Introduction
Pancreatic cancer has a high mortality rate, with
the worst 1- and 5-year survival rates of all cancers. In 2015, it
is estimated that there will be 48,960 new cases of pancreatic
cancer and an ~40,560 individuals are expected to succumb to this
disease, according to the Surveillance, Epidemiology and End
Results database (1). The incidence
of infiltrating pancreatic ductal cell adenocarcinoma (PDAC) is
increasing, especially in age groups >50 years (2). Due to the late presentation of symptoms,
PDAC is often diagnosed at an advanced stage. The long-term
survival rates cannot be improved with radical resection. Although
numerous biomarkers of PDAC are awaiting progression to the clinic,
there has been a concerted effort to explore tumor markers that may
be valuable in diagnosing PDAC earlier, improving prognosis, and
subsequently progressing through to clinical pharmacology
experiments. The approaches of the potential biomarker studies
include genomics, proteomics and metabolomics (3). Identification of biomarkers with a
relatively high susceptibility and specificity for PDAC is
necessary.
Metastasis to distant sites in the body, via blood
vessels and lymphatic ducts results in poor prognosis. Cancer cell
spread is facilitated by the secretion of enzymes that can degrade
the main structural elements of the basement membrane (BM) and
extracellular cell matrix (ECM). Heparin sulfate proteoglycans
(HSPGs) are an important constituent of the BM, and these interact
with collagen IV, laminin, and numerous cytokines, chemokines, and
growth factors. Heparanase (HPSE) is often secreted by cancer
cells, and degrades the heparin sulfate (HS) chain of HSPGs, thus
damaging the anionic and mechanical barriers surrounding the cells
and promoting the proteolysis of cytokines, including members of
the vascular endothelial growth factor (VEGF) family (4). The VEGF family consists of VEGFs-A-E,
placental growth factor and snake venom VEGF. VEGF-A predominantly
promotes angiogenesis through interactions with VEGF receptor-1,2
(VEGFR-1,2), whereas VEGF-C induces lymphangiogenesis by attaching
to VEGF receptor-3 (VEGFR-3) (5).
PDAC is known to be predominantly hypovascular and
its blood flow volume reaches only one-third of the pancreatic
tissues of a healthy organ (6,7). Lymph
node metastasis is an independent risk factor of PDAC. VEGF-C is of
importance in increasing lymphatic vessel density (LVD), and
VEGFR-3 expression is highly restricted to lymphatic endothelial
cells (8,9). On the basis of these previous studies,
it seems that VEGF-C has a greater role in PDAC than VEGF-A. For
this reason, the present study has selected HPSE and VEGF-C as
experimental indicators to investigate whether HPSE can regulate
VEGF-C expression and its role in invasion of pancreatic carcinoma
cells in vitro, and to analyze the clinicopathological
characteristics of PDAC patients. The present study was approved by
the ethics committee of Qilu Hospital, Shandong University (Jinan,
China) and written informed consent was obtained from all
patients.
Materials and methods
Cell culture and HPSE expression
vector construction
Human primary pancreatic adenocarcinoma BxPC-3 cells
were purchased from American Type Culture Collection (Manassas, VA,
USA) and maintained in Dulbecco's modified Eagle's medium (DMEM;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10%
fetal calf serum (Gibco; Thermo Fisher Scientific, Inc.), 50 U/ml
penicillin and 50 µg/ml streptomycin (Sigma-Aldrich, St. Louis, MO,
USA). A GV230 plasmid was purchased, containing the gene for
enhanced green fluorescent protein (EGFP) and the XhoI/Kpnl
restriction sites (GeneChem Co., Ltd., Shanghai, China). Total RNA
was isolated from BxPC-3 with TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) and then treated with RQ1 RNAse-Free DNase
(2 units DNase/1 µg RNA; Promega Corporation, Sydney, Australia).
Following reverse transcription of 5 µg total RNA using oligo(dT)
primers (Thermo Fisher Scientific, Inc.), the resulting cDNA was
then amplified by polymerase chain reaction (PCR) using Pfu DNA
polymerase (Thermo Fisher Scientific, Inc.) and the following HPSE
primers: Forward 5′-TCCTGCGTACCTGAGGTTTG-3′, and reverse
5′-CCATTCCAACCGTAACTTCTCCT-3′. The following PCR protocol was used:
95°C initial denaturation 3 min, 35 cycles (95°C subsequent
denaturation 45 sec, 61°C annealing 50 sec, 72°C extension 3 min),
and 72°C final extension 10 min. Glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) was used as a reference gene, using the same
PCR conditions. The PCR products were separated using 1.5% agarose
gel electrophoresis and visualized with 6X DNA Loading Buffer
(Beyotime Institute of Biotechnology, Haimen, China) under
ultraviolet light. The amplified HPSE DNA fragment was purified
from the corresponding band in the agarose gel and incubated with
XhoI/Kpnl at 37°C for 2 h. The processed HPSE fragment was then
cloned into GV230 using T4 DNA ligase (Thermo Fisher Scientific,
Inc.). The recombinant plasmid was transformed into DH5α (Beyotime
Institute of Biotechnology) and confirmed by both PCR of the
bacterial solution and enzymatic digestion. The PCR detection used
the following HPSE primers: Forward
5′-CGCGTAGTGATGCCATGTAACTGAAT-3′, and reverse
5′-CGCTTCGATCCCAAGAAGGAATCAAC-3′. The PCR program was 95°C for 3
min, 30 cycles (95°C 45 sec; 59°C 45 sec; 72°C 50 sec), 72°C for 6
min.
Transient transfection and
semiquantitative PCR
Three pairs of small interfering RNAs (siRNAs)
targeting HPSE mRNA were designed and chemically synthesized
(GenePharma Co., Ltd., Shanghai, China). These siRNA sequences are
as follows: Sense 5′-GCUUCGAGUAUACCUUCAUTT-3′, and anti-sense
5′-AUGAAGGUAUACUCGAAGCTT-3′ for siRNA-1 (targeting the 1425–1446
encoding region of human HPSE); sense 5′-GUCCAACUCAAUGGUCUAATT-3′,
and anti-sense 5′-UUAGACCAUUGAGUUGGACTT-3′ for siRNA-2 (targeting
the 1612–1633 encoding region of HPSE); sense
5′-CUUGCCAGCUUUCUCAUAUTT-3′, and anti-sense
5′-AUAUGAGAAAGCUGGCAAGTT-3′ for siRNA-3 (targeting the 1704–1725
encoding region). Additionally, a negative control siRNA and
siGAPDH were used as controls, and negative control-FAM was used to
evaluate the transfection efficiency of siRNA. BxPC-3 cells were
inoculated in 6-well plates and then transfected with the
GV230/HPSE plasmid and 3 pairs of siRNAs in the presence of
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) when 80–90%
confluent. Cells were divided into seven groups as follows: siRNA-1
group, siRNA-2 group, siRNA-3 group, GV230/HPSE group, GV230 group,
blank control group, and siGAPDH group. Following transfection and
24 h of cell culture, total RNA and cDNA were prepared as described
earlier. The HPSE expression status in every group was evaluated by
PCR using specific primers. Semiquantitative analysis performed
using the data generated from electrophoresis was carried out using
Quantity One software (version 4.6.2; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). Utilizing the absorbance value of the HPSE band
normalized to GAPDH, the siRNA with the best interference
efficiency was selected for use in experiments in the present
study.
Reverse transcription-quantitative PCR
(RT-qPCR)
The cDNA sample of each group (siRNA group,
GV230/HPSE group, GV230 group, blank control group), generated by
reverse transcription, was diluted to working concentration (1 µl
cDNA HPSE in 50 µl ddH2O; 1 µl cDNA VEGF-C in 50 µl ddH2O and 1 µl
cDNA GAPDH in 100 µl ddH2O). The following PCR primers were used in
the present study: Forward, 5′-TCCTGCGTACCTGAGGTTTG-3′, and
reverse, 5′-CCATTCCAACCGTAACTTCTCCT-3′ for HPSE (product, 169 bp);
forward, 5′-ATGTGTGTCCGTCTACAGATGT-3′ and reverse,
5′-GGAAGTGTGATTGGCAAAACTGA-3′ for VEGF-C (product, 160 bp) and
forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′ for GAPDH (product, 197 bp). RT-qPCR
was performed using a Mastercycler Ep Realplex (Eppendorf, Hamburg,
Germany) using SYBR Green Real-time PCR Master Mix (Toyobo Co.,
Ltd., Osaka, Japan) with the following settings: Initial
denaturation at 95°C, followed by 40 cycles of denaturation for 15
sec at 95°C, annealing for 15 sec at 63°C and extension for 45 sec
at 72°C. Melting curves were used to exclude nonspecific
amplification and the emergence of primer dimers. The comparative
cycle threshold method (2-ΔΔCq) was applied to present the gene of
interest relative to the internal reference gene.
Immunoblotting assay
At 48 h following transfection, cells were dissolved
in RIPA Lysis Buffer (Beyotime Institute of Biotechnology)
containing 1 mmol/l phenylmethanesulfonyl fluoride, incubated at
4°C for 2 h and then centrifuged at 14,000 × g for 20 min. The
protein concentration of the supernatant was measured using a BCA
Protein Assay Kit (Thermo Fisher Scientific, Inc.). Proteins (100
mg loading) were separated by electrophoresis on 10%
SDS-polyacrylamide gel and transferred to polyvinylidene fluoride
membrane (PVDF; Merck Millipore, Darmstadt, Germany) by wet
electroblotting (Bio-Rad Laboratories, Inc.). The PVDF membrane for
each sample was incubated with rabbit monoclonal antibodies
(anti-HPSE, anti-VEGF and anti-β-actin), purchased from Abcam
(Cambridge, UK). β-actin was used to confirm equal loading of every
sample. The secondary antibody used was horseradish
peroxidase-labeled goat-anti-rabbit IgG (Sigma-Aldrich).
Immunoreactive bands were visualized by enhanced chemiluminescence
(Merck Millipore), exposed in the luminescent image analyzer
(FluorChemE; ProteinSimple; Bio-Techne, San Jose, CA, USA) and
semiquantified using Quantity One software (Bio-Rad Laboratories,
Inc.).
Transwell invasion assay
Invasion assays utilized 24-well cell Transwell®
inserts, coated with Matrigel® (Corning Inc., Corning, NY, USA). A
total of 1×107 cells/ml from each group (siRNA group, GV230/HPSE
group, blank control group) suspended in 200 µl DMEM (Thermo Fisher
Scientific, Inc.) containing 2% fetal calf serum were seeded in the
upper part of each chamber, and the lower section was filled with
500 µl of complete medium. Following 48 h of incubation,
non-invading cells on the upper surface of the Transwell® membrane
(8 µm pores, polyethylene membrane) were removed with a cotton
swab, and the attached cells on lower surface of the filter were
fixed with methanol, stained with Giemsa and counted in at least
four fields under an Olympus IX71 inverted fluorescence microscope
(Olympus Corp., Tokyo, Japan). The invasion assay was performed in
triplicate.
Sample collection and RT-qPCR analysis
of patient tissues
34 PDAC specimens were acquired from 20 male and 14
female patients (age range, 42–78 years; median age, 61 years)
during September 2012 to December 2013 at Qilu Hospital. According
to the American Joint Committee on Cancer staging system, there
were 9 stage I, 11 stage II, 11 stage III, and 3 stage IV ductal
cell adenocarcinomas. Among these specimens, 11 tumors were
well-differentiated, 13 tumors were moderately-differentiated, and
10 tumors were poorly-differentiated. Patients were diagnosed by
imaging-based findings, such as contrast-enhanced computed
tomography, endoscopic ultrasonography and magnetic resonance
imaging. No patients received chemotherapy or radiation therapy
prior to surgery, however, percutaneous transhepatic cholangial
drainage (PTCD) was allowed. Five patients underwent
pancreaticoduodenectomy, 7 patients underwent pylorus-preserving
pancreatoduodenectomy, 8 with distal pancreatectomy plus
splenectomy, 8 with laparoscopic exploration or exploratory
laparotomy and 6 with bilioenteric anastomosis or gastrojejunostomy
(some with chemical impairment of celiac ganglion). Immediately
following surgical removal, specimens were snap-frozen and stored
in liquid nitrogen container until use. The frozen tissues were
homogenized in TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and total RNA was isolated according to the
manufacturer's guideline. RT-qPCR was performed as described
earlier. Pathological reports were obtained to analyze the
association between prognosis factors for PDAC and HPSE and VEGF-C
mRNA expression.
Statistical analysis
Results are presented as mean ± standard error.
Student's t test was used for statistical analysis of the
semiquantitative PCR, 2-ΔΔCq values from RT-qPCR experiments,
immunoblotting and invasion assays. The Mann-Whitney U test was
used to assess the correlation of HPSE and VEGF-C expression and
clinicopathological parameters. Spearman rank correlation analysis
was used to justify the relationship of HPSE to VEGF-C in PDAC.
P<0.05 was taken to represent a statistically significant
differences in all tests.
Results
Construction of GV230/HPSE expression
vector
The PCR product of HPSE and GV230 plasmid linearized
by XhoI/KpnI digestion were subjected to 1.5% agarose gel
electrophoresis (Fig. 1A and B). The
correct recombinant plasmid was confirmed by both PCR of the
bacterial solution and enzymatic digestion of restriction sites
(Fig. 1C and D). Furthermore,
according to the sequencing report, HPSE full-length gene was
successfully cloned into eukaryotic expression vector without base
mutations, mismatches or frame shifts.
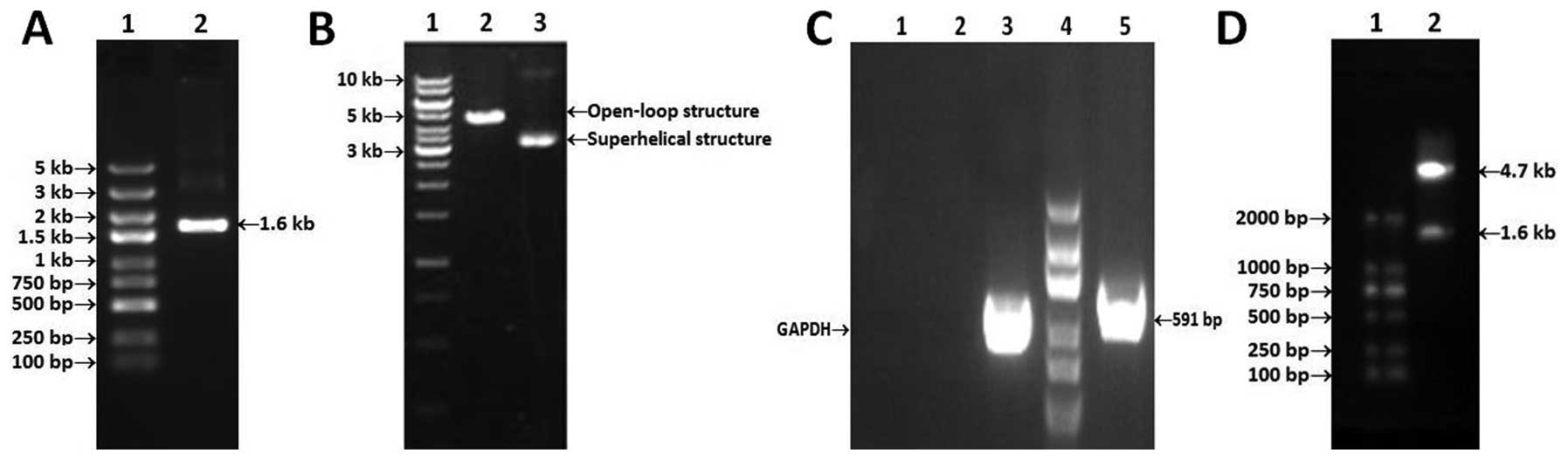 | Figure 1.Construction of GV230/HPSE. (A)
Electrophoresis of PCR products on 1.5% agarose gels. Lane 1, DNA
ladder marker(5 kb-100 bp; lane 2, PCR product of HPSE (1.6 kb).
(B) Electrophoresis of GV230 vacant plasmid and product after
restricted enzyme digestion. Lane 1, DNA ladder maker (10 kb-250
bp); lane 2, Linear plasmid after digestion with XhoI/Kpnl (4.7
kb); lane 3, GV230 plasmid extracted from DH5a. (C) PCR product of
DNA isolated from bacterial solution. Lane 1, negative control
(ddH2O); lane 2, negative control (linear plasmid self-ligation);
lane 3, amplified GAPDH fragment of DH5a; lane 4, DNA ladder marker
(2 kb-100 bp); lane 5, HPSE identifying band (591 bp). (D)
Digestion product of recombinant plasmid GV230/HPSE. Lane 1, DNA
ladder marker (2 kb-100 bp), lane 2, GV230 fragment and HPSE
fragment. |
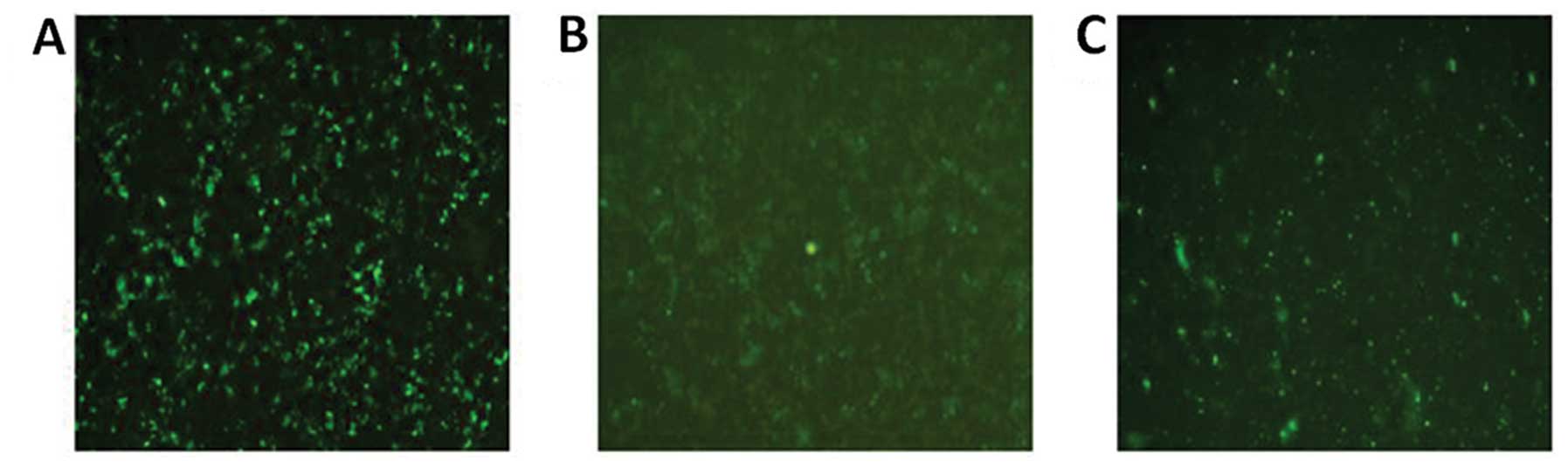
Transfection efficiencies of GV230,
GV230/HPSE and siRNA
At 48 h following transfection, fluorescence images
(Fig. 2A–C) were captured using an
Olympus IX71 inverted fluorescence microscope (Olympus Corp.).
After calculating the proportion of fluorescent cells to total
cells in several fields, the approximate transfection efficiencies
of the GV230 plasmid, GV230/HPSE recombinant plasmid and siRNA were
calculated as 46.14±3.8%, 33.89±6.65% and 58.06±4.3%,
respectively.
The influences of GV230/HPSE and
HPSE-siRNAs on mRNA levels of HPSE and VEGF-C in vitro
Images of the agarose gel PCR results were acquired
(Fig. 3A), and the absorbance value
of the HPSE band was normalized to that of GAPDH using Quantity One
software. The relative absorbancies were 0.048±0.022 (siRNA-1
group), 0.186±0.112 (siRNA-2 group), 0.136±0.077 (siRNA-3 group),
0.935±0.159 (GV230/HPSE group), 0.448±0.047 (GV230 group) and
0.441±0.101 (blank control group). The siRNA-1 with the best
interference effect was selected for use in subsequent trials.
BxPC-3 cells were divided into four groups: siRNA group, GV230/HPSE
group, GV230 group, blank control group. RT-qPCR was performed and
the 2-ΔΔCq value of each sample was calculated relative to the
blank control, to reflect the relative gene expression levels
following treatment (Fig. 3B and C)
(10,11). It was ascertained that no primer
dimerisation and non-specific amplification occurred, according to
melting curves (Fig. 3C and D). As
shown in Table I, there was a 10.7-
and 3.24-fold (All P<0.01) elevation of HPSE mRNA and VEGF-C
mRNA in GV230/HPSE group, respectively, whereas siRNA group
exhibited decreased mRNA expression (−2.45-fold, P<0.01;
−1.84-fold, P<0.01).
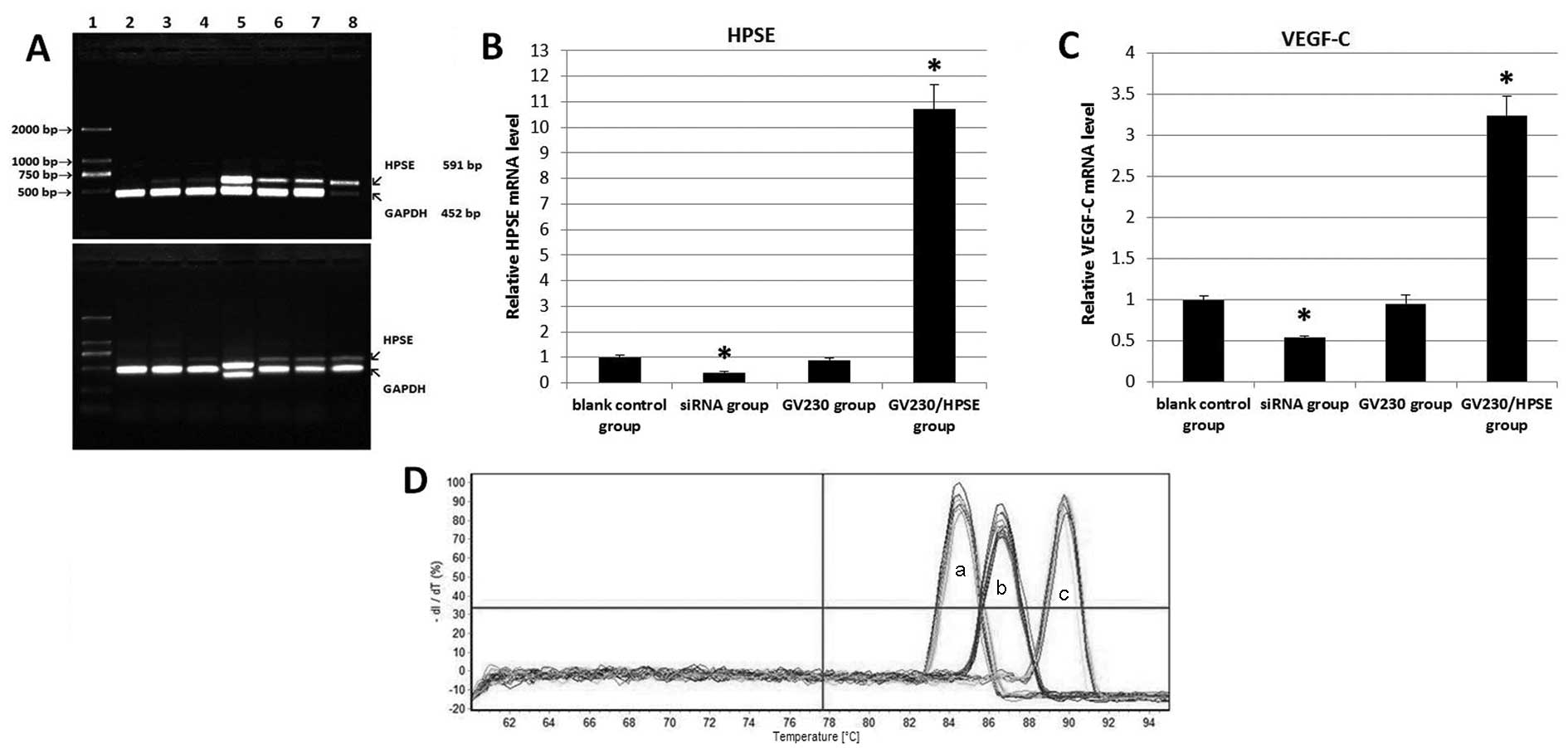 | Figure 3.The influences of GV230/HPSE and
HPSE-siRNAs on mRNA levels of HPSE and VEGF-C. (A) Semiquantitative
PCR of HPSE and GAPDH. Two repeats of the same assay are presented.
Lane 1: DNA ladder marker (2 kb-100 bp); lane 2, siRNA-1 group;
lane 3, siRNA-2 group; lane 4,siRNA-3 group; lane 5, GV230/HPSE
group; lane 6, GV230 group; lane 7, blank control group; lane 8,
siGAPDH group. (B and C) RT-qPCR results of relative expression of
HPSE and VEGF in treated BxPC-3 cells. (D) Melting curves for the
RT-qPCR assays (a: HPSE, b: VEGF-C, c: GAPDH). *P<0.05. HPSE,
heparanase; VEGF, vascular endothelial growth factor; siRNA, small
interfering RNA; RT-qPCR, reverse transcription quantitative
PCR. |
 | Table I.Comparison of relative mRNA
levels. |
Table I.
Comparison of relative mRNA
levels.
|
| HPSE mRNA | VEGF-C mRNA |
|---|
|
|
|
|
|---|
| Group |
2−ΔΔCq | ta | Pa |
2−ΔΔCq | ta | Pa |
|---|
| Blank control
group | 1.000±0.094 |
|
| 1.000±0.043 |
|
|
| siRNA group | 0.408±0.038 |
−8.970 | <0.01 | 0.542±0.016 |
−17.479 | <0.01 |
| GV230 group | 0.892±0.065 |
−1.677 |
0.169 | 0.951±0.114 |
−0.715 |
0.514 |
| GV230/HPSE group | 10.707±0.948 | 17.642 | <0.01 | 3.239±0.233 | 16.344 | <0.01 |
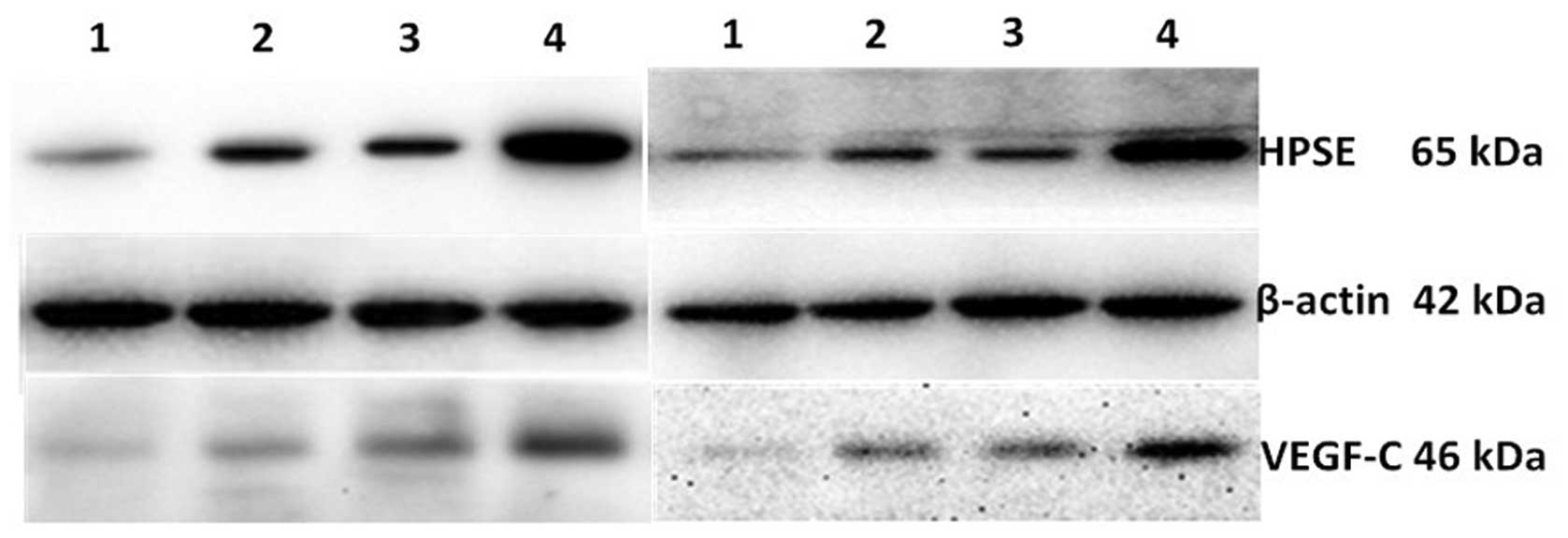
Expression of HPSE and VEGF-C
proteins
Western blot images were acquired using FluorChemE
(Fig. 4) and scanned by Quantity One
to calculate the relative optical density of HPSE and VEGF bands.
Due to the relatively low expression of VEGF-C and HPSE in BxPC-3,
increased protein loading and exposure time were necessary and
therefore increased background was visible. HPSE and VEGF-C had
significantly increased expression in cells transfected with
GV230/HSPE when compared to control cells (1.245±0.077 and
0.307±0.016 fold respectively; P<0.01). Cells treated with siRNA
targeted at HSPE has significantly reduced expression of HPSE and
VEGF-C compared to the untreated control cells groups when compared
to the blank control group (0.161±0.024 and 0.094±0.004,
respectively).
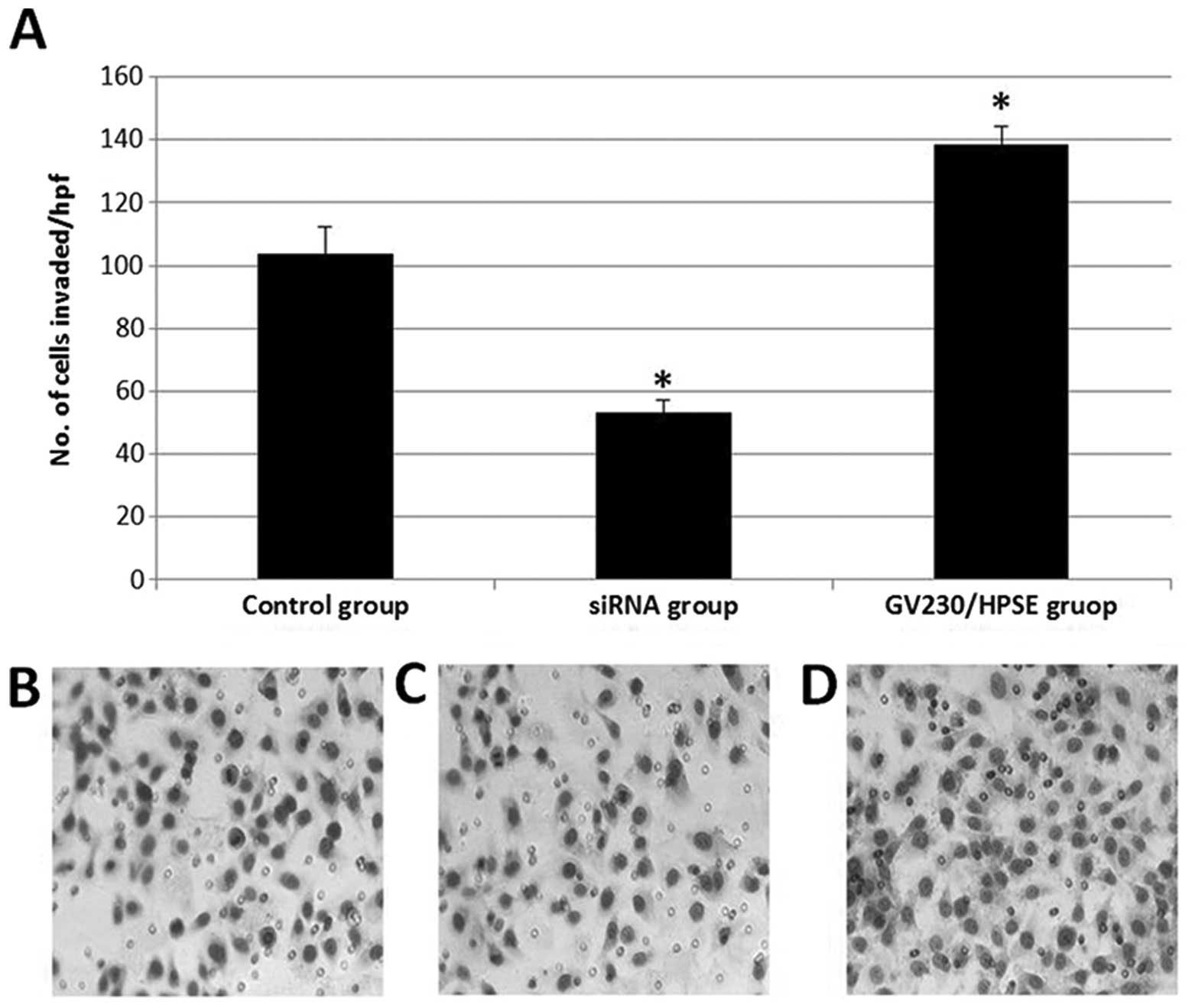
BxPC-3 cells invasion upon HPSE
overexpression and suppression
Compared to the blank control group, 138±5 cells in
the GV230/HPSE group and 53±4 cells in the siRNA group invaded
through the Matrigel and Transwell® membrane (Fig. 5A–D). This indicates that
overexpression of HPSE induces BxPC-3 cell invasion, whereas
knockdown of HPSE by siRNA reduces cell invasion.
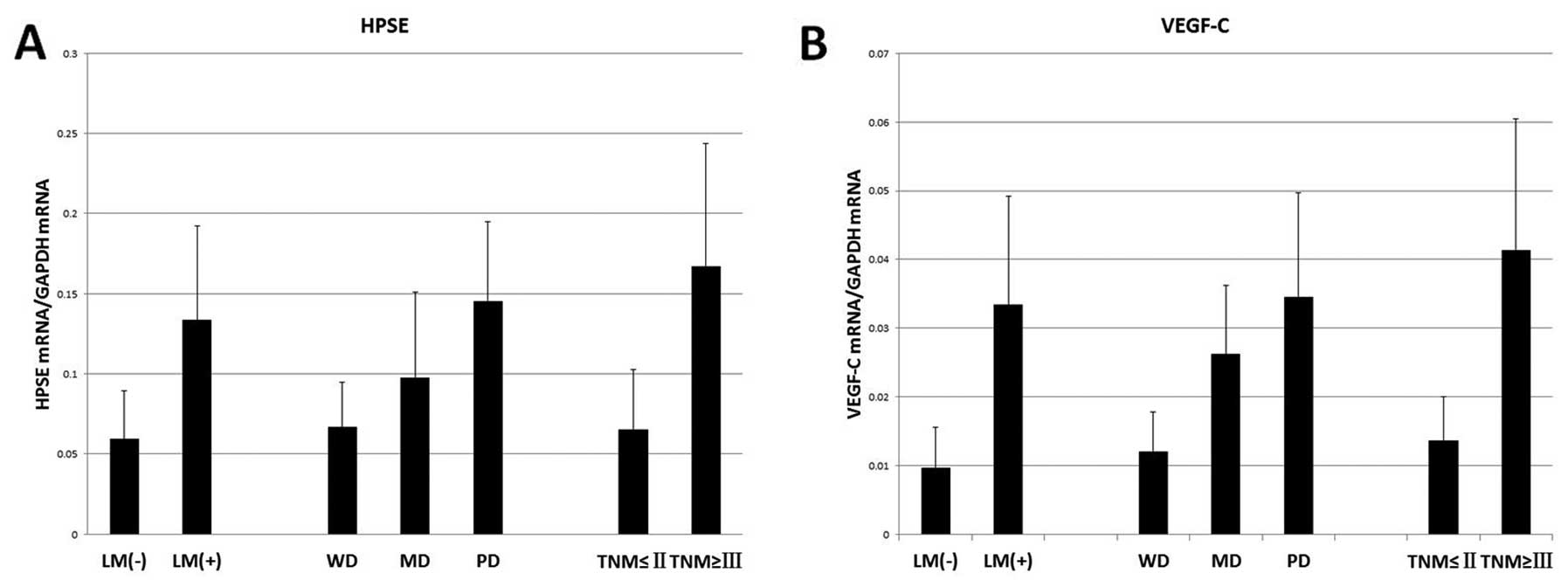
Expression of HPSE and VEGF-C mRNA in
human pancreatic cancer tissue and their association with
prognostic factors of postoperative PDAC patients
The lymph node metastasis status, tumor
differentiation grade and tumor-node-metastasis (TNM) stage were
selected as the predictors of prognosis (12,13). The
diameter and lymphatic metastasis status of unresectable pancreatic
mass were estimated by imaging tests combined with operative
findings. According to Mann-Whitney U test, earlier lymphatic
metastasis, poorer differentiation and more advanced tumor stages
were more frequently noted in PDAC patients with high HPSE or
VEGF-C expression than in patients with low HPSE or VEGF-C
expression (Table II, Fig. 6). Furthermore, Spearman rank
correlation analysis of 2-ΔΔ Cq values indicated a positive
correlation between HPSE and VEGF-C (r=0.812, P<0.01).
 | Table II.Correlation between mRNA levels and
clinicopathological characteristics. |
Table II.
Correlation between mRNA levels and
clinicopathological characteristics.
| Clinicopatheological
characteristic | Samples, no. | HPSE
(2−ΔΔCq) | PHPSEa | VEGF-C
(2−ΔΔCq) | PVEGF-Ca |
|---|
| Lymph node
metastasis |
|
|
|
|
|
| No | 12 | 0.0594±0.0299 | <0.01 | 0.0097±0.0059 | <0.01 |
|
Yes | 22 | 0.1336±0.0587 | <0.01 | 0.0334±0.0157 | <0.01 |
| Pathology
staging |
|
|
|
|
|
| Well
differentiated | 11 | 0.0668±0.0279 | <0.01 | 0.0119±0.0058 | <0.01 |
|
Moderately differentiated | 13 | 0.0977±0.0531 | <0.01 | 0.0262±0.0100 | <0.01 |
| Poorly
differentiated | 10 | 0.1455±0.0496 | <0.01 | 0.0346±0.0151 | <0.01 |
| TNM staging |
|
|
|
|
|
| Stage I
and II | 20 | 0.0656±0.0372 | <0.01 | 0.0136±0.0064 | <0.01 |
| Stage
III and IV | 14 | 0.1672±0.0767 | <0.01 | 0.0413±0.0191 | <0.01 |
Discussion
PDAC is among the leading causes of gastrointestinal
cancer fatalities in China and worldwide. The 5-year survival rate
is reported to be 6.8–25%, and the median survival time of
postoperative patients ranges from 8 to 12 months (14). Patients with lymph node metastasis
have shorter survival times, with the majority of individuals
surviving ≤2 years. VEGF-C is expressed in several cancers, such as
gastrointestinal malignant tumor, ovarian cancer, prostate cancer
and breast cancer (15–19). Some clinical follow-up studies have
associated the negative correlation of VEGF-C expression and the
5-year survival rate. Additionally, upregulation of VEGF-C is
identified in those who have multiple and distant lymph node
metastasis (20). These previous
studies have demonstrated that HPSE, together with VEGF-C, play an
essential role in the metastasis of PDAC. The present study sought
to elucidate the underlying association between the two proteins
using in vitro and tumor tissues experiments, including
siRNA and gene overexpression techniques. The GV230 eukaryotic
expression vector containing the pUC promoter and EGFP gene allowed
the upregulation of HPSE and estimation of transfection efficiency.
Although the GV230/HPSE vector produced less fluorescent brightness
and lower transfection efficiencies compared to the empty vector,
due to the recombinant size (6.3 kb) and the sharing of one
promoter (HPSE and EGFP), this had no significant impact on the
test results. Our results demonstrate a 10.7- and 3.24-fold
increase of HPSE mRNA and VEGF-C mRNA, respectively, in the
GV230/HPSE group, and a 2.45-and 1.84-fold decrease in siRNA group
were detected by RT-qPCR. Compared to control cells, a 2.84-fold
increase in HPSE protein and a 1.70-fold of VEGF-C protein was
expressed in GV230/HPSE cells, while a 2.72- and 1.91-fold
reduction of proteins, respectively, were observed in the siRNA
group. Thus, we conclude that HPSE modulates VEGF-C expression,
although, the rangeability of HPSE is higher than VEGF-C. HPSE and
other cytokines may influence VEGF-C at the same time, including
components of VEGF autocrine signaling pathway (21,22).
Additionally, other uncontrollable factors in the present study may
have weakened the regulation effect, such as the transfection
efficiency and loading errors. Zetser et al (23) demonstrated that HPSE overexpression in
human embryonic kidney 293, MDA-MB-435 human breast carcinoma, and
rat C6 glioma cells generated a 3- to 6-fold increase in VEGF
protein and mRNA levels. This increase may consist predominantly of
variations in VEGF-A and VEGF-C. The recombinant plasmid used in
the present study contains full length of HPSE-mRNA CDS and encodes
secretory HPSE, a signal peptide (amino acids 1–35 located at the
-NH2 terminus of the 50 kDa subunit). The signal peptide oriented
precursor protein is positioned and processed at endoplasmic
reticulum and Golgi apparatus, and the resulting HPSE in its native
conformation is stored in the lysosome and secreted when necessary
(24). It has been previously
demonstrated that elevation of VEGF in cells is associated with
upregulated HPSE secretion, demonstrated by using several
artificial variants of HPSE, such as deletion of the signal
peptide, which generates a variant of HPSE that fails to get
secreted, is resistant to proteolysis, and lacks enzymatic activity
(25). Another HPSE variant is
targeted to cell membrane by introducing the platelet derived
growth factor receptor (PDGFR) transmembrane domain at the
HPSE-COOH terminus. No significant change in VEGF expression was
observed in cells expressing non-secretory HPSE, therefore VEGF
upregulation requires HPSE secretion, but not its enzymatic
function (24).
A number of previous studies have reported
non-enzymatic activity of HPSE, as follows: i) Extrinsic addition
of HPSE stimulates Akt-dependent endothelial cell migration
(26); ii) extracellular
signal–regulated kinase activation enhances the adhesive capability
of certain cell lines, mediated by β1-integrin, Akt and Pyk2
(27–29); iii) inducing peritumoral angiogenesis
and lymphangiogenesis by regulating VEGF expression; iv)
Upregulating tissue factor, and interacting with the tissue factor
pathway inhibitor on the cell surface, resulting in increased
endothelial and tumor cell surface coagulation activity (30). Furthermore, Zetser et al
(23) provided evidence that HPSE
enhanced p38 phosphorylation and is actively involved in regulation
of VEGF gene expression via Src activation. Src is a cytosolic
tyrosine kinase, regulated by several extracellular signal
molecules and is important in the occurrence and development of
tumors (31). P38 is the member of
the mitogen-activated protein kinase family and participates in
signaling initiated by a wide variety of extracellular stimuli. It
is implicated in cell biological behaviors ranging from apoptosis,
proliferation, differentiation, and to some extent, tumor cell
survival and metastasis (32). We
suggest that HPSE promotes lymphangiogenesis and facilitates
invasion of pancreatic carcinoma cells by acting as a critical
cytokine implicated in signal pathways that include VEGF-C, p38 and
Src. Further trials are required to identify the HPSE receptor
located in the cell membrane or cytoplasm. The metastatic potential
of PDAC cells needs to be further studied in transplanted tumor
models.
Based on the 34 ductal cell adenocarcinomas examined
in the present study, patients with relatively high mRNA levels of
HPSE and VEGF-C possess more advanced tumor stages, especially
lymphatic metastasis, which is a sign of poor prognosis. In
addition, a positive correlation (r=0.812) demonstrates that the
elevation of VEGF-C may be partly attributed to the induction of
HPSE. Therefore, HPSE may serve as a novel target for cancer
chemotherapy. Several HPSE inhibitors have been discovered to date,
ranging from HS analogues to neutralizing antibodies. PI-88 is a
chemically synthesized compound with a non-cleavable structure that
can competitively inhibit HS-related angiogenesis. As a potent
counterpart of HS, PI-88 exerts significant anti-tumor,
anti-angiogenic, and anti-metastatic activity in numerous animal
models. The activities mentioned above are attributed to
intervening in HS recognition with angiogenic factors, such as VEGF
and basic fibroblast growth factor, as well as suppression of HPSE
activity. Other HS analogues with the same pharmacological
activity, such as necuparanib and roneparstat, are already in early
stage clinical development (4).
In conclusion, the present study suggests that joint
detection of HPSE and VEGF-C may be of significance as a diagnostic
marker and therapeutic target for patients with pancreatic
cancer.
Acknowledgements
The present study was funded by the Shandong
Provincial Natural Science Foundation, China (no.
2008BSB14026).
References
|
1
|
National Cancer Institute: Surveillance,
Epidemiology, and End Results Program. SEER Stat Fact Sheets:
Pancreas Cancer. http://seer.cancer.gov/statfacts/html/pancreas.htmlAccessed.
April 08–2015
|
|
2
|
Han H and Von Hoff DD: SnapShot:
Pancreatic cancer. Cancer Cell. 23:424. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Winter JM, Yeo CJ and Brody JR:
Diagnostic, prognostic, and predictive biomarkers in pancreatic
cancer. J Surg Oncol. 107:15–22. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hammond E, Khurana A, Shridhar V and
Dredge K: The Role of Heparanase and Sulfatases in the Modification
of Heparan Sulfate Proteoglycans within the Tumor Microenvironment
and Opportunities for Novel Cancer Therapeutics. Front Oncol.
4:1952014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takahashi H and Shibuya M: The vascular
endothelial growth factor (VEGF)/VEGF receptor system and its role
under physiological and pathological conditions. Clin Sci (Lond).
109:227–241. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hosoki T: Dynamic CT of pancreatic tumors.
AJR Am J Roentgenol. 140:959–965. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Matsubara H, Itoh A, Kawashima H, Kasugai
T, Ohno E, et al: Dynamic quantitative evaluation of
contrast-enhanced endoscopic ultrasonography in the diagnosis of
pancreatic diseases. Pancreas. 40:1073–1079. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Omoto I, Matsumoto M, Okumura H, Uchikado
Y, Setoyama T, Kita Y, Owaki T, Kijima Y, Shinchi H, Ishigami S, et
al: Expression of vascular endothelial growth factor-C and vascular
endothelial growth factor receptor-3 in esophageal squamous cell
carcinoma. Oncol Lett. 7:1027–1032. 2014.PubMed/NCBI
|
|
9
|
de Oliveira ATT, Reis RM, Afonso J,
Martinho O, Matos D, Carvalho AL, Vazquez VL, Silva TB,
Scapulatempo C, Saad SS, et al: Lymphangiogenic VEGF-C and VEGFR-3
expression in genetically characterised gastrointestinal stromal
tumours. Histol Histopathol. 26:1499–1507. 2011.PubMed/NCBI
|
|
10
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Neuzillet C, Sauvanet A and Hammel P:
Prognostic factors for resectable pancreatic adenocarcinoma. J Visc
Surg. 148:e232–e243. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Argon A, Nart D, Oruc N, Coker A and
Ozutemiz O: The prognostic significance of clinicopathological
features and apoptosis inhibitor proteins in pancreas ductal
adenocarcinoma. Acta Gastroenterol Belg. 77:229–234.
2014.PubMed/NCBI
|
|
14
|
Jung KW, Kim MH, Lee TY, Kwon S, Oh HC,
Lee SS, Seo DW and Lee SK: Clinicopathological aspects of 542 cases
of pancreatic cancer: A special emphasis on small pancreatic
cancer. J Korean Med Sci. 22(Suppl): S79–S85. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gou HF, Chen XC, Zhu J, Jiang M, Yang Y,
Cao D and Hou M: Expressions of COX-2 and VEGF-C in gastric cancer:
Correlations with lymphangiogenesis and prognostic implications. J
Exp Clin Cancer Res. 30:142011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kozlowski M, Naumnik W, Niklinski J,
Milewski R, Dziegielewski P and Laudanski J: Vascular endothelial
growth factor C and D expression correlates with lymph node
metastasis and poor prognosis in patients with resected esophageal
cancer. Neoplasma. 58:311–319. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang KJ and Sui LH: The relevance and
role of vascular endothelial growth factor C, matrix
metalloproteinase-2 and E-cadherin in epithelial ovarian cancer.
Med Oncol. 29:318–323. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun GG, Wang YD, Cui DW, Cheng YJ and Hu
WN: EMP1 regulates caspase-9 and VEGFC expression and suppresses
prostate cancer cell proliferation and invasion. Tumour Biol.
35:3455–3462. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ciobanu M, Eremia IA, Crăiţoiu S,
Mărgăritescu CL, Stepan A, Pătraşcu V, Georgescu CC, Cernea D and
Dumitrescu D: Lymphatic microvessels density, VEGF-C, and VEGFR-3
expression in 25 cases of breast invasive lobular carcinoma. Rom J
Morphol Embryol. 54:925–934. 2013.PubMed/NCBI
|
|
20
|
Kurahara H, Takao S, Maemura K, Shinchi H,
Natsugoe S and Aikou T: Impact of vascular endothelial growth
factor-C and -D expression in human pancreatic cancer: Its
relationship to lymph node metastasis. Clin Cancer Res.
10:8413–8420. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Perrot-Applanat M and Di Benedetto M:
Autocrine functions of VEGF in breast tumor cells Adhesion,
survival, migration and invasion. Cell Adhes Migr. 6:547–553. 2012.
View Article : Google Scholar
|
|
22
|
Decio A, Taraboletti G, Patton V, Alzani
R, Perego P, Fruscio R, Jürgensmeier JM, Giavazzi R and Belotti D:
Vascular endothelial growth factor c promotes ovarian carcinoma
progression through paracrine and autocrine mechanisms. Am J
Pathol. 184:1050–1061. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zetser A, Bashenko Y, Edovitsky E,
Levy-Adam F, Vlodavsky I and Ilan N: Heparanase induces vascular
endothelial growth factor expression: Correlation with p38
phosphorylation levels and Src activation. Cancer Res.
66:1455–1463. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zetser A, Levy-Adam F, Kaplan V,
Gingis-Velitski S, Bashenko Y, Schubert S, Flugelman MY, Vlodavsky
I and Ilan N: Processing and activation of latent heparanase occurs
in lysosomes. J Cell Sci. 117:2249–2258. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Levy-Adam F, Abboud-Jarrous G, Guerrini M,
Beccati D, Vlodavsky I and Ilan N: Identification and
characterization of heparin/heparan sulfate binding domains of the
endoglycosidase heparanase. J Biol Chem. 280:20457–20466. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yuan L, Hu J, Luo Y, Liu Q, Li T, Parish
CR, Freeman C, Zhu X, Ma W, Hu X, et al: Upregulation of heparanase
in high-glucose-treated endothelial cells promotes endothelial cell
migration and proliferation and correlates with Akt and
extracellular-signal-regulated kinase phosphorylation. Mol Vis.
18:1684–1695. 2012.PubMed/NCBI
|
|
27
|
Riaz A, Ilan N, Vlodavsky I, Li JP and
Johansson S: Characterization of heparanase-induced
phosphatidylinositol 3-kinase-AKT activation and its integrin
dependence. J Biol Chem. 288:12366–12375. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sotnikov I, Hershkoviz R, Grabovsky V,
Ilan N, Cahalon L, Vlodavsky I, Alon R and Lider O: Enzymatically
quiescent heparanase augments T cell interactions with VCAM-1 and
extracellular matrix components under versatile dynamic contexts. J
Immunol. 172:5185–5193. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Goldshmidt O, Zcharia E, Cohen M, Aingorn
H, Cohen I, Nadav L, Katz BZ, Geiger B and Vlodavsky I: Heparanase
mediates cell adhesion independent of its enzymatic activity. FASEB
J. 17:1015–1025. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nadir Y and Brenner B: Heparanase-A Link
between Coagulation, Angiogenesis, and Cancer. Rambam Maimonides
Med J. 3:e00022012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Irby RB and Yeatman TJ: Role of Src
expression and activation in human cancer. Oncogene. 19:5636–5642.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mittelstadt PR, Salvador JM, Fornace AJ Jr
and Ashwell JD: Activating p38 MAPK: New tricks for an old kinase.
Cell Cycle. 4:1189–1192. 2005. View Article : Google Scholar : PubMed/NCBI
|