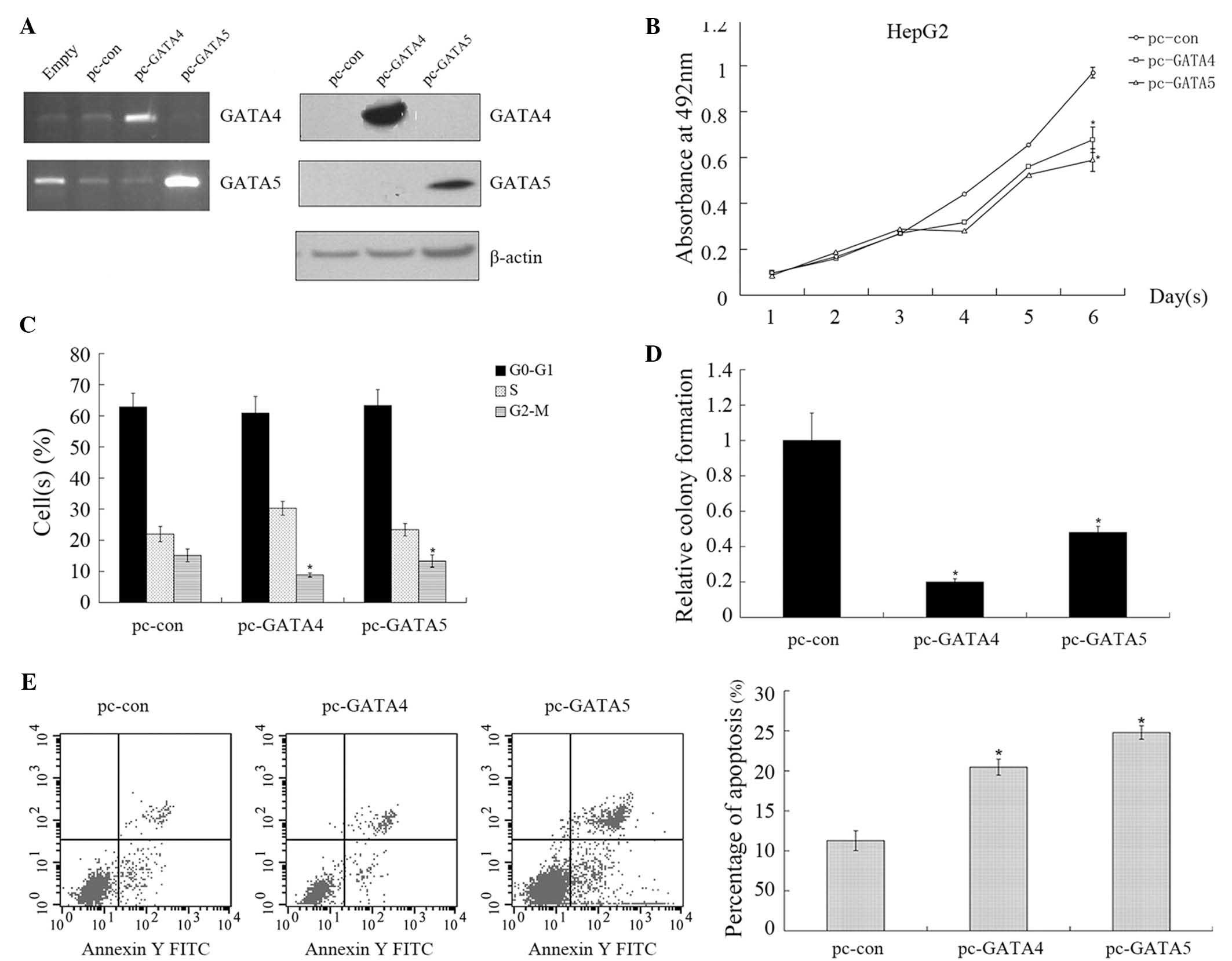 | Figure 3.GATA4 and GATA5 expression reduces
cell viability, inhibits colony formation and induces apoptosis in
hepatocellular carcinoma cells. (A) HepG2 cells were transfected
with control, pcMV-GATA4 or pcMV-GATA5, respectively for 72 h.
Total cellular RNA or protein was extracted for reverse
transcription-polymerase chain reaction and western blot analyses,
respectively. (B) HepG2 cells were transfected with control,
pcMV-GATA4 or pcMV-GATA5 for 72 h and subjected to an MTS assay.
(C) HepG2 cells were transfected with control, pcMV-GATA4 or
pcMV-GATA5 for 72 h and subjected to propidium iodide staining and
flow cytometry. (D) HepG2 cells were transfected with control,
pcMV-GATA4 or pcMV-GATA5 for 72 h, treated with gentamycin sulfate
for 14 d, and subsequently subjected to a colony formation assay.
Quantification of colonies is expressed as the mean ± standard
error, relative to the control transfectants (pc-con) of three
independent experiments (*P<0.05). (E) HepG2 cells were
transfected with control, pcMV-GATA4 or pcMV-GATA5 for 72 h and
subjected to apoptosis detection and flow cytometry. The results
indicated that GATA4 and GATA5 inhibited cell proliferation, and
induced cell cycle arrest and apoptosis, *P<0.05. pc, pCMV
vector; con, control; FITC, fluorescein isothiocyanate. |
GATA4 and GATA5 inhibit Wnt/β-catenin
activity in HCC cells
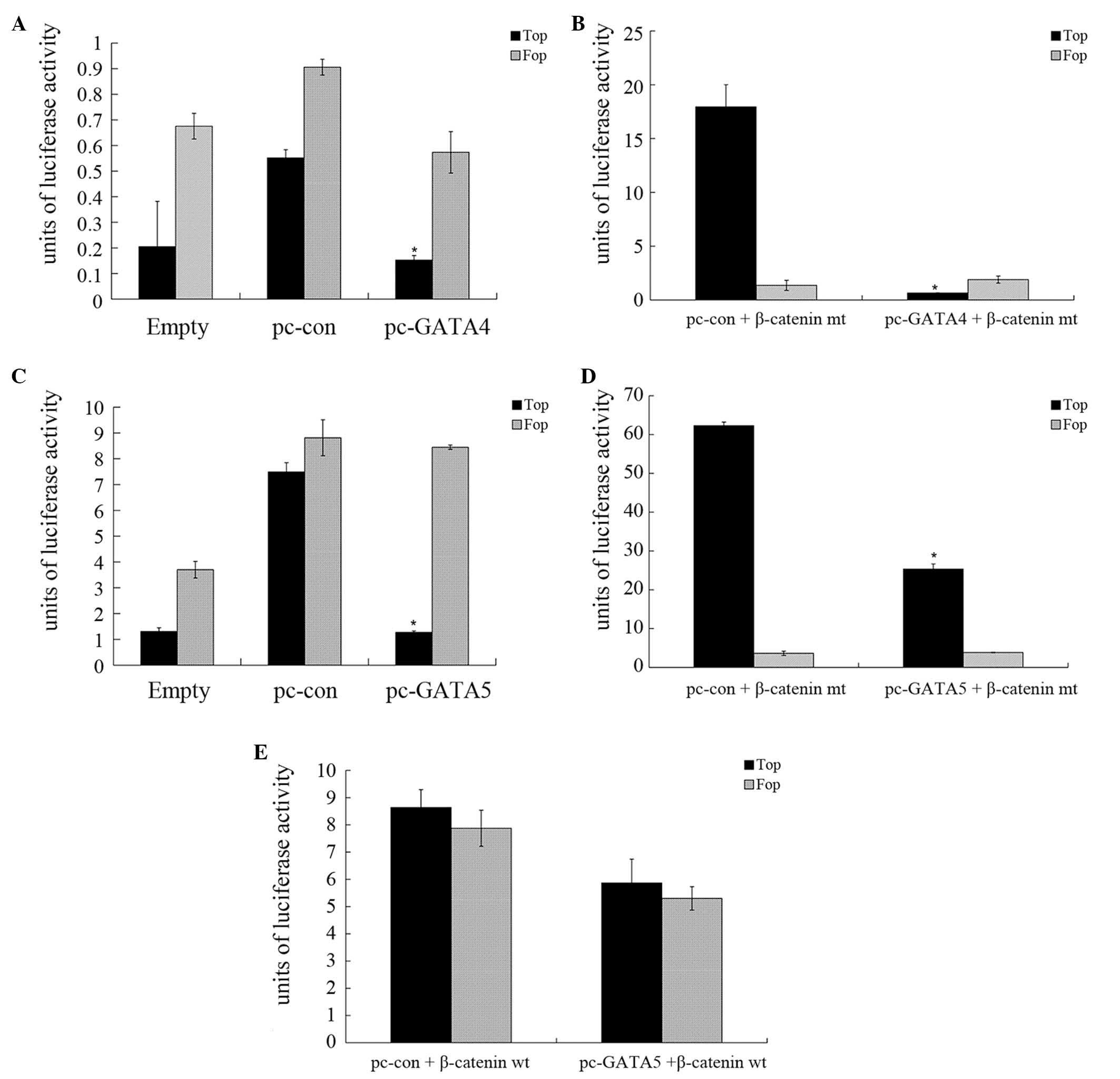
The present study also investigated whether GATA4 or
GATA5 affected Wnt/β-catenin signaling in HepG2 cells, by
performing a luciferase reporter assay. The results revealed that
the luciferase activity of TCF in pCMV-GATA4-transfected HepG2
cells was reduced compared with that of the control HepG2 cells
(Fig. 4A; P<0.05). The luciferase
activity of TCF in pCMV-GATA4-transfected HepG2 cells following
co-transfection with a mutated β-catenin plasmid was significantly
reduced compared with those transfected with the control vector
(Fig. 4B; P<0.05).
GATA5-transfection luciferase reporter assays demonstrated similar
results (Fig. 4C and D). However, the
luciferase activity in cells co-transfected with pCMV-GATA5 and
wild-type β-catenin exhibited no significant changes compared with
that of the control cells (Fig. 4E),
indicating that GATA4 and GATA5 inhibited the activity of the
Wnt/β-catenin pathway in HCC.
Discussion
In the current study, the expression of GATA4 and
GATA5 and the methylation status of their gene promoters in HCC
cells were initially determined. It was demonstrated that the
expression of GATA4 and GATA5 mRNA was lost, and the GATA4 and
GATA5 promoter regions were hypermethylated in certain HCC cell
lines. In addition, ex vivo data revealed that the GATA5
promoter was frequently methylated in HCC tissues. Restoration of
GATA4 and GATA5 expression in HCC cells significantly reduced tumor
cell proliferation and colony formation, as well as inducing
apoptosis. In conclusion, the results of the present study suggest
that GATA4 and GATA5 may have a role in the suppression of HCC
development.
A previous study revealed that loss of GATA4 and
GATA5 expression was due to gene promoter methylation in gastric
and ovarian cancer, which was also confirmed in lung cancer,
esophageal cancer and pancreatic cancer (14–16). In
the current study, it was demonstrated that the methylation of
GATA4 and GATA5 promoters were in accordance with these previous
studies. To the best of our knowledge, the present study is the
first investigation into GATA4 and GATA5 methylation in HCC. GATA4
was revealed to be significantly downregulated in HCC cell lines,
and its promoter was hypermethylated or partially methylated in 3/7
HCC cell lines. In addition, GATA5 was significantly downregulated
in HCC cell lines and its promoter was hypermethylated or partially
methylated in 4/7 HCC cell lines and 13/38 (34.2%) cases of primary
liver tumors. The results of the present study implied that
aberrant methylation of the GATA4 and GATA5 promoter regions
results in a loss of GATA4 and GATA5 expression in HCC cell lines.
However, HCC cell lines that have methylated and unmethylated
alleles express weak and strong levels of GATA4 and GATA5,
indicating the heterogeneous population present in the cell
culture. The present study also demonstrated that GATA5 promoter
methylation was more frequent in older patients, indicating that
GATA5 promoter methylation may be an independent risk factor for
older HCC patients.
GATA4 and 5, as transcription factors, are involved
in normal cell growth and also in adrenal gland, lung, ovary and
colorectal cancer development (12).
Functionally, they activate the promoter of tumor suppressor genes
and serve as tumor suppressors, the lost expression of which
accelerates the development of colon cancer. For example, colon
cells with GATA4 and 5 expression had reduced colony formation,
proliferation, migration, invasion and anchor growth (12). In a previous study, GATA4 and GATA5
expression was lost in HepG2 cells and GATA4 and GATA5 transfection
experiments inhibited tumor cell growth and colony formation, but
induces tumor cell apoptosis. These data indicate that GATA4 and
GATA5 were able to inhibit liver cancer cell proliferation. Under
the same conditions, GATA5 inhibits growth and induce apoptosis to
a greater extent compared with GATA4. The present study also
revealed that GATA4 and GATA5 may be involved in the regulation of
the Wnt signaling pathway. Whether the expression of significant
molecules in the Wnt signaling pathway, such as β-catenin, are
regulated by GATA4 and GATA5 will require further investigation in
future studies.
The demethylation agent 5-AZA is an effective
anticancer therapy (24,25). In the current study, 5-AZA treatment
rescued the expression of GATA5 mRNA by reversing GATA5
hypermethylation in HCC cell lines. This finding indicated that
epigenetic therapy may be useful in at least a subset of HCC
patients. However, further studies are required to verify this
possibility.
In conclusion, the current study demonstrated that
loss of GATA4 and GATA5 expression occurred primarily due to
promoter methylation in HCC tissues and cells. Restoration of GATA4
and GATA5 expression inhibited tumor cell viability and colony
formation, as well as inducing apoptosis. Future studies may be
required to investigate the underlying mechanisms of GATA4 and
GATA5 antitumor activities in HCC.
Acknowledgements
The present study was supported in part by grants
from the China National Science Foundation (grant no. 81201957) and
from the National ST Major Project for Infectious Diseases of China
(grant no. 2012ZX10002-017).
References
|
1
|
Parkin DM: Global cancer statistics in the
year 2000. Lancet Oncol. 2:533–543. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thorgeirsson SS and Grisham JW: Molecular
pathogenesis of human hepatocellular carcinoma. Nat Genet.
31:339–346. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Um TH, Kim H, Oh BK, Kim MS, Kim KS, Jung
G and Park YN: Aberrant CpG island hypermethylation in dysplastic
nodules and early HCC of hepatitis B virus-related human multistep
hepatocarcinogenesis. J Hepatol. 54:939–947. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nishida N, Nagasaka T, Nishimura T, Ikai
I, Boland CR and Goel A: Aberrant methylation of multiple tumor
suppressor genes in aging liver, chronic hepatitis and
hepatocellular carcinoma. Hepatology. 47:908–918. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Molkentin JD: The zinc finger-containing
transcription factors GATA-4, −5, and −6. Ubiquitously expressed
regulators of tissue-specific gene expression. J Biol Chem.
15(275): 38949–38952. 2000. View Article : Google Scholar
|
|
7
|
Patient RK and McGhee JD: The GATA family
(vertebrates and invertebrates). Curr Opin Genet Dev. 12:416–422.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Laverriere AC, MacNeill C, Mueller C,
Poelmann RE, Burch JB and Evans T: GATA-4/5/6, a subfamily of three
transcription factors transcribed in developing heart and gut. J
Biol Chem. 269:23177–23184. 1994.PubMed/NCBI
|
|
9
|
Fujikura J, Yamato E, Yonemura S, Hosoda
K, Masui S, Nakao K, Miyazaki Ji J and Niwa H: Differentiation of
embryonic stem cells is induced by GATA factors. Genes Dev.
16:784–789. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gao X, Sedgwick T, Shi YB and Evans T:
Distinct functions are implicated for the GATA-4, −5, and −6
transcription factors in the regulation of intestine epithelial
cell differentiation. Mol Cell Biol. 18:2901–2911. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Akiyama Y, Watkins N, Suzuki H, Jair KW,
van Engeland M, Esteller M, Sakai H, Ren CY, Yuasa Y, Herman JG and
Baylin SB: GATA-4 and GATA-5 transcription factor genes and
potential downstream antitumor target genes are epigenetically
silenced in colorectal and gastric cancer. Mol Cell Biol.
23:8429–8439. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hellebrekers DM, Lentjes MH, van den Bosch
SM, Melotte V, Wouters KA, Daenen KL, Smits KM, Akiyama Y, Yuasa Y,
Sanduleanu S, et al: GATA4 and GATA5 are potential tumor
suppressors and biomarkers in colorectal cancer. Clin Cancer Res.
15:3990–3997. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lassus H, Laitinen MP, Anttonen M,
Heikinheimo M, Aaltonen LA, Ritvos O and Butzow R: Comparison of
serous and mucinous ovarian carcinomas: Distinct pattern of allelic
loss at distal 8p and expression of transcription factor GATA-4.
Lab Invest. 81:517–526. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peters I, Gebauer K, Dubrowinskaja N,
Atschekzei F, Kramer MW, Hennenlotter J, Tezval H, Abbas M, Scherer
R, Merseburger AS, et al: GATA5 CpG island hypermethylation is an
independent predictor for poor clinical outcome in renal cell
carcinoma. Oncol Rep. 31:1523–1530. 2014.PubMed/NCBI
|
|
15
|
Guo M, House MG, Akiyama Y, Qi Y, Capagna
D, Harmon J, Baylin SB, Brock MV and Herman JG: Hypermethylation of
the GATA gene family in esophageal cancer. Int J Cancer.
119:2078–2083. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wen XZ, Akiyama Y, Pan KF, Liu ZJ, Lu ZM,
Zhou J, Gu LK, Dong CX, Zhu BD, Ji JF, et al: Methylation of GATA-4
and GATA-5 and development of sporadic gastric carcinomas. World J
Gastroenterol. 16:1201–1208. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Martin J, Afouda BA and Hoppler S:
Wnt⁄beta-catenin signalling regulates cardiomyogenesis via GATA
transcription factors. J Anat. 216:92–107. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Afouda BA, Martin J, Liu F, Ciau-Uitz A,
Patient R and Hoppler S: GATA transcription factors integrate Wnt
signalling during heart development. Development. 135:3185–3190.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jia Y, Yang Y, Liu S, Herman JG, Lu F and
Guo M: SOX17 antagonizes WNT/β-catenin signaling pathway in
hepatocellular carcinoma. Epigenetics. 5:743–749. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mayer MP: A new set of useful cloning and
expression vectors derived from pBlueScript. Gene. 163:41–46. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo M, Akiyama Y, House MG, Hooker CM,
Heath E, Gabrielson E, Yang SC, Han Y, Baylin SB, Herman JG and
Brock MV: Hypermethylation of the GATA genes in lung cancer. Clin
Cancer Res. 10:7917–7924. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Herman JG, Graff JR, Myöhänen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: A novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kristensen LS, Nielsen HM and Hansen LL:
Epigenetics and cancer treatment. Eur J Pharmacol. 625:131–142.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Meng F, Sun G, Zhong M, Yu Y and Brewer
MA: Anticancer efficacy of cisplatin and trichostatin A or
5-aza-2′-deoxycytidine on ovarian cancer. Br J Cancer. 108:579–586.
2013. View Article : Google Scholar : PubMed/NCBI
|