Introduction
Ovarian cancer is one of the most prevalent cancers
observed in females and is the leading cause of mortality among
gynecological malignancies (1). The
5-year survival rate of patients with ovarian cancer has remained
relatively consistent over the past few decades, despite advances
in surgical techniques and medical treatment (1). Increased efforts are required to achieve
a greater understanding of the molecular mechanisms underlying
ovarian cancer, and to aid the development of novel diagnostic and
therapeutic strategies.
The human genome produces a large number of long
non-coding RNA (lncRNA) transcripts that have structural,
regulatory and other unknown functions (2). In recent years, lncRNAs have gained
increasing attention from researchers. Different from the small
non-coding RNAs (18–200 nucleotides), lncRNAs are generally defined
as being ≥200 nucleotides in length (3). With the advance of high-throughput
sequencing technologies, it has been predicted that thousands of
human lncRNAs may exist (4). Despite
only a handful of lncRNAs being functionally characterized to date,
lncRNAs are believed to serve a role in almost all processes of
cellular and molecular biology, including the initiation and
progression of cancer (5).
BC200 RNA is a 200-nucleotide non-coding RNA that is
specifically expressed in neurons of the human nervous system
(6,7).
As a translational modulator, BC200 is implicated in the regulation
of local synaptodendritic protein synthesis in neurons through its
interaction with certain translational machinery components
(8). However, the neuron-specific
control of BC200 expression is dysregulated during carcinogenesis
in non-neuronal human tissues (9). It
has been reported that BC200 RNA is expressed in carcinomas of the
breast, cervix, esophagus, lungs, ovaries, parotid gland and
tongue, but not in corresponding normal tissues (9). Similar to the majority of lncRNAs, the
role of BC200 in cancer remains unclear.
In the present study, BC200 expression was detected
in ovarian cancer tissues and normal ovary tissues through the use
of quantitative polymerase chain reaction (qPCR). Furthermore, the
biological functions of BC200 were evaluated using RNA interference
in two ovarian cancer cell lines.
Materials and methods
Clinical samples
A total of 22 ovarian tissue samples, including 10
normal ovarian samples and 12 epithelial ovarian cancer samples,
were obtained from 22 patients who had previously undergone surgery
at the First and Third Affiliated Hospitals of Harbin Medical
University (Harbin, China), between May 2012 and December 2014. Of
the 12 epithelial ovarian cancer samples, 8 were serous, 2
endometrial and 2 mucinous. Normal ovarian samples were obtained
from patients who had previously received an ovariotomy due to the
presence of endometrial or cervical cancer. Therefore, the normal
ovarian tissues and epithelial ovarian cancer tissues were not
matched from the same patient. Patients in the two groups (normal
and cancer) were selected from the same hospital, and were matched
to cases in terms of the same age (±5 years) and date of hospital
admission. All samples were flash-frozen in liquid nitrogen and
stored at −80°C until RNA extraction. The diagnosis of each sample
was confirmed by at least two pathologists. No patient had received
any therapy prior to surgery. The research was performed according
to The Code of Ethics of the World Medical Association (Declaration
of Helsinki). All patients provided informed consent prior to their
inclusion in the study, and the study was initiated only after
approval by the Ethical Committee of Harbin Medical University.
Cell lines
The human ovarian cancer cell lines, SKOV3 and
A2780, used in the present study were purchased from the Cell
Resource Center, Institute of Basic Medical Sciences, Chinese
Academy of Medical Sciences (Beijing, China). The cells were
maintained in Dulbeccos modified Eagle's medium (DMEM; HyClone; GE
Healthcare Life Sciences, Logan, UT, USA), with 10% fetal bovine
serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc., Waltham,
MA, USA), 100 µg/ml streptomycin and 100 IU/ml penicillin at 37°C
in a humidified atmosphere containing 5% CO2.
RNA extraction and qPCR
Total RNA from the frozen tissue samples and cell
lines was extracted with TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.), following the manufacturers protocol. The cDNA
was synthesized from 1 µg of total RNA using EasyScript™ Reverse
Transcriptase (Beijing TransGen Biotech Co., Ltd., Beijing, China).
BC200 levels were quantified by qPCR using SYBR Green PCR MasterMix
(Beijing TransGen Biotech Co., Ltd.) and CFX96 Touch™ Real-Time PCR
Detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Primers (Shanghai GenePharma Co., Ltd., Shanghai, China) specific
for BC200 were designed as follows: Forward,
5-AGACCTGCCTGGGCAATATAGC-3 and reverse,
5-GTTGTTGCTTTGAGGGAAGTTACG-3. BC200 levels were normalized to
β-actin. Primers specific for β-actin were designed as follows:
Forward, 5-CCCTGGCACCCAGCAC-3 and reverse, 5-GCCGATCCACACGGAGTAC-3.
PCR conditions were as follows: Denaturation at 95°C for 5 min; and
then 39 cycles of amplification at 95°C for 10 sec/cycle and 60°C
for 30 sec. Melting curve analyses were performed using the PCR
products and progressive heating from 65°C to 95°C.
Transfection of small interfering RNA
(siRNA)
Three siRNAs (siRNA1, siRNA2 and siRNA3) targeting
BC200, and a scrambled siRNA used as negative control (NC), were
purchased from Shanghai GenePharma Co., Ltd. siRNA oligonucleotides
(50 nmol/l) were transfected into the SKOV3 and A2780 cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.),
following the manufacturers protocol. Target sequences for BC200
siRNAs were as follows: siRNA1, 5-AATAAGCGTAACTTCCCTCAAAG-3;
siRNA2, 5-AACTTCCCTCAAAGCAACAACCC-3; and siRNA3,
5-AAGCGTAACTTCCCTCAAAGCAA-3.
Cell counting kit-8 (CCK-8) assay
At 24 h post-transfection, the cells were seeded in
a 96-well plate at a density of 1×104 cells/well, and
subsequently incubated in DMEM supplemented with 10% FBS at 37°C
for 3 days. During the 3-day incubation, cell proliferation was
evaluated using the CCK-8 kit (Beyotime Institute of Biotechnology,
Beijing, China) at 0 (after adherence to the wall for 4 h), 24, 48
and 72 h. The CCK-8 reagent was added to the cell culture medium at
10 µl/well. Following incubation for 2 h, absorbance was determined
at a wavelength of 450 nm.
Luminescent cell viability assay
At 24 h post-transfection, the cells in the 96-well
plate were treated with graded carboplatin (0, 50 and 100µg/ml;
Qilu Pharmaceutical Co., Ltd., Jinan, China) for 48 h. Cellular
viability was measured using the CellTiter-Glo® Luminescent Cell
Viability assay (Promega Corporation, Madison, WI, USA). The
96-well plate was briefly equilibrated to room temperature for ~30
min. The CellTiter-Glo® reagent (100 µl) was then added to each
well. Following this, the media and reagent were mixed for 2 min on
an orbital shaker and left to incubate at room temperature for 10
min prior to recording luminescence, using a SpectraMax® M5
microplate reader (Molecular Devices, Inc., Sunnyvale, CA, USA).
Luminescence values were normalized to a dimethyl sulfoxide
control, and final values were presented as a relative
percentage.
Cell migration and invasion
assays
A cell migration assay was performed using a
Transwell chamber (Costar; Corning Incorporated, Corning, NY, USA),
and an invasion assay was performed using the BioCoat Matrigel
Invasion Chamber (BD Biosciences, Franklin Lakes, NJ), following
the manufacturers protocol. The diameter of the pore was 8.0 µm.
Following transfection with siRNA for 24 h, 5×104 cells
(SKOV3 and A2780, individually) were plated in the upper chamber in
serum-free media. The bottom chamber contained DMEM with 10% FBS.
The chamber was incubated at 37°C for 24 h to allow for cell
migration and invasion, and subsequently the bottom of the chamber
insert was fixed and stained with hematoxylin. Cells on the stained
membrane were counted in five randomly selected ×200 magnification
fields under light microscopy. The average cell number was
calculated and used to represent the invasive and migratory
ability.
Statistical analysis
All data are presented as the mean ± standard
deviation. Comparisons between groups were tested by Students
t-test or a one-way analysis of variance using GraphPad Prism 5.0
software (GraphPad Software, La Jolla, CA, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
BC200 is downregulated in ovarian
cancer
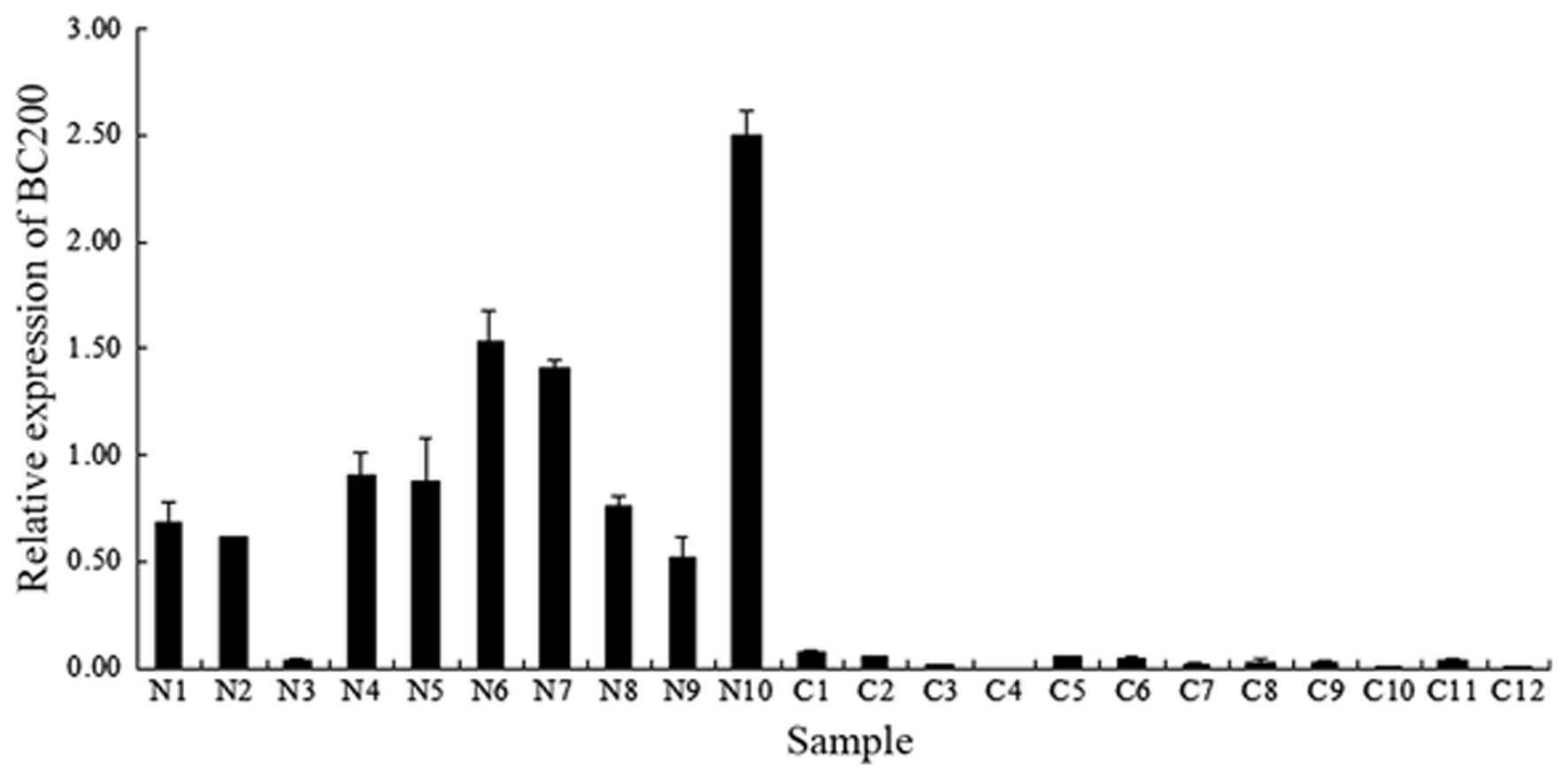
During the present study, the level of BC200 was
compared between normal ovarian samples and ovarian cancer samples
by means of qPCR. As presented in Fig.
1, the relative expression level of BC200 was greater in the
majority of normal ovarian samples (9/10), whereas in the ovarian
cancer samples, BC200 was significantly downregulated (12/12). The
average level of BC200 in the normal tissues was >30 times
higher than that observed in the ovarian cancer tissues. This
downregulation of BC200 may therefore be associated with the
initiation and progression of ovarian cancer.
siRNA is effective in knocking down
BC200 in SKOV3 and A2780 cells
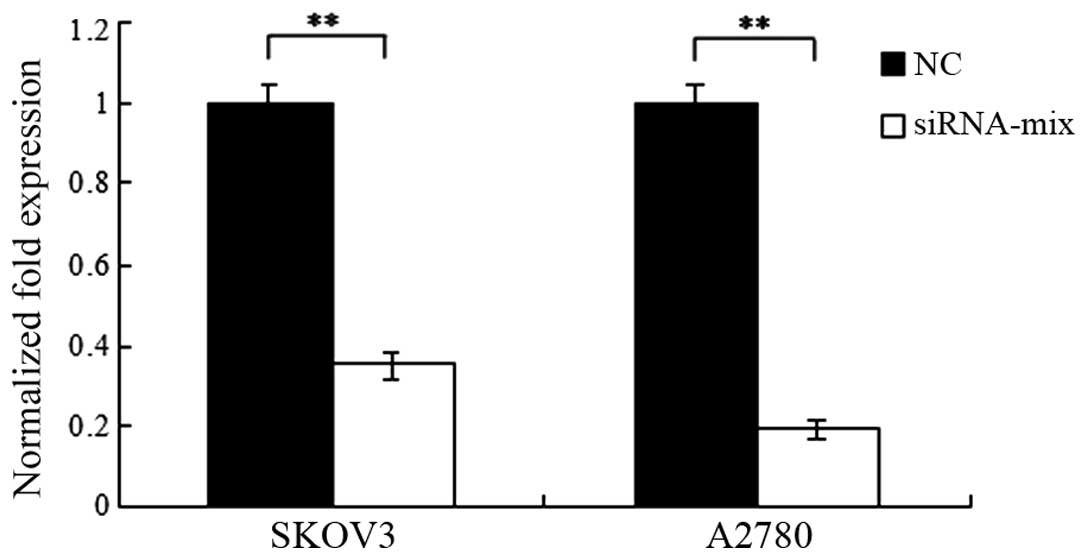
To observe the efficiency of siRNAs targeting BC200,
the level of BC200 was analyzed by qPCR at 48 h post-transfection
with the addition of each siRNA. The results demonstrated that all
three siRNAs, and the mixture of the three siRNAs, were effective
to knock down BC200 in the two cell lines (P<0.01; Fig. 2). The siRNA-mix was the most effective
and was thus used for the following experiments.
BC200 inhibits the proliferative
ability of ovarian cancer cells
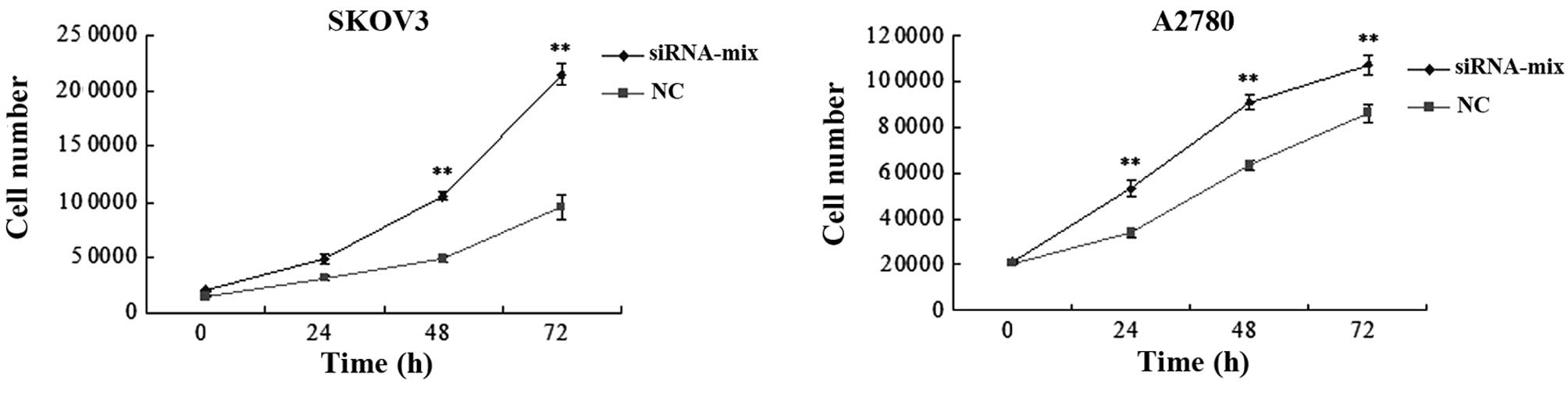
A CCK-8 assay was used to establish the
proliferative ability of siRNA-transfected and NC SKOV3 and A2780
cells. In the two cell lines, the siRNA-transfected cells grew
faster than the NC cells (Fig. 3). A
significant difference was observed at 48 and 72 h in the SKOV3
cells and at 24, 48 and 72 h in the A2780 cells (P<0.01).
Knockdown of BC200 therefore promotes the proliferative ability of
ovarian cancer cells, indicating that BC200 may inhibit cell
proliferation through an unknown pathway.
BC200 is associated with cell death
induced by carboplatin
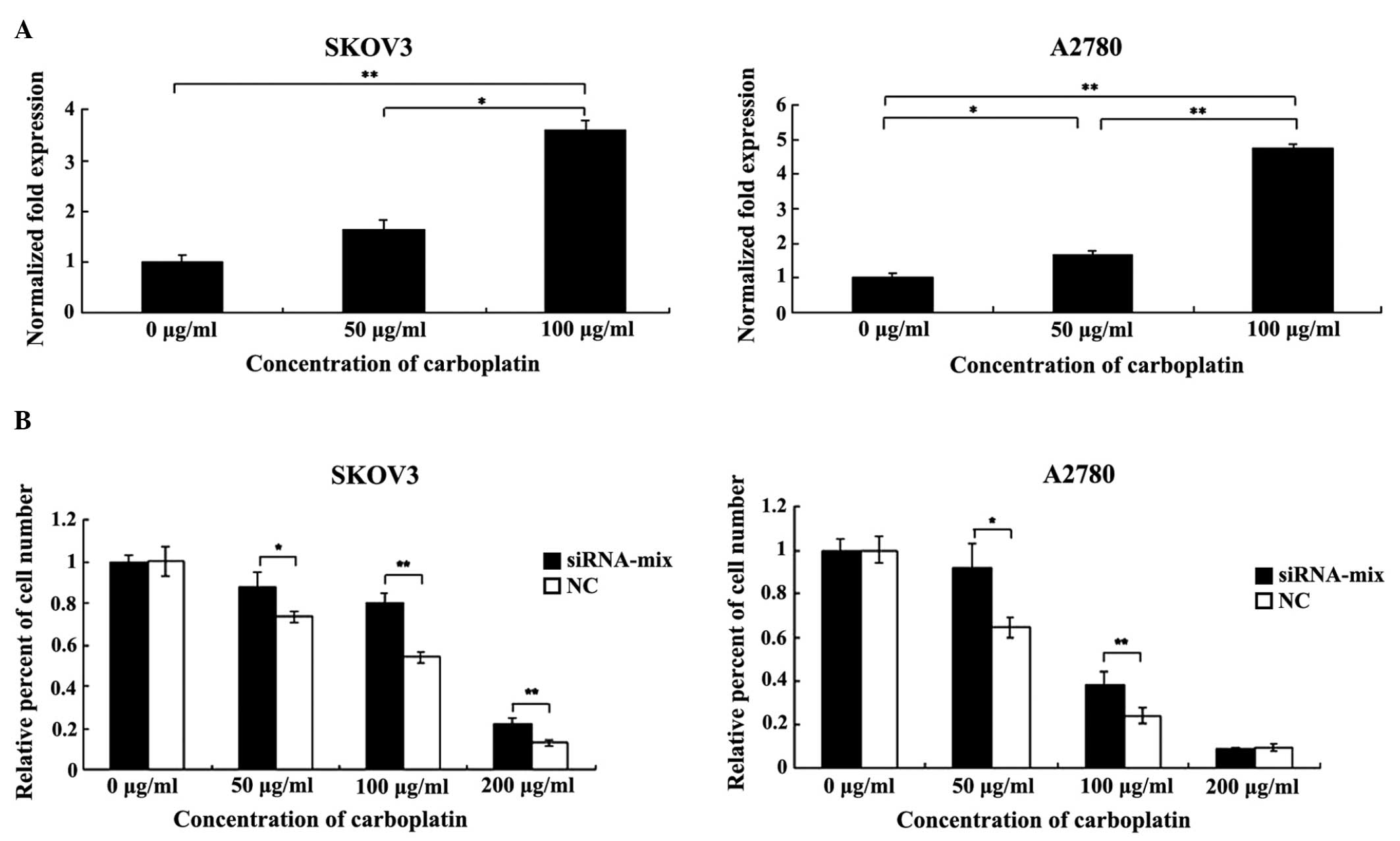
Initial experiments were performed to determine the
range of carboplatin concentration that elicited growth inhibition
and cell death. The SKOV3 and A2780 cells were treated with graded
carboplatin (0, 50 and 100 µg/ml) for 48 h, and following this, the
level of BC200 was detected by qPCR. Notably, the level of BC200
was upregulated in the ovarian cancer cells when carboplatin was
added to the medium in a dose-dependent manner (P<0.05 and
P<0.01; Fig. 4A). Subsequently, a
cell viability assay was performed to analyze the role of BC200 in
sensitizing the cells to carboplatin treatment. In the ovarian
cancer cells treated with carboplatin, the siRNA-transfected cells
exhibited higher cell viability in comparison to the NC cells
(P<0.05 and P<0.01; Fig. 4B),
indicating that the downregulation of BC200 contributed to the
chemoresistance of the ovarian cancer cells to carboplatin.
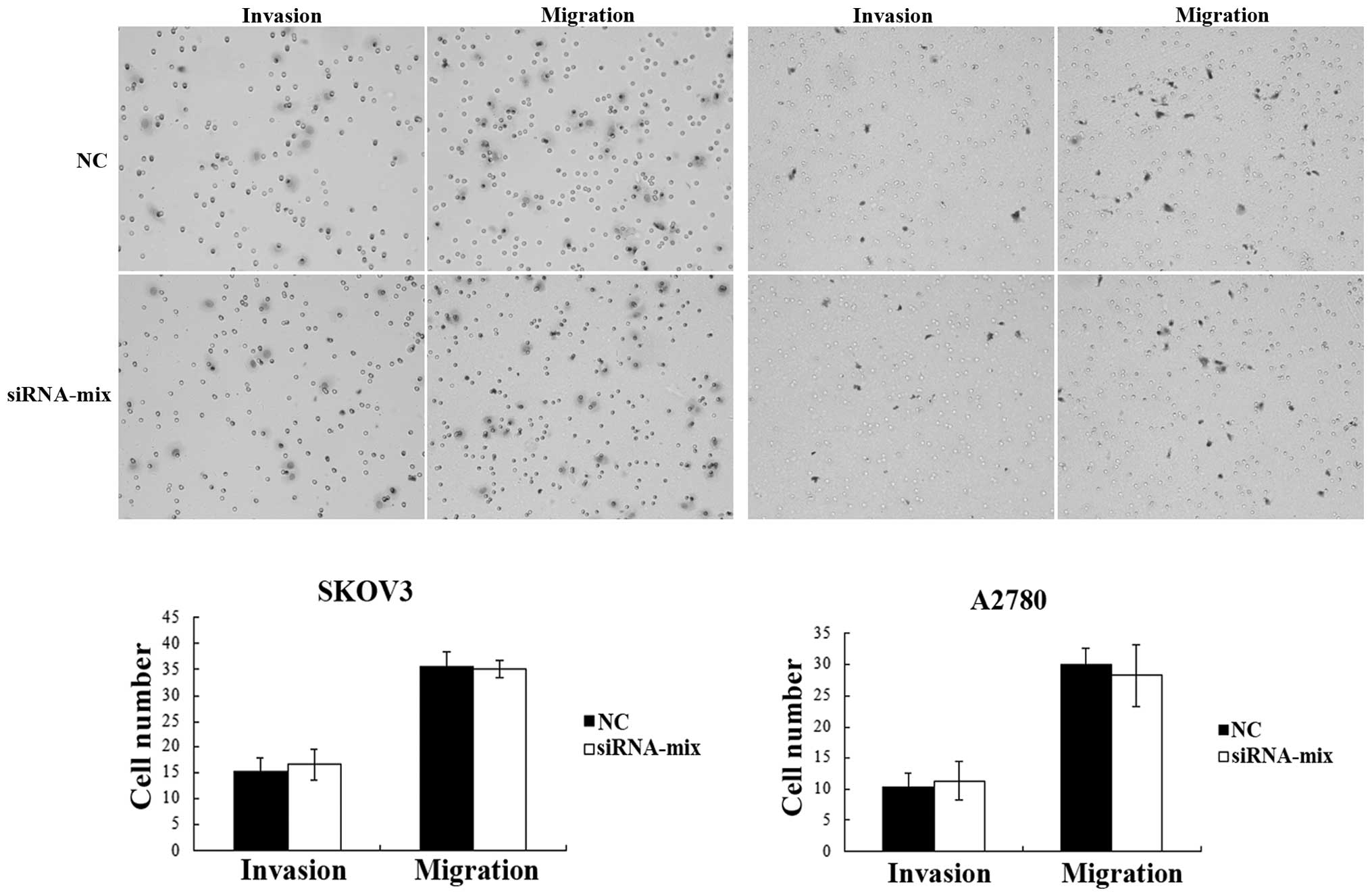
BC200 has no effect on the invasive
and migratory ability of ovarian cancer cells
To investigate the effect of BC200 on cell mobility,
invasion and migration, assays were performed on the
siRNA-transfected and NC SKOV3 and A2780 cells using Transwell
chambers with and without Matrigel. The results demonstrated that
there was no difference in the average number of cells observed on
the stained membrane between the two groups for each cell line
(P>0.05; Fig. 5), indicating that
BC200 had no effect on the invasive and migratory ability of the
ovarian cancer cells.
Discussion
In recent years, lncRNAs have gained increasing
attention as a novel class of molecules demonstrating a functional
role in carcinogenesis (10,11). Previous studies have demonstrated that
the expression of lncRNAs is dysregulated in a variety of human
cancers, and these aberrant lncRNAs may serve key roles during the
initiation and progression of cancer (12–14).
However, lncRNA research remains premature when compared to what is
understood about protein-coding RNA and microRNA. In fact, the
biological functions of the majority of lncRNAs are currently
unknown. To date, several lncRNAs have been identified to be
abnormally expressed in ovarian cancer, including imprinted
maternally expressed transcript, X-inactive specific transcript,
long stress-induced non-coding transcript 5, Pvt1 oncogene, serum
resistance-associated gene and host genes 2 (15–20). These
lncRNAs are involved in various biological processes of ovarian
cancer, including chemotherapy, proliferation, apoptosis and
metastasis (21–24). Previous studies have suggested the
potential role of lncRNAs in ovarian cancer to a certain extent
(15–24), however, further investigation is
required to explain their respective functions and mechanisms of
action.
BC200 is an lncRNA that is reported to be associated
with ovarian cancer. However, the function and mechanism of BC200
in cancer is not fully understood. In the present study, the
expression of BC200 in ovarian cancer tissues and normal ovarian
tissues was compared. The results demonstrated that BC200 was
significantly downregulated in the ovarian cancer tissues,
suggesting that the reduction of BC200 may be associated with the
occurrence of ovarian cancer, and thus, BC200 may potentially be
used as a diagnostic marker. However, Chen et al (9) reported that BC200 RNA was detected at
low levels in ovarian cancer tissues and was undetectable in normal
ovarian tissues. The possible causes of this conflict may have been
due to the following reasons: Firstly, the study by Chen et
al only detected BC200 in one pair of ovarian tissues from the
same patient, whereas the results from the current study were based
on detection in 10 normal ovarian samples and 12 ovarian cancer
samples. Secondly, the study by Chen et al used matched
normal and tumor tissues from the same patients, whereas the normal
and tumor tissues in the present study were obtained from different
patients. Despite results from an internal control possibly holding
more weight, one limitation is that it cannot be ensured that
matched ‘normal’ tissue really is normal. Malignant tumors
generally lack a clear boundary, and whether the tissue is normal
or not cannot be determined by the naked eye. Thirdly, the study by
Chen et al used northern blot hybridization to detect BC200,
while the present study used qPCR. Additionally, high levels of
BC200 in normal ovarian tissues highlight the uncertainty of the
theory that BC200 expression is neuron-specific. In addition to the
primate nervous system, BC200 is also expressed in germ cells,
including oocytes, and in cultured immortal cell lines of
non-neural origin (25,26).
Based on the results of the present study, we
hypothesize that BC200 may function as a tumor suppressor in
ovarian cancer. BC200 expression was significantly decreased in
ovarian cancer when compared with normal ovarian tissues.
Furthermore, the downregulation of BC200 contributed to ovarian
cancer cell proliferation, and inhibited the sensitivity of ovarian
cancer cells to carboplatin. The effect of BC200 on chemoresistance
was supported by additional evidence. SKOV3 is a platinum-resistant
cell line, whilst A2780 is a platinum-sensitive cell line. Notably,
the level of BC200 was significantly higher in the A2780 cells than
in the SKOV3 cells (data not shown). To date, lncRNAs, including
maternally expressed 3 (MEG3) and papillary thyroid carcinoma
susceptibility candidate 3 (PTCSC3), have been reported to function
as tumor suppressors. Downregulation of MEG3 has been observed in
various types of human cancer, and the lncRNA has been demonstrated
to inhibit cancer cell proliferation and induce apoptosis (27,28).
PTCSC3 is a newly identified, thyroid-specific lncRNA that is
significantly downregulated in thyroid cancer. PTCSC3 can induce
growth inhibition, cell cycle arrest and increased apoptosis
(29,30). BC200 was previously speculated to be
associated with invasion and migration in breast cancer. It was
reported that BC200 was expressed at high levels in invasive
carcinomas of the breast, and may potentially serve as a molecular
tool in the diagnosis and/or prognosis of breast cancer (31). However, the results of the present
study demonstrated that BC200 has no effect on the migration and
invasion ability of ovarian cancer cells.
In conclusion, the expression of BC200 was observed
to be reduced in ovarian cancer tissues when compared with normal
ovarian tissues, and the inhibition of BC200 was demonstrated to
promote the proliferative ability of ovarian cancer cells.
Furthermore, carboplatin induced the expression of BC200, and BC200
increased the sensitivity of ovarian cancer cells to carboplatin.
The results of the current study identified a partial function of
BC200 in ovarian cancer, and also provided a novel target for
mechanistic and clinical treatment studies. Continued investigation
is required to further expand our understanding of the function and
mechanism of action of BC200 in ovarian cancer.
Acknowledgements
This study was supported by the Heilongjiang
Provincial Health Bureau (grant no. 2012-518), the National Natural
Science Foundation of China (grant nos. 81302061 and 81241092) and
the 973 Program earlier research project (grant no.
2012CB526705).
Glossary
Abbreviations
Abbreviations:
|
lncRNAs
|
long non-coding RNAs
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
FBS
|
fetal bovine serum
|
|
NC
|
negative control
|
|
CCK-8
|
cell counting kit-8
|
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carninci P, Kasukawa T, Katayama S, Gough
J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al:
FANTOM Consortium; RIKEN Genome Exploration Research Group and
Genome Science Group (Genome Network Project Core Group): The
transcriptional landscape of the mammalian genome. Science.
309:1559–1563. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Michalak P: RNA world - the dark matter of
evolutionary genomics. J Evol Biol. 19:1768–1774. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs, Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tiedge H, Chen W and Brosius J: Primary
structure, neural-specific expression, and dendritic location of
human BC200 RNA. J Neurosci. 13:2382–2390. 1993.PubMed/NCBI
|
|
8
|
Wang H, Iacoangeli A, Popp S, Muslimov IA,
Imataka H, Sonenberg N, Lomakin IB and Tiedge H: Dendritic BC1 RNA,
Functional role in regulation of translation initiation. J
Neurosci. 22:10232–10241. 2002.PubMed/NCBI
|
|
9
|
Chen W, Böcker W, Brosius J and Tiedge H:
Expression of neural BC200 RNA in human tumours. J Pathol.
183:345–351. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brunner AL, Beck AH, Edris B, Sweeney RT,
Zhu SX, Li R, Montgomery K, Varma S, Gilks T, Guo X, et al:
Transcriptional profiling of long non-coding RNAs and novel
transcribed regions across a diverse panel of archived human
cancers. Genome Biol. 13:R752012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tano K, Mizuno R, Okada T, Rakwal R,
Shibato J, Masuo Y, Ijiri K and Akimitsu N: MALAT-1 enhances cell
motility of lung adenocarcinoma cells by influencing the expression
of motility-related genes. FEBS Lett. 584:4575–4580. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tanos V, Prus D, Ayesh S, Weinstein D,
Tykocinski ML, De-Groot N, Hochberg A and Ariel I: Expression of
the imprinted H19 oncofetal RNA in epithelial ovarian cancer. Eur J
Obstet Gynecol Reprod Biol. 85:7–11. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Benoît MH, Hudson TJ, Maire G, Squire JA,
Arcand SL, Provencher D, Mes-Masson AM and Tonin PN: Global
analysis of chromosome X gene expression in primary cultures of
normal ovarian surface epithelial cells and epithelial ovarian
cancer cell lines. Int J Oncol. 30:5–17. 2007.PubMed/NCBI
|
|
17
|
Silva JM, Boczek NJ, Berres MW, Ma X and
Smith DI: LSINCT5 is over expressed in breast and ovarian cancer
and affects cellular proliferation. RNA Biol. 8:496–505. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guan Y, Kuo WL, Stilwell JL, Takano H,
Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, et al:
Amplification of PVT1 contributes to the pathophysiology of ovarian
and breast cancer. Clin Cancer Res. 13:5745–5755. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hussein-Fikret S and Fuller PJ: Expression
of nuclear receptor coregulators in ovarian stromal and epithelial
tumours. Mol Cell Endocrinol. 229:149–160. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rangel LB, Sherman-Baust CA, Wernyj RP,
Schwartz DR, Cho KR and Morin PJ: Characterization of novel human
ovarian cancer-specific transcripts (HOSTs) identified by serial
analysis of gene expression. Oncogene. 22:7225–7232. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mizrahi A, Czerniak A, Levy T, Amiur S,
Gallula J, Matouk I, Abu-lail R, Sorin V, Birman T, de Groot N, et
al: Development of targeted therapy for ovarian cancer mediated by
a plasmid expressing diphtheria toxin under the control of H19
regulatory sequences. J Transl Med. 7:692009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang KC, Rao PH, Lau CC, Heard E, Ng SK,
Brown C, Mok SC, Berkowitz RS and Ng SW: Relationship of XIST
expression and responses of ovarian cancer to chemotherapy. Mol
Cancer Ther. 1:769–776. 2002.PubMed/NCBI
|
|
23
|
Liu SP, Yang JX, Cao DY and Shen K:
Identification of differentially expressed long non-coding RNAs in
human ovarian cancer cells with different metastatic potentials.
Cancer Biol Med. 10:138–141. 2013.PubMed/NCBI
|
|
24
|
Gao Y, Meng H, Liu S, Hu J, Zhang Y, Jiao
T, Liu Y, Ou J, Wang D, Yao L, et al: LncRNA-HOST2 regulates cell
biological behaviors in epithelial ovarian cancer through a
mechanism involving microRNA let-7b. Hum Mol Genet. 24:841–852.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Watson JB and Sutcliffe JG: Primate
brain-specific cytoplasmic transcript of the Alu repeat family. Mol
Cell Biol. 7:3324–3327. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
McKinnon RD, Danielson P, Brow MA, Bloom
FE and Sutcliffe JG: Expression of small cytoplasmic transcripts of
the rat identifier element in vivo and in cultured cells. Mol Cell
Biol. 7:2148–2154. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu
WQ, Xie WP and Hou YY: Long non-coding RNA MEG3 inhibits NSCLC
cells proliferation and induces apoptosis by affecting p53
expression. BMC Cancer. 13:4612013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou Y, Zhang X and Klibanski A: MEG3
noncoding RNA, A tumor suppressor. J Mol Endocrinol. 48:R45–R53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fan M, Li X, Jiang W, Huang Y, Li J and
Wang Z: A long non-coding RNA, PTCSC3, as a tumor suppressor and a
target of miRNAs in thyroid cancer cells. Exp Ther Med.
5:1143–1146. 2013.PubMed/NCBI
|
|
30
|
Jendrzejewski J, He H, Radomska HS, Li W,
Tomsic J, Liyanarachchi S, Davuluri RV, Nagy R and de la Chapelle
A: The polymorphism rs944289 predisposes to papillary thyroid
carcinoma through a large intergenic noncoding RNA gene of tumor
suppressor type. Proc Natl Acad Sci USA. 109:8646–8651. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Iacoangeli A, Lin Y, Morley EJ, Muslimov
IA, Bianchi R, Reilly J, Weedon J, Diallo R, Böcker W and Tiedge H:
BC200 RNA in invasive and preinvasive breast cancer.
Carcinogenesis. 25:2125–2133. 2004. View Article : Google Scholar : PubMed/NCBI
|