Introduction
Bladder cancer (BC) is the ninth most commonly
observed cancer globally, with an estimated 386,300 novel cases and
150,200 mortalities in 2011 (1).
Transitional cell carcinoma (TCC) is the most predominant type,
accounting for 90% of all cases of diagnosed BC (1). Clinical studies have divided TCC
patients into two distinct subtypes: Non-muscle-invasive TCCs
(NMI-TCCs) and muscle-invasive TCCs (MI-TCCs). In total, 70% of
patients present with NMI-TCC, while the remaining 30% present with
MI-TCC. MI-TCCs are less common than NMI-TCCs, but are associated
with an increased mortality rate (2).
Therefore, the present study primarily focused on MI-TCC due to its
poor prognosis and survival rates.
The occurrence and development of cancer frequently
involves the accumulation of genomic alterations (3), therefore it is important to identify
these alterations using various techniques. A number of researchers
have employed next-generation sequencing technologies, including
whole-genome, whole-exome and whole-transcriptome approaches. This
enables the investigation of the main types of alterations to the
somatic cancer genome, including point mutations, copy number
alterations, microbial infections, small insertions and deletions,
and chromosomal rearrangements), providing insights for improving
our understanding of cancer biology, diagnosis and therapy in acute
myeloid leukemia (4,5), breast cancer (6–8), melanoma
(9), lung cancer (10,11),
kidney cancer (12) and ovarian clear
cell carcinoma (13), among
others.
Previous research based on candidate gene approaches
has suggested that BC may represent a heterogeneous disease
(2). The two subgroups of TCCs may
possess differential genetic backgrounds: NMI-TCCs frequently carry
mutations in the fibroblast growth factor receptor 3 (FGFR3) and
RAS genes, while MI-TCCs often carry mutations in the tumor protein
p53 (TP53) and retinoblastoma 1 (RB1) genes (2). These findings provided significant
insights into potential diagnoses and therapeutic applications,
nevertheless, to the best of our knowledge, a comprehensive
analysis of TCC has not been performed. Gui et al (14) identified genetic aberrations of
unknown chromatin remodeling genes [Ubiquitously transcribed
tetratricopeptide repeat, X chromosome, mixed-lineage leukemia
protein 3, CREBBP-EP300, nuclear receptor corepressor 1, AT rich
interactive domain 1A (SWI-like) and chromodomain helicase DNA
binding protein 6] in TCC by whole-exome sequencing and proposed
that aberrant chromatin regulation may be a hallmark of BC. Li
et al (15) employed
single-cell sequencing analysis to investigate the genetic
properties of bladder tumor alterations at the single-cell level
and to assess the evolution of bladder cancer at a cell-population
level. Guo et al (16)
observed frequent alterations in stromal antigen 2 and extra
spindle pole bodies homolog 1, and recurrent fusion involving FGFR3
and transforming, acidic coiled-coil containing protein 3. These
genes were identified to be involved in the sister chromatid
cohesion and segregation (SCCS) process by whole-genome and
whole-exome sequencing, and evidence was provided that genetic
alterations affecting the SCCS process may be involved in bladder
tumorigenesis, meaning that a novel therapeutic possibility for
bladder cancer was identified.
The identification of these novel disease-associated
genes in TCC has exhibited a significant effect on the diagnosis
and treatment of BC; however, there may be a large number of novel
genes that have not yet been identified. Thus, the present study
employed whole-exome sequencing methods to detect somatic mutations
in two MI-TCC patients. The aim of the present study was to screen
the MI-TCC patients systematically to identify any previously
unidentified MI-TCC-associated genes. A total of 565 somatic
mutation candidates were detected in the sequenced exomes of the
two MI-TCC patients, and 8 nonsynonymous mutation genes were
validated, including copine VII (CPNE7), serine/arginine repetitive
matrix 5, HECT, C2 and WW domain-containing E3 ubiquitin protein
ligase 1 (HECW1), zinc finger protein (ZNF)792, ZNF273,
trichohyalin (TCHH), RNA binding motif protein, X-linked-like 3
(RBMXL3) and acyl-CoA synthetase medium-chain family member 2A
(ACSM2A). These novel mutation genes may be associated with the
mechanism of bladder tumorigenesis and development. Identification
of these genes may have therapeutic implications and may assist
with the development of future treatments for bladder cancer.
Materials and methods
Sample description and
preparation
Samples were taken from two patients who had been
newly diagnosed with primary MI-TCC of the bladder at the First
Affiliated Hospital of Soochow University (Suzhou, China),
according to the 2004 World Health Organization/International
Society of Urological Pathology grading system (17). Each subject was provided with
appropriate information prior to recruitment for the present study,
according to the regulations of the Institutional Ethics Review
Boards. Cancerous tissue samples and normal controls
(morphologically adjacent healthy bladder tissue) were rapidly
frozen in liquid nitrogen following collection and were stored at
−80°C until subsequent study. The pathological type of bladder
cancer was observed to be high-grade muscle invasive urothelial
carcinoma (T2), and was microscopically validated by two
independent pathologists. In the present study, only TCCs with
malignant cell purities >80% were selected for DNA extraction
and subsequent sequencing.
Genomic DNA extraction and whole-exome
sequencing
Genomic DNA from tumor and matched para-carcinoma
normal tissue samples for the two patients with MI-TCC was isolated
using a DNA extraction kit (Wizard Genomic DNA Purification Kit®;
Promega Corp., Madison, WI, USA), and DNA fragment libraries were
sheared and constructed according to the manufacturer's protocols,
provided using the 5500 SOLiD™ Fragment Library Core kit (Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Subsequently, the exome capture procedure was performed according
to the manufacturer's protocols with the A14060 TargetSeq™ Exome
Enrichment kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.).
Enriched DNA fragment libraries were subsequently
sequenced on the 3730 DNA Analyzer (Applied Biosystems; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
protocols, and 200-bp paired-end reads were generated.
Whole-exome sequencing read mapping
and detection of somatic mutations
High-quality paired-end reads were gap aligned to
the National Center for Biotechnology Information human reference
genome (hg19) using Burrows-Wheeler Aligner (BWA; http://bio-bwa.sourceforge.net/) following
elimination of whole-exome sequencing reads, including low-quality
reads and polymerase chain reaction (PCR) duplicates with >5
unknown bases (18). Local
realignment of the BWA-aligned reads was subsequently corrected
with the Genome Analysis Toolkit (GATK; https://www.broadinstitute.org/gatk/index.php)
(19). The raw lists of potential
somatic substitutions were obtained by VarScan (v2.2; http://varscan.sourceforge.net/) based on the
GATK-alignments (20,21). In order to remove germline variants,
somatic mutations were referenced using the dbSNP database (version
135; http://www.ncbi.nlm.nih.gov/SNP/).
The somatic mutation candidates were subsequently submitted and
annotated using ANNOVAR tool (http://annovar.openbioinformatics.org/en/latest/)
(22). For these software packages, a
number of rules must be complied with: i) All samples must be
covered sufficiently (5–100×) at the genomic position; ii) the
average base reads for a given genomic position in tumor samples
should be ≥2; iii) the variants should be supported by at least 10%
of the total reads in the tumors, and no high-quality
variant-supporting reads are allowed in normal controls (14); and iv) the variant P-values in the
tumors should be ≤0.05. The shared single nucleotide polymorphisms
(SNPs) and insertion/deletions (INDELs) were screened and selected
from the sequence outcomes of the two samples, following filtering
using the aforementioned criteria.
In order to filter out identical mutations, the SNP
data sets were referenced against the Standard Nucleotide Basic
Local Alignment Search Tool (BLAST;
blast.cbi.nlm.nih.gov/Blast.cgi) and SNP BLAST
(blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch
&LINK_LOC=blasthom). The remaining mutations were subjected to
subsequent analyses. Bam files were aligned to the Integrative
Genomics Viewer 2.0 tool (www.broadinstitute.org/igv/) (23,24).
Validation of somatic substitutions by
Sanger sequencing
Sanger sequencing based on PCR amplification was
used to validate the non-silent somatic variants. PCR primers for
putative somatic variants were designed by Primer Premier 5.0
software (PREMIER Biosoft International, Palo Alto, CA, USA). If
variants were successfully confirmed in tumors by reverse
transcription (RT)-PCR, identical primer pairs were used to
validate the putative mutation by Sanger sequencing in 61 MI-TCC
formalin-fixed, paraffin-embedded (FFPE) samples. Total RNA was
isolated from tissue samples using Invitrogen TRIzol reagent
(Thermo Fisher Scientific, Waltham, MA, USA), according to the
manufacturer's protocol. RT-PCR was carried out using the OneStep
RT-PCR kit (catalog no. 210212; Qiagen, Hilden, Germany), 500 ng
total RNA template. The total volume of the reaction was 50 µl and
the following PCR cycling parameters were used: 50°C for 30 min to
reverse transcribe the RNA; 95°C for 15 min; 94°C for 45 sec, 56°C
for 1 min and 72 °C for 1 min, for 28 cycles; and then 72°C for 10
min. Equal volumes of PCR products were resolved on agarose gel,
visualized with ethidium bromide, and photographed. Images were
analyzed with AlphaImager 2200 (Alpha Innotech Co., San Leandro,
CA, USA).
Immunohistochemistry (IHC) of the
putative mutated gene
A total of 10 matched tissue samples, including
cancer and adjacent normal tissues, were obtained from
pathologically validated MI-TCC surgical specimens and were
immediately fixed in 4% 3-heptanone in Gibco phosphate-buffered
saline (PBS; Thermo Fisher Scientific) for 24 h, and then embedded
in paraffin. Subsequently, HECW1 staining was accomplished with
EnVision™ IHC (Dako, Glostrup, Denmark) protocols. The 5-µm thick
paraffin-embedded tissue sections were prepared and subsequently
deparaffinized using xylene (Beyotime Institute of Biotechnology,
Haimen, China) and rehydrated with graded ethanol (Beyotime
Institute of Biotechnology). Antigen retrieval was performed at
95°C in an oven for 45 min. The slides were dewaxed using xylene
and rehydrated with an ethanol series. The slides were then treated
with 3% hydrogen peroxide in methanol for 10 min at room
temperature to inactivate endogenous peroxidase activity, following
by washing with PBS 3 times for 3 min each. The samples were
incubated with rabbit anti-human polyclonal HECW1 antibody
(dilution, 1:200; catalog no. ab-121264; Abcam, Cambridge, UK)
overnight at 4°C, followed by washing with PBS 3 times for 3 min
each. The sections were incubated with goat anti-rabbit IgG
secondary antibody (dilution, 1:200; catalog no. ab-97051; Abcam)
for 30 min at room temperature, washed with PBS 3 times for 3 min
each and stained with color reagent 3,3′-diaminobenzidine, followed
by rinsing in water and counterstaining with hematoxylin (Beyotime
Institute of Biotechnology). The slides were mounted using
permanent mounting medium. Each set of experiments was performed in
triplicate and completed under identical experimental
conditions.
For evaluation of HECW1 expression in MI-TCC and
normal tissues, the positive staining cell counting method was
employed. A total of 10 visual fields were randomly selected for
each sample and examined by a light microscope (IX53; Olympus,
Tokyo, Japan) under high magnification (×400). Bladder cancer cells
that were immunoreactive to anti-HECW1 demonstrated brown staining
in the nucleus and cytoplasm. Samples were scored by counting five
high-power fields per slice and represented the average of three
independent experiments. A score of ≥10% brown stained cells in the
total number of five high-power fields was considered to indicate
the expression of HECW1, whereas <10% brown stained cells
indicated no or negative expression.
Statistical analysis
Data were compared using Fisher's exact test in SPSS
version 13.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
An extensive repertoire of somatic
mutations is identifiable using whole-genome sequencing
Whole-exome sequencing of genomic DNA from tissue
samples of two individuals exhibiting MI-TCC (stage, ≥T2) and their
matched para-carcinoma normal tissue samples was performed. Using
the SOLiD™ 5500 platform, 75-bp short-read sequences were
generated. An average coverage depth of 63.29× was gained for all
the samples sequenced, with 88.64% of the targeted bases being
sufficiently covered.
Following the use of bioinformatic algorithms and
stringent criteria to validate somatically acquired genetic
variation from the raw sequencing data, 121 predicated candidate
somatic mutations were identified, and 34 somatic substitutions and
3 INDELS were confirmed. C:G>T:A transitions were the
predominant mutation spectrum in the two MI-TCC samples. A total of
6 nonsynonymous mutation genes were identified (CPNE7, RBMXL3,
ACSM2A, HECW1, ZNF273 and TCHH) by comparing the standard
nucleotide BLAST and SNP BLAST. The 6 nonsynonymous mutation genes
were subjected to rigorous validation by RT-PCR and Sanger
sequencing in 61 FFPE MI-TCC cases. Mutations obtained from
whole-exome sequencing outcomes were compared with those obtained
from Sanger sequencing outcomes in the 6 nonsynonymous mutation
genes, in order to validate the recurrently mutated genes that may
be involved in TCC tumorigenesis. It was identified that there were
no mutations at the same point or near to 5 of the nonsynonymous
mutation genes (CPNE7, RBMXL3, ACSM2A, ZNF273, TCHH). However, 4
missense and 1 nonsense point mutations were identified in the
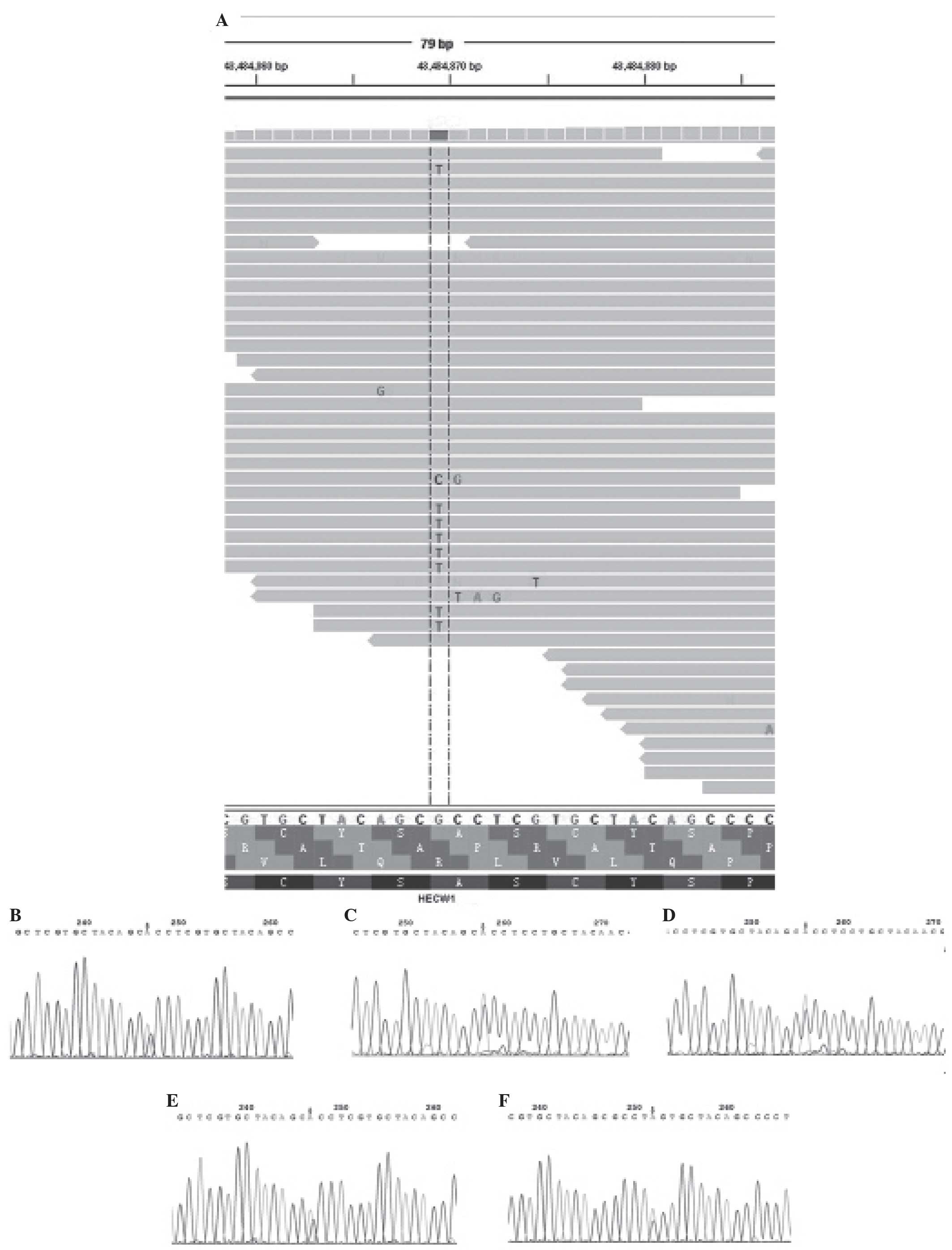
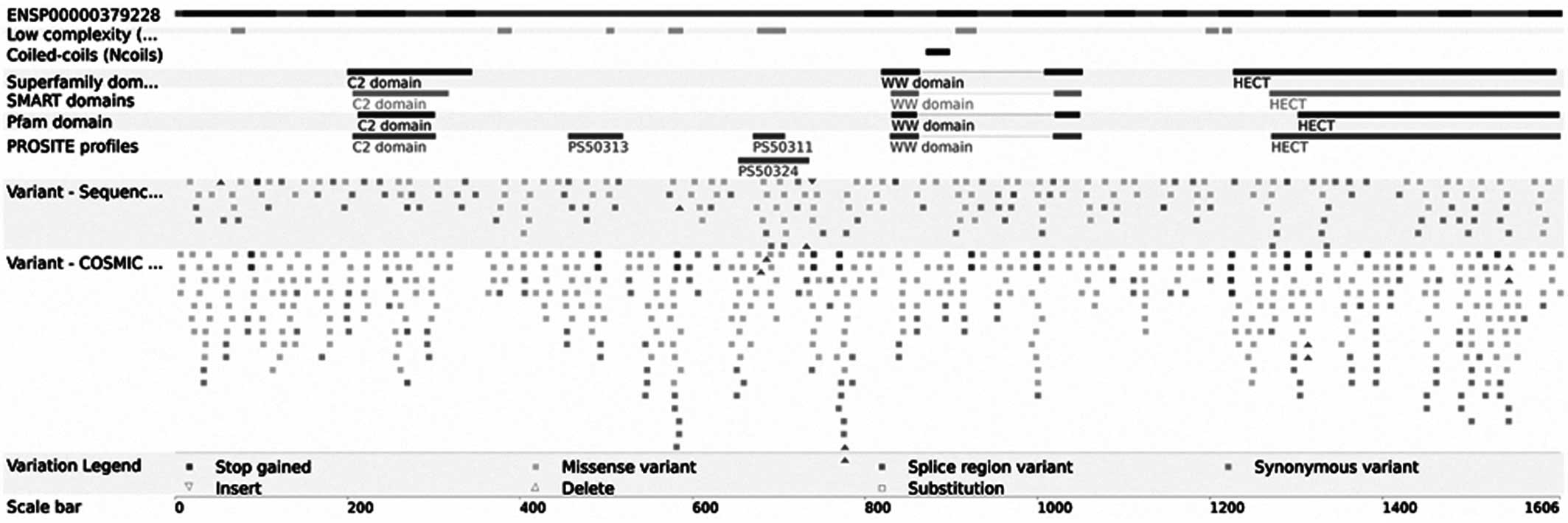
HECW1 gene, which was altered in 6.6% of TCC cases (Fig. 1). In addition, it was identified that
the HECW1 mutant gene was located at exon 11 on chromosome 7, and
the mutation point aggregate was in PS5011 (cysteine-rich) and
PS50324 (serine-rich) of exon 11
(asia.ensembl.org/Homo_sapiens/Transcript/ProteinSummary?db=core;
g=ENSG00000002746; r=7:43112599–43566001; t=ENST00000395891)
(Fig. 2).
HECW1 expression is detectable in
bladder tissue specimens
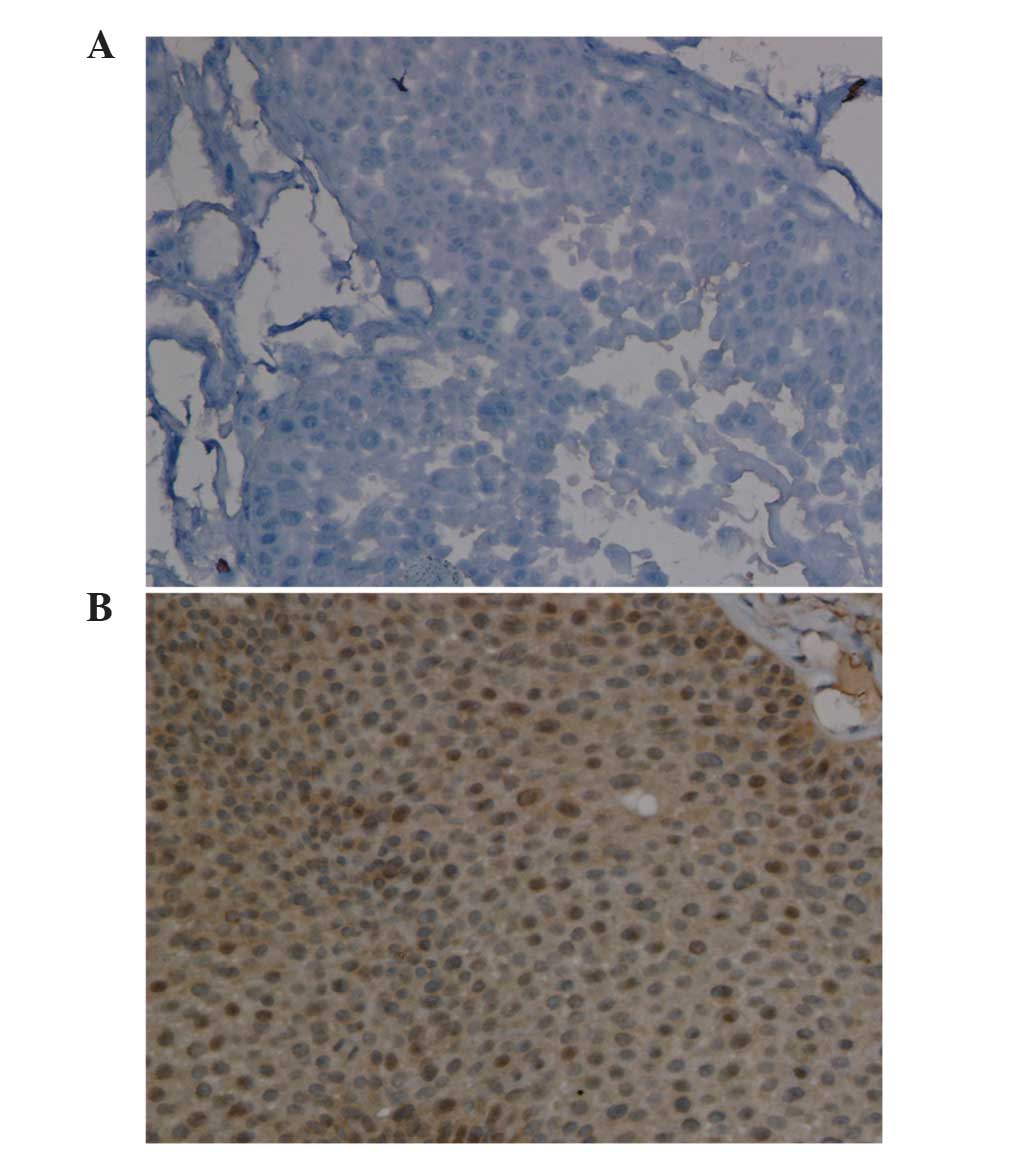
Immunostaining analysis revealed that HECW1 protein
was localized to the cytoplasm and nucleus, and was highly
expressed in MI-TCC bladder tissue specimens, with a total positive
rate of 100% (10/10 cases). However, HECW1 protein exhibited almost
no expression in all normal bladder urothelium samples (Fig. 3). Fisher's exact test revealed that
HECW1 expression was statistically significant in MI-TCC specimens
compared with control tissues (P<0.001).
Discussion
In the present study, the sequencing and analysis of
two MI-TCC cancer genomes was performed using whole-exome
sequencing technology. Unlike whole-genome and whole-transcriptome
genetic analysis, the present study did not detect copy number
alterations, chromosomal translocations or epigenetic changes.
However, a number of novel mutated genes were identified, which may
be relevant for TCC. C:G>T:A transitions were the most common
substitution spectrum in the two MI-TCC samples, as has previously
been reported in TCC cases (14) and
a number of other human cancers (10,25,26). The
six somatic mutations that were discovered in the present study
were single base changes, and none of them had previously been
identified in the TCC genome. The novel nonsynonymous mutation
genes that were detected did not include the well-known bladder
cancer genes (FGFR3, RAS, TP53 and RB1) and contrasted with the
findings of previous studies (14–16). This
discrepancy may be due to differing experimental conditions, and
algorithm and filter use. Subsequently, the mutation results were
validated by Sanger sequencing and a recurrent mutant HECW1 gene
was identified. HECW1 mutations were detected at exon 11 on
chromosome 7 in more than one MI-TCC genome, suggesting that these
mutations did not occur randomly and potentially possessed a
significant role in the pathogenesis of TCC. In the present study,
the remaining five nonsynonymous mutation genes were not validated
by Sanger sequencing. Small quantities of tissue samples may have
contributed to these results. Therefore, additional studies with
increased sample sizes are required to achieve conclusive
results.
HECW1, also known as NEDD4-like ubiquitin protein
ligase 1 (NEDL1), which encodes HECT-type E3 ubiquitin ligase, is
primarily detected in human neuronal tissues (27,28) and
may regulate the bone morphogenetic protein signaling pathway
during embryonic development and bone remodeling (29). HECW1 may combine misfolded superoxide
dismutase 1, translocon-associated protein-δ, and dishevelled-1 to
form an ubiquitinated protein complex that mutually affect their
functions and may be included in potentially cytotoxic protein
aggregates, leading to motor neuron death in familial amyotrophic
lateral sclerosis (ALS) (27).
Notably, Zhang et al (30)
identified that human NEDL1 transgenic mice may exhibit ALS-like
symptoms. The HECW1 gene has been identified to be significantly
upregulated in favorable neuroblastoma compared with unfavorable
neuroblastoma (27). HECW1 cooperates
with p53 to enhance its transcriptional pro-apoptotic activity,
which may be capable of inducing apoptosis in cancerous cells
possessing wild-type p53 independent of E3 ligase activity
(31). Furthermore, small interfering
RNA-mediated knockdown of endogenous HECW1 reduced the
transcription of p53 target genes induced by Adriamycin (ADR) and
allowed the resistance of U2OS cells to ADR (28). In addition, HECW1 has been observed to
negatively regulate ErbB4 activity leading to suppression of its
expression and functioning in breast cancer (32). The results of these previous studies
indicate that HECW1 may act as a tumor suppressor gene in
neuroblastoma, osteosarcoma and breast cancer. This may provide a
novel insight for the investigation of HECW1 chemosensitivity in
bladder cancer.
In the present study, it was identified that HECW1
protein was expressed at significantly increased levels in MI-TCC
compared with normal bladder urothelium. This was in complete
contrast to the expression of HECW1 in neuroblastoma, osteosarcoma
and breast cancer. Therefore, the present study proposed that the
HECW1 gene may have an alternative role in TCC, which may be
opposite to its role in neuroblastoma and breast cancer. However,
the role of the HECW1 gene mutation in bladder cancer, and whether
this mutation affected the expression and functioning of the
protein remain to be elucidated.
In conclusion, the present study successfully used a
next-generation whole-exome sequencing approach to identify novel
candidate genes that may be relevant for bladder cancer
pathogenesis. The novel mutant HECW1 gene has been identified to
have a significant role and may act as a tumor suppressor in
various tumors other than bladder cancer. The results of the
present study demonstrated the power of whole-exome sequencing in
identifying mutational genes in cancer. It is predicted that this
technology may lead to the identification of previously unknown
mutant genes that may have potential as future therapeutic
targets.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81272839, 81300537
and 81472401), the Outstanding Medical Academic Leader Program of
Jiangsu Province (grant no. LJ201138), the Key Discipline of
Medicine of Jiangsu Province (grant nos. XK201151 and BL2014038)
and the Key Laboratory Foundation of Suzhou (grant no.
SZS201001).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu XR: Urothelial tumorigenesis: A tale of
divergent pathways. Nat Rev Cancer. 5:713–725. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Haimovich AD: Methods, challenges, and
promise of next-generation sequencing in cancer biology. Yale J
Biol Med. 84:439–446. 2011.PubMed/NCBI
|
|
4
|
Mardis ER, Ding L, Dooling DJ, Larson DE,
McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath
SD, et al: Recurring mutations found by sequencing an acute myeloid
leukemia genome. N Engl J Med. 361:1058–1066. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ley TJ, Mardis ER, Ding L, Fulton B,
McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S,
Hickenbotham M, et al: DNA sequencing of a cytogenetically normal
acute myeloid leukaemia genome. Nature. 456:66–72. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stephens PJ, McBride DJ, Lin ML, Varela I,
Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie
LJ, et al: Complex landscapes of somatic rearrangement in human
breast cancer genomes. Nature. 462:1005–1010. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ding L, Ellis MJ, Li S, Larson DE, Chen K,
Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, et al:
Genome remodelling in a basal-like breast cancer metastasis and
xenograft. Nature. 464:999–1005. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shah SP, Morin RD, Khattra J, Prentice L,
Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al:
Mutational evolution in a lobular breast tumour profiled at single
nucleotide resolution. Nature. 461:809–813. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pleasance ED, Cheetham RK, Stephens PJ,
McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordóñez GR,
Bignell GR, et al: A comprehensive catalogue of somatic mutations
from a human cancer genome. Nature. 463:191–196. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pleasance ED, Stephens PJ, O'Meara S,
McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman
C, et al: A small-cell lung cancer genome with complex signatures
of tobacco exposure. Nature. 463:184–190. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Campbell PJ, Stephens PJ, Pleasance ED,
O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S,
Hardy C, et al: Identification of somatically acquired
rearrangements in cancer using genome-wide massively parallel
paired-end sequencing. Nat Genet. 40:722–729. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu X, Hou Y, Yin X, Bao L, Tang A, Song L,
Li F, Tsang S, Wu K, Wu H, et al: Single-cell exome sequencing
reveals single-nucleotide mutation characteristics of a kidney
tumor. Cell. 148:886–895. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jones S, Wang TL, Shih IEM, Mao TL,
Nakayama K, Roden R, Glas R, Slamon D, Diaz LA Jr, Vogelstein B, et
al: Frequent mutations of chromatin remodeling gene ARID1A in
ovarian clear cell carcinoma. Science. 330:228–231. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao
S, Wu R, Chen C, Li X, Zhou L, et al: Frequent mutations of
chromatin remodeling genes in transitional cell carcinoma of the
bladder. Nat Genet. 43:875–878. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Y, Xu X, Song L, Hou Y, Li Z, Tsang S,
Li F, Im KM, Wu K, Wu H, et al: Single-cell sequencing analysis
characterizes common and cell-lineage-specific mutations in a
muscle-invasive bladder cancer. Gigascience. 1:122012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo G, Sun X, Chen C, Wu S, Huang P, Li Z,
Dean M, Huang Y, Jia W, Zhou Q, et al: Whole-genome and whole-exome
sequencing of bladder cancer identifies frequent alterations in
genes involved in sister chromatid cohesion and segregation. Nat
Genet. 45:1459–1463. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
El Gendy H, Madkour B, Abdelaty S, Essawy
F, Khattab D, Hammam O, El Kholy A and Nour HH: Galectin 3 for the
diagnosis of bladder cancer. Arab J Urol. 12:178–181. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The genome analysis toolkit: A MapReduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Koboldt DC, Chen K, Wylie T, Larson DE,
McLellan MD, Mardis ER, Weinstock GM, Wilson RK and Ding L:
VarScan: Variant detection in massively parallel sequencing of
individual and pooled samples. Bioinformatics. 25:2283–2285. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Koboldt DC, Zhang Q, Larson DE, Shen D,
McLellan MD, Lin L, Miller CA, Mardis ER, Ding L and Wilson RK:
VarScan 2: Somatic mutation and copy number alteration discovery in
cancer by exome sequencing. Genome Res. 22:568–576. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang K, Li M and Hakonarson H: ANNOVAR:
Functional annotation of genetic variants from high-throughput
sequencing data. Nucleic Acids Res. 38:e1642010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pflueger D, Terry S, Sboner A, Habegger L,
Esgueva R, Lin PC, Svensson MA, Kitabayashi N, Moss BJ, MacDonald
TY, Cao X, et al: Discovery of non-ETS gene fusions in human
prostate cancer using next-generation RNA sequencing. Genome Res.
21:56–67. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Robinson JT, Thorvaldsdóttir H, Winckler
W, Guttman M, Lander ES, Getz G and Mesirov JP: Integrative
genomics viewer. Nat Biotechnol. 29:24–26. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ding L, Ley TJ, Larson DE, Miller CA,
Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan
MD, et al: Clonal evolution in relapsed acute myeloid leukaemia
revealed by whole-genome sequencing. Nature. 481:506–510. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Greenman C, Stephens P, Smith R, Dalgliesh
GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C,
et al: Patterns of somatic mutation in human cancer genomes.
Nature. 446:153–158. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Miyazaki K, Fujita T, Ozaki T, Kato C,
Kurose Y, Sakamoto M, Kato S, Goto T, Itoyama Y, Aoki M and
Nakagawara A: NEDL1, a novel ubiquitin-protein isopeptide ligase
for dishevelled-1, targets mutant superoxide dismutase-1. J Biol
Chem. 279:11327–11335. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Donovan P and Poronnik P: Nedd4 and
Nedd4-2: Ubiquitin ligases at work in the neuron. Int J Biochem
Cell Biol. 45:706–710. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cui Y, He S, Xing C, Lu K, Wang J, Xing G,
Meng A, Jia S, He F and Zhang L: SCFFBXL15 regulates BMP
signalling by directing the degradation of HECT-type ubiquitin
ligase Smurf1. EMBO J. 30:2675–2689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang L, Haraguchi S, Koda T, Hashimoto K
and Nakagawara A: Muscle atrophy and motor neuron degeneration in
human NEDL1 transgenic mice. J Biomed Biotechnol. 2011:8310922011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Y, Ozaki T, Kikuchi H, Yamamoto H,
Ohira M and Nakagawara A: A novel HECT-type E3 ubiquitin protein
ligase NEDL1 enhances the p53-mediated apoptotic cell death in its
catalytic activity-independent manner. Oncogene. 27:3700–3709.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Y, Zhou Z, Alimandi M and Chen C: WW
domain containing E3 ubiquitin protein ligase 1 targets the
full-length ErbB4 for ubiquitin-mediated degradation in breast
cancer. Oncogene. 28:2948–2958. 2009. View Article : Google Scholar : PubMed/NCBI
|