Introduction
Colorectal cancer (CRC) is one of the most common
type of cancer in the world, and its incidence is continuously
increasing in China (1). Global
cancer statistics indicate that CRC remains to be the fourth
leading cause of cancer-associated mortalities worldwide,
responsible for >600,000 mortalities annually (2). The major therapeutic approaches for CRC
are surgery, neoadjuvant radiotherapy and adjuvant chemotherapy.
Carcinoembryonic antigen and cancer antigen 19–9 are widely used
biomarkers for CRC diagnosis. Endoscopy and blood screening have
been proved to effectively reduce the incidence and mortality of
CRC (3). However, the prognosis of
CRC patients remains poor, with a 50–59% 5-year survival rate
(2).
MicroRNAs (miRNAs/miRs) are small non-coding RNA
molecules that are 20–24 nucleotides in length, and regulate 60% of
coding genes by preventing mRNA molecule translation and/or
promoting degradation. Increasing evidence has revealed that miRNAs
are critical in various diseases, including cancer. Novel functions
and mechanisms by which miRNAs regulate genes are constantly being
investigated (4–6).
miRNAs that promote carcinogenesis are termed
oncomiRs (7). Numerous oncomiRs are
overexpressed in CRC compared with noncancerous tissues, including
miR-494, miR-21, miR-23a and miR-130b (8–11). These
oncomiRs effect CRC cell proliferation, apoptosis, migration and
invasion (12). However, another type
of miRNA, termed tumor suppressive miRs (tsmiRs), are significantly
downregulated in human colorectal adenocarcinoma tissue samples
compared with the adjacent normal colorectal tissues, including
miR-339-5p, miR-27a and miR-139-5p (13–15).
miR-517a is considered to be a novel oncomiR and was observed to be
elevated in human hepatocellular carcinoma (HCC) (16); expression of miR-517a increased
proliferation, migration, and invasion of HCC cells in vitro
(16). Furthermore, hsa-miR-517a
exhibited significant inverse correlation with cyclin-dependent
kinase 2a (CDKN2A) in glioblastoma; low expression of CDKN2A was
associated with worse prognosis (17). A recent study reported that
manipulation of miR-517a-3p expression changed lung cancer cell
proliferation, migration and invasion capacity (18). Furthermore, miR-517a-3p accelerated
lung cancer cell proliferation migration and invasion through
inhibiting forkhead box J3 (FOXJ3) expression (18). However, the clinical significance of
miR-517a in CRC, as well as its associated molecular pathways
involved in the development and progression of CRC, have yet to be
elucidated.
The present study aimed to explore the clinical
significance of miR-517a, and its role in CRC cell migration and
invasion. The present study demonstrated that overexpression of
miR-517a is observed in CRC tissues compared with noncancerous
tissues, and its overexpression correlates with poor prognostic
features. Furthermore, miR-517a appears to be an independent
prognostic marker for predicting the survival of patients with CRC.
In vitro experiments indicated that miR-517a promotes CRC
cell migration and invasion. Mechanistically, the current results
demonstrate that miR-517a may potentiate the invasive behavior of
CRC cells by inhibiting FOXJ3.
Materials and methods
Clinical samples and cell lines
A total of 90 pairs of CRC and matched adjacent
non-tumor tissue samples were obtained during colorectomies from
the Department of Pathology, the Second Affiliated Hospital of
Xi'an Jiaotong University (Xi'an, China) between January 2007 and
January 2009. The clinical specimens were frozen and stored at
−80°C for reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) and western blot analysis. Samples were
paraformaldehyde-fixed (Nanjing SenBeiJia Biological Technoogy Co.,
Ltd., Nanjing, China) and paraffin-embedded (Meryer Chemical
Technology Co., Ltd., Shanghai, China). for immunohistochemical
staining. The demographic features and clinicopathological
parameters are indicated in Table I.
All specimens had confirmed pathological diagnosis of CRC and were
classified according to the International Union Against Cancer and
American Joint Committee on Cancer criteria (7th edition) (19). Patients did not receive preoperative
chemotherapy or embolization. All samples were used after obtaining
informed consent. The Xi'an Jiaotong University Ethics Committee
approved all protocols according to the Declaration of Helsinki (as
revised in Tokyo 2004) (20).
 | Table I.Clinical association analysis of
miR-517a expression in patients with colorectal cancer. |
Table I.
Clinical association analysis of
miR-517a expression in patients with colorectal cancer.
|
|
| Patients, n |
|
|---|
|
|
|
|
|
|---|
| Feature | Total patients, n
(n=90) | Low miR-517a | High miR-517a | P-value |
|---|
| Age, years |
|
|
| 0.490 |
| ≤60 | 27 | 15 | 12 |
|
|
>60 | 63 | 30 | 33 |
|
| Gender |
|
|
| 0.389 |
| Male | 54 | 29 | 25 |
|
|
Female | 36 | 16 | 20 |
|
| Tumor grade |
|
|
| 0.468 |
|
G1/G2 | 67 | 35 | 32 |
|
|
G3/G4 | 23 | 10 | 13 |
|
| Size, cm |
|
|
| 0.660 |
|
<5 | 32 | 17 | 15 |
|
| ≥5 | 58 | 28 | 30 |
|
| Tumor invasion |
|
|
| 0.011a |
|
T1/T2 | 20 | 15 | 5 |
|
|
T3/T4 | 70 | 30 | 40 |
|
| Lymph node
metastases |
|
|
| 0.038a |
|
Absent | 63 | 36 | 27 |
|
|
Present | 27 | 9 | 18 |
|
| Distant
metastasis |
|
|
| 0.020a |
|
Absent | 71 | 40 | 31 |
|
|
Present | 19 | 5 | 14 |
|
| TNM stage |
|
|
| 0.001a |
|
I/II | 54 | 35 | 19 |
|
|
III/IV | 36 | 10 | 26 |
|
In the present study, two CRC cell lines (HCT-116
and SW480) were used. In a pre-experiment, a noncancerous colon
epithelial cell line (HCEC) was used as a control to evaluate the
expression of miR-517a in HCT-116 and SW480 cells (data not shown).
These cell lines were purchased from the Institute of Biochemistry
and Cell Biology, Chinese Academy of Sciences, where they were
established. Cells were cultured in complete Dulbecco's modified
Eagle medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) containing 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.) with 100 U/ml penicillin and 100 µg/ml
streptomycin (Sigma-Aldrich, St. Louis, MO, USA) at 37°C in a
humidified incubator containing 5% CO2.
RT-qPCR
The mir-Vana miRNA Isolation kit (Ambion; Thermo
Fisher Scientific, Inc.) was used to isolate total RNA from frozen
patient samples and cell lines, according to the manufacturer's
protocol (21). The RNA was treated
with DNase (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was
synthesized using the Universal cDNA Synthesis kit II (Exiqon,
Vedbaek, Denmark). The RT-qPCR analyses were performed in the ABI
PRISM 7300 Sequence Detection System (Applied Biosystems; Thermo
Fisher Scientific, Inc.) using ExiLENT SYBR Green Master Mix
(Exiqon) for miR-517a. The reactions were incubated at 95°C for 60
sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 34 sec.
RNU6B (U6) was measured as an internal control for miRNA. All
samples were normalized to internal controls and fold changes were
calculated based on relative quantification (2−ΔΔCq).
The primers for miR-517a and U6 were purchased from Exiqon. The
following primer sequences were used: miR-517a stem-loop RT primer,
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACACTCTA; miR-517a forward,
CGGCGGATCGTGCATCCCTTTA; miR-517a reverse, GTGCAGGGTCCGAGGT; U6
forward, CTCGCTTCGGCAGCACA; U6 reverse, AACGCTTCACGAATTTGCGT.
Target prediction
To determine the molecular mechanisms by which
miR-517a inhibits CRC cell migration and invasion, predicted target
genes of miR-517a were retrieved and analyzed using the publicly
available databases, TargetScan 6.2 (www.targetscan.org) and miRanda (www.microrna.org).
Immunohistochemical staining
The paraffin-embedded tissue samples from
postoperative patients were cut in 5-cm sections. Then the samples
were deparaffinized in xylene (ZSGB-BIO, Beijing, China) and
rehydrated using a series of graded alcohol. Slides were blocked
with 10% goat serum (ZSGB-BIO) prior to incubation with rabbit
anti-human polyclonal FOXJ3 (dilution, 1:100; cat no. SAB4500907;
Sigma-Aldrich) antibody. The samples were incubated overnight with
the primary antibody at 4°C, and then incubated with goat
anti-rabbit secondary antibody (dilution, 1:1,000; cat no.
ZDR-5403; ZSGB-BIO) at room temperature for 2h followed by
3,3′-diaminobenzidine-labeled secondary antibody. Subsequently,
sections were counterstained with hematoxylin (ZSGB-BIO). Protein
staining was evaluated under a light microscope (BX-51; Olympus,
Tokyo, Japan) at x400 magnification.
Cell transfection
miRNA vectors, including miR-517a expression vector
(HmiR0330-MR04), scrambled control vector for miR-517a
(CmiR0001-MR04), miR-517a inhibitor (HmiR-AN0575-AM04) and negative
control for the miR-517a inhibitor (CmiR-AN0001-AM04), were
purchased from Genecopoeia (Guangzhou, China). Cells were
transfected with the aforementioned vectors using Lipofectamine
2000, according to the manufacturer's instructions (Invitrogen;
Thermo Fisher Scientific, Inc.).
Transwell assays
Transwell migration assays were performed in 12-well
plates with 8-µm BioCoat control inserts (BD Biosciences, Bedford,
MA, USA). Cells (1–2×105) that were suspended in 500 µl
serum-free DMEM were seeded in the upper well. DMEM medium with 10%
FBS was added to the lower well. After 12 h incubation, the
membranes were removed, cells on the side facing the upper well
were wiped with a cotton swab, and the membranes were stained with
crystal violet (MCE China, Shanghai, China). At least 6
representative images of each well were captured (BX-51; Olympus,
Tokyo, Japan) and cell numbers were counted using ImageJ software
(version 1.46r; National Institutes of Health, Bethesda, MD, USA).
A BioCoat Matrigel invasion chamber (BD Biosciences) was used for
Transwell invasion assays; otherwise, the same protocols were used
for Transwell migration assays. The experiments were performed in
triplicate.
Western blot analysis
Total proteins were extracted from cells with
radioimmunoprecipitation lysis buffer (Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA) and centrifuged at 14,000 × g for 10
min, separated by 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (Sigma-Aldrich) at 165 V for 1.5 h and then
transferred onto a PVDF membrane (Roche, Indianapolis, IN, USA).
The membrane was blocked with 5% skimmed milk (Shanghai Haoran Bio
Technologies Co., Ltd., Shanghai, China and incubated with the
appropriate antibody overnight at 4°C. The following primary
antibodies were used: Rabbit anti-human polyclonal FOXJ3 (dilution,
1:1,000; cat no. SAB4500907; Sigma-Aldrich) and mouse anti-human
monoclonal GAPDH (dilution, 1:5,000; cat no. G8140; United States
Biological, Swampscott, MA, USA). Horseradish peroxidase-conjugated
donkey anti-rabbit and goat anti-mouse secondary antibodies (cat
nos. sc-2313 and sc-2005, respectively; Santa Cruz Biotechnology,
Inc.) were used at room temperature for 2 h with a dilution of
1:1,000–1:5,000 and expression as detected using enhanced
chemiluminescence reagents (Thermo Fisher Scientific, Inc.).
Statistical analysis
Results are expressed as the mean ± standard error
of the mean. The quantitative data were compared between two groups
using the two-tailed Student's t-test. Categorical data were
analyzed using the Pearson's χ2 test. The Kaplan-Meier
method and log-rank test were used to compare the cumulative
recurrence and survival rates. The independent factors influencing
the survival and recurrence of CRC patients were determined using
the Cox proportional hazards model. Correlation analysis was tested
by the Spearman's rank correlation coefficient. P<0.05 was
considered to indicate statistically significant differences.
Results
Clinical significance of miR-517a in
CRC specimens
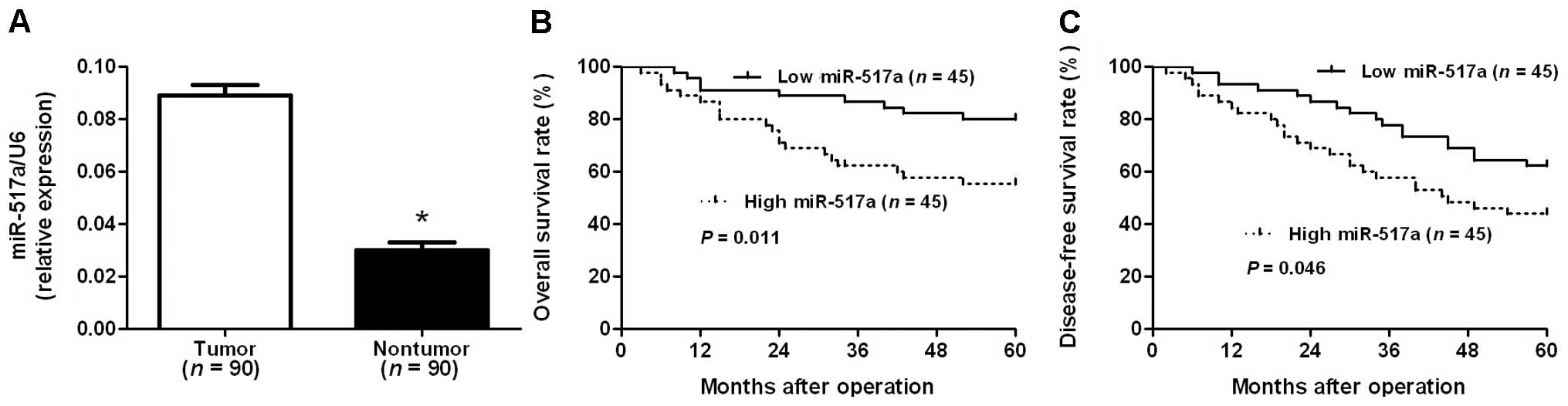
The expression of miR-517a was determined by RT-qPCR
and normalized against an endogenous control (U6 RNA) in 90 pairs
of CRC and adjacent non-tumor tissues. The data indicated that the
expression of miR-517a in CRC tissues was significantly higher than
in adjacent non-tumor tissues (P<0.001; Fig. 1A). The expression of miR-517a was
considered to be low (n=45) or high (n=45) according to the cutoff
value, which was defined as the median of the cohort. As indicated
in Table I, clinical association
analysis revealed that high expression of miR-517a was prominently
associated with poor prognostic features, including high tumor
invasion, lymph node metastases, distant metastasis and advanced
TNM stage (P<0.05). Furthermore, CRC patients with high
expression of miR-517a were associated with poor overall and
disease-free survival (P=0.011 and 0.046, respectively; Fig. 1B and C). Notably, miR-517a expression,
in addition to TNM stage, is an independent factor for predicting
the survival of patients with CRC (P<0.05; Table II). These data indicate that miR-517a
acts as a potent biomarker for predicting the prognosis of patients
with CRC.
 | Table II.Multivariate Cox regression analysis
of 5-year overall and disease-free survival of 90 patients with
colorectal cancer. |
Table II.
Multivariate Cox regression analysis
of 5-year overall and disease-free survival of 90 patients with
colorectal cancer.
|
| Overall
survival | Disease-free
survival |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Tumor invasion
(T1/T2 vs. T3/T4) | 1.511 | 0.998–2.410 | 0.051 | 1.523 | 0.969–2.395 | 0.068 |
| Lymph node
metastases (absent vs. present) | 1.437 | 0.939–2.200 | 0.095 | 1.395 | 0.894–2.178 | 0.143 |
| Distant metastasis
(absent vs. present) | 1.928 | 1.161–3.204 | 0.011a | 1.385 | 0.911–2.106 | 0.127 |
| TNM stage (I/II vs.
III/IV) | 1.993 | 1.290–3.080 | 0.002a | 1.623 | 1.034–2.548 | 0.035a |
| miR-517a expression
(low vs. high) | 2.394 | 1.466–3.909 |
<0.001a | 1.754 | 1.033–2.978 | 0.038a |
miR-517a promotes cell migration and
invasion in CRC
A noncancerous colon epithelial cell line (HCEC) was
used as a control to evaluate the expression of miR-517a in HCT-116
and SW480 cells in a pre-experiment. The levels of miR-517a
expression in the HCT-116 and SW480 cells were approximately 4.6–
and-1.5 fold higher compared with the HCEC cells (data not shown).
To investigate the potential role of miR-517a in CRC, the
expression of miR-517a was altered by transfecting CRC cells with
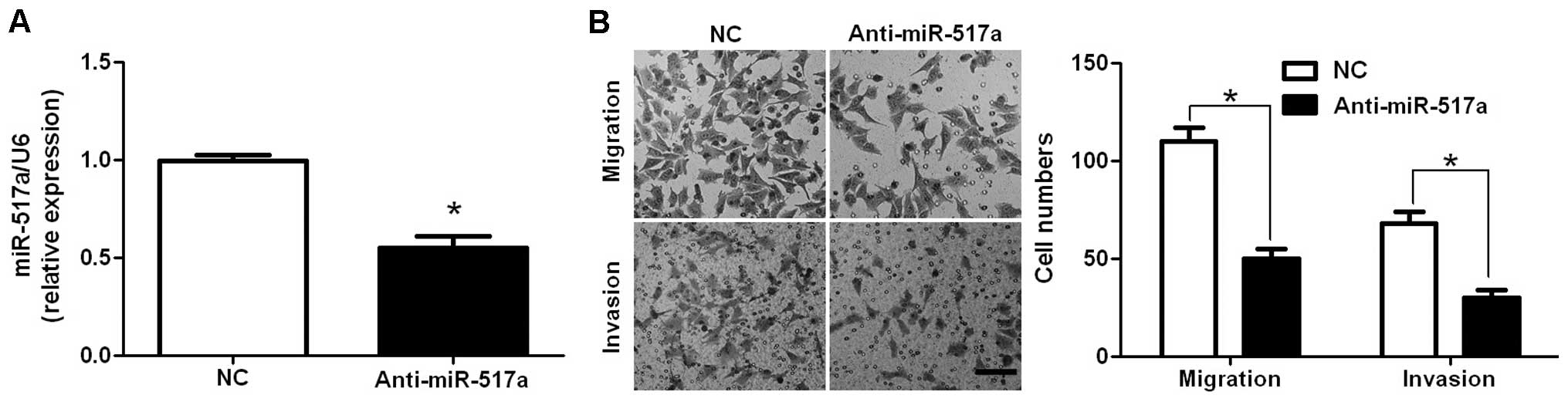
miR-517a mimics and inhibitors. As assessed by RT-qPCR, the
expression of miR-517a was significantly downregulated by miR-517a
inhibitors in HCT-116 cells (P=0.011; Fig. 2A). Transwell migration assays were
performed to analyze the effect of altering miR-517a levels on cell
migration. It was determined that downregulation of miR-517a led to
a significant reduction of cell migration in HCT-116 cells
(P=0.018; Fig. 2B). Furthermore, as
determined by Transwell invasion assays, the number of invaded
HCT-116 cells was significantly reduced following downregulation of
miR-517a (P=0.032; Fig. 2B). Next,
SW480 cells, which showed a significant 5.43–fold reduction in
miR-517a expression compared with HCT-116 cells under normal
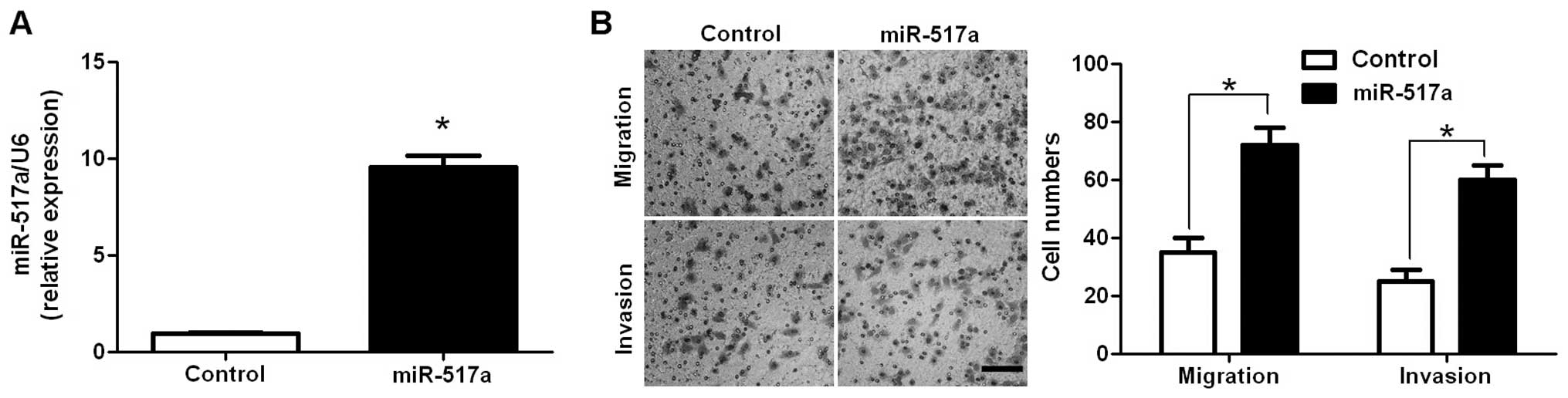
conditions, were used for gain-of-function experiments.
miR-517a-overexpressing SW480 cells were established and confirmed
by RT-qPCR (P=0.005; Fig. 3A). As
expected, upregulation of miR-517a significantly increased the
number of migrated and invaded SW480 cells (P=0.008 and 0.019,
respectively; Fig. 3B). Thus,
miR-517a appears to promote cell migration and invasion in CRC.
miR-517a inversely regulates FOXJ3
abundance in CRC
A previous study reported that miR-517a-3p
accelerates cell proliferation, migration and invasion through
inhibiting FOXJ3 expression in human lung cancer (18). In addition, TargetScan and miRanda
databases were used to identify FOXJ3 as one of the targets of
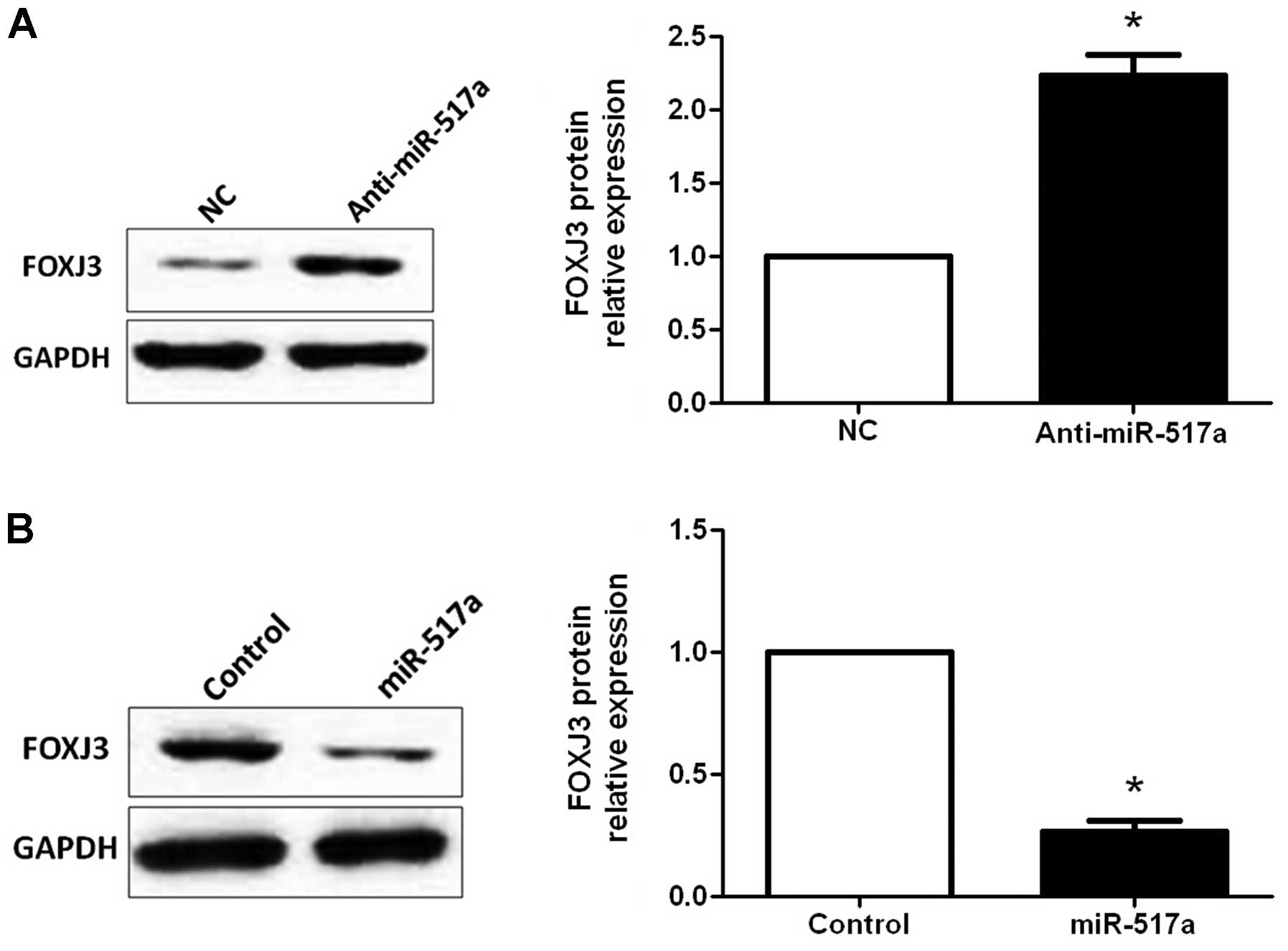
miR-517a. Therefore, HCT-116 cells transfected with negative
control or miR-517a inhibitors were subjected to western blotting
for FOXJ3. As assessed by immunoblotting, downregulation of
miR-517a significantly increased the expression of FOXJ3 protein in
HCT-116 cells (P=0.014; Fig. 4A). By
contrast, upregulation of miR-517a significantly decreased the
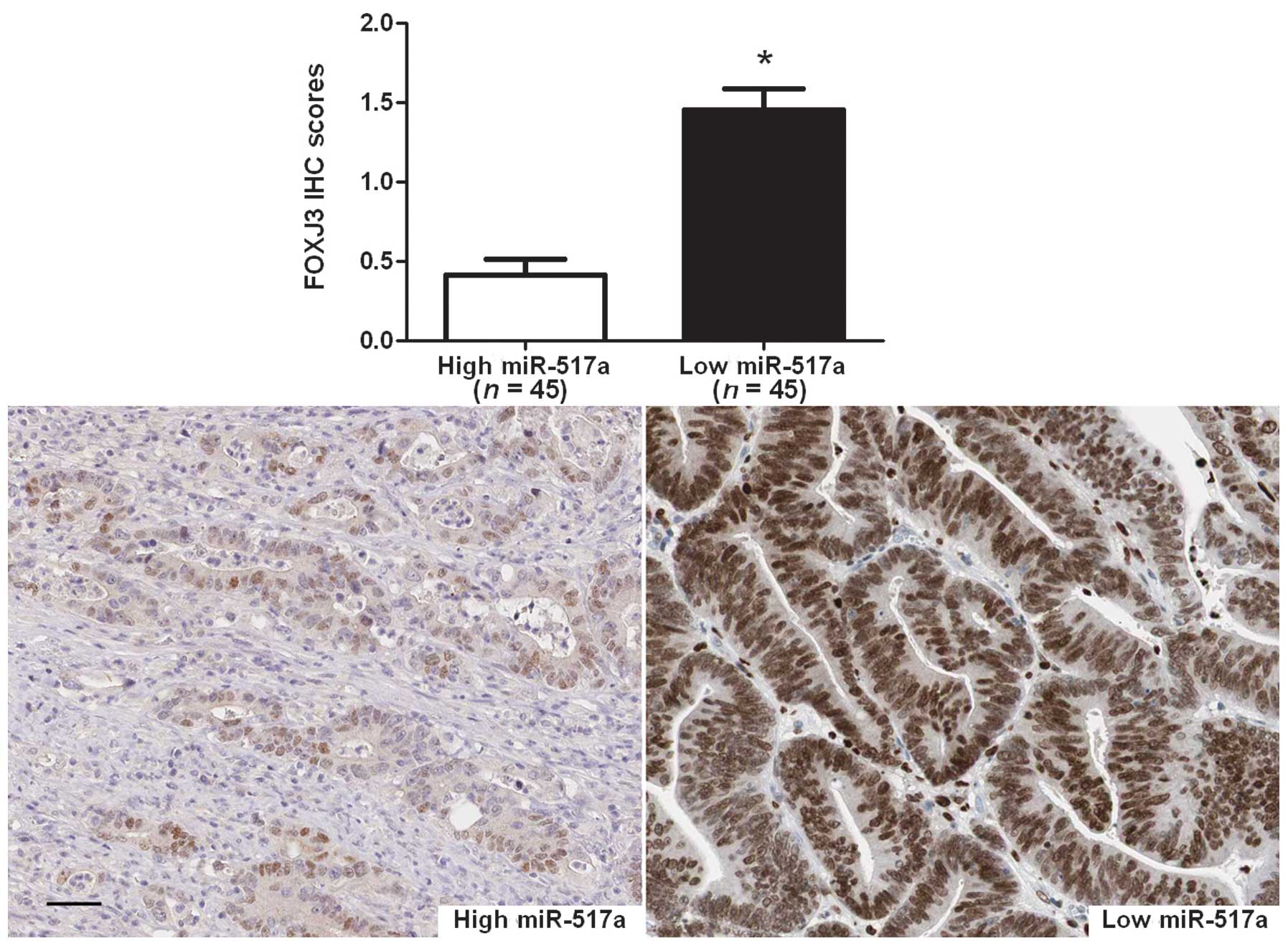
expression of FOXJ3 protein in SW480 cells (P=0.004; Fig. 4B). Next, the correlation between FOXJ3
and miR-517a expression were evaluated in CRC tissues. The
expression of FOXJ3 in miR-517a high-expressing tumors was
significantly lower than those in miR-517a low-expressing tumors
(P<0.001; Fig. 5). Furthermore,
Spearman's rank correlation analysis indicated that miR-517a was
inversely correlated with FOXJ3 expression in CRC tissues
(r=−0.458; P=0.012). Taken together, these data indicate that
miR-517a suppresses FOXJ3 expression in CRC.
Discussion
Increasing evidence has indicated that the
deregulation and dysfunction of miRNAs is key in the development
and progression of CRC (22,23). Increased expression of oncomiRs and
decreased expression of tsmiRs have been observed in CRC tissues as
compared with adjacent non-tumor tissues (24). However, miRNAs that are correlated
with the recurrence and metastasis of CRC remain poorly understood.
Considering the findings of previous studies, the present study
aimed to explore the clinical significance of miR-517a and its role
in CRC cell migration and invasion. The current data demonstrated
that the expression of miR-517a in CRC tissues was significantly
higher than in adjacent noncancerous tissues. Clinical analysis
determined that high expression of miR-517a was significantly
correlated with high tumor invasion, lymph node metastases, distant
metastasis and advanced TNM stage in CRC. Furthermore, Kaplan-Meier
and multivariate Cox regression analysis demonstrated that miR-517a
was an independent prognostic marker for the predicting survival of
patients with CRC. Together, these results suggest that miR-517a is
critical for the prognostic determination in CRC.
miR-517a has been proposed to be involved in tumor
metastasis of HCC and lung cancer (16,18).
Therefore, the role of miR-517a in CRC was further investigated.
The present study identified that downregulation of miR-517a
inhibited cell migration and invasion in HCT-116 cells.
Furthermore, upregulation of miR-517a increased the number of
migrated and invaded SW480 cells. These data suggest that miR-517a
promotes cell migration and invasion in CRC. To determine the
molecular mechanisms by which miR-517a inhibits CRC cell migration
and invasion, predicted target genes of miR-517a were retrieved and
analyzed using publicly available databases (TargetScan and
miRanda). FOXJ3, which is known to be a key transcription factor of
mitochondrial biogenesis and was identified to upregulate myocyte
enhancer factor 2C (25,26), was predicted as one of the targets of
miR-517a. A recent study reported that miR-517a-3p accelerates cell
proliferation, migration and invasion through inhibiting FOXJ3
expression in human lung cancer (18). Thus, the regulatory effect of miR-517a
on FOXJ3 in CRC cells was investigated. The data indicated that
downregulation of miR-517a increased the expression level of FOXJ3
protein in HCT-116 cells. By contrast, upregulation of miR-517a
reduced FOXJ3 expression in SW480 cells. Furthermore, a significant
inverse correlation between miR-517a and FOXJ3 expression was
observed in CRC tissues. Accordingly, we propose that miR-517a may
promote cell migration and invasion by targeting FOXJ3 in CRC.
In conclusion, the present study determined that
miR-517a is overexpressed in CRC and its high expression is
associated with poor prognostic features. Furthermore, high
expression of miR-517a appears to be a prognostic marker for
predicting poor survival of CRC patients. In vitro
experiments demonstrated that miR-517a facilitates CRC cell
migration and invasion. Mechanistically, we propose that miR-517a
may promote CRC cell mobility by suppressing FOXJ3. Taken together,
we propose that miR-517a may potentially act as a clinical
biomarker and may also be a therapeutic target in CRC.
Acknowledgements
The present study was supported by a grant from the
National Natural Science Foundation of China (no. 81171356).
References
|
1
|
Li L and Ma BB: Colorectal cancer in
Chinese patients: Current and emerging treatment options. Onco
Targets Ther. 7:1817–1828. 2014.PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van Hees F, Saini SD, Lansdorp-Vogelaar I,
Vijan S, Meester RG, de Koning HJ, Zauber AG and van Ballegooijen
M: Personalizing colonoscopy screening for elderly individuals
based on screening history, cancer risk, and comorbidity status
could increase cost effectiveness. Gastroenterology. 149:1425–1437.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J
and Sung JJ: MicroRNA dysregulation in gastric cancer: A new player
enters the game. Oncogene. 29:5761–5771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tu K, Zheng X, Dou C, Li C, Yang W, Yao Y
and Liu Q: MicroRNA-130b promotes cell aggressiveness by inhibiting
peroxisome proliferator-activated receptor gamma in human
hepatocellular carcinoma. Int J Mol Sci. 15:20486–20499. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tu K, Li C, Zheng X, Yang W, Yao Y and Liu
Q: Prognostic significance of miR-218 in human hepatocellular
carcinoma and its role in cell growth. Oncol Rep. 32:1571–1577.
2014.PubMed/NCBI
|
|
8
|
Sun HB, Chen X, Ji H, Wu T, Lu HW, Zhang
Y, Li H and Li YM: miR494 is an independent prognostic factor and
promotes cell migration and invasion in colorectal cancer by
directly targeting PTEN. Int J Oncol. 45:2486–2494. 2014.PubMed/NCBI
|
|
9
|
Liu K, Li G, Fan C, Zhou X, Wu B and Li J:
Increased expression of microRNA-21 and its association with
chemotherapeutic response in human colorectal cancer. J Int Med
Res. 39:2288–2295. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jahid S, Sun J, Edwards RA, Dizon D,
Panarelli NC, Milsom JW, Sikandar SS, Gümüs ZH and Lipkin SM:
miR-23a promotes the transition from indolent to invasive
colorectal cancer. Cancer Discov. 2:540–553. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Colangelo T, Fucci A, Votino C, Sabatino
L, Pancione M, Laudanna C, Binaschi M, Bigioni M, Maggi CA, Parente
D, et al: MicroRNA-130b promotes tumor development and is
associated with poor prognosis in colorectal cancer. Neoplasia.
15:1218–1231. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ahmed FE: miRNA as markers for the
diagnostic screening of colon cancer. Expert Rev Anticancer Ther.
14:463–485. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang C, Liu J, Wang X, Wu R, Lin M,
Laddha SV, Yang Q, Chan CS and Feng Z: MicroRNA-339-5p inhibits
colorectal tumorigenesis through regulation of the MDM2/p53
signaling. Oncotarget. 5:9106–9117. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bao Y, Chen Z, Guo Y, Feng Y, Li Z, Han W,
Wang J, Zhao W, Jiao Y, Li K, et al: Tumor suppressor microRNA-27a
in colorectal carcinogenesis and progression by targeting SGPP1 and
Smad2. PLoS One. 9:e1059912014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang L, Dong Y, Zhu N, Tsoi H, Zhao Z, Wu
CW, Wang K, Zheng S, Ng SS, Chan FK, et al: microRNA-139-5p exerts
tumor suppressor function by targeting NOTCH1 in colorectal cancer.
Mol Cancer. 13:1242014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Toffanin S, Hoshida Y, Lachenmayer A,
Villanueva A, Cabellos L, Minguez B, Savic R, Ward SC, Thung S,
Chiang DY, et al: MicroRNA-based classification of hepatocellular
carcinoma and oncogenic role of miR-517a. Gastroenterology.
140:1618–1628, e16. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Feng J, Kim ST, Liu W, Zhang Z, Zhu Y,
Berens M, Sun J and Xu J: An integrated analysis of germline and
somatic, genetic and epigenetic alterations at 9p21.3 in
glioblastoma. Cancer. 118:232–240. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jin J, Zhou S, Li C, Xu R, Zu L, You J and
Zhang B: MiR-517a-3p accelerates lung cancer cell proliferation and
invasion through inhibiting FOXJ3 expression. Life Sci. 108:48–53.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tong LL, Gao P, Wang ZN, Song YX, Xu YY,
Sun Z, Xing CZ and Xu HM: Is the seventh edition of the UICC/AJCC
TNM staging system reasonable for patients with tumor deposits in
colorectal cancer? Ann Surg. 255:208–213. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
World Medical Association: World Medical
Association Declaration of Helsinki: Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chang RM, Yang H, Fang F, Xu JF and Yang
LY: MicroRNA-331-3p promotes proliferation and metastasis of
hepatocellular carcinoma by targeting PH domain and leucine-rich
repeat protein phosphatase. Hepatology. 60:1251–1263. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Muhammad S, Kaur K, Huang R, Zhang Q, Kaur
P, Yazdani HO, Bilal MU, Zheng J, Zheng L and Wang XS: MicroRNAs in
colorectal cancer: Role in metastasis and clinical perspectives.
World J Gastroenterol. 20:17011–17019. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tokarz P and Blasiak J: The role of
microRNA in metastatic colorectal cancer and its significance in
cancer prognosis and treatment. Acta Biochim Pol. 59:467–474.
2012.PubMed/NCBI
|
|
24
|
Bonfrate L, Altomare DF, Di Lena M,
Travaglio E, Rotelli MT, De Luca A and Portincasa P: MicroRNA in
colorectal cancer: New perspectives for diagnosis, prognosis and
treatment. J Gastrointestin Liver Dis. 22:311–320. 2013.PubMed/NCBI
|
|
25
|
Landgren H and Carlsson P: FoxJ3, a novel
mammalian forkhead gene expressed in neuroectoderm, neural crest
and myotome. Dev Dyn. 231:396–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Alexander MS, Shi X, Voelker KA, Grange
RW, Garcia JA, Hammer RE and Garry DJ: Foxj3 transcriptionally
activates Mef2c and regulates adult skeletal muscle fiber type
identity. Dev Biol. 337:396–404. 2010. View Article : Google Scholar : PubMed/NCBI
|