Introduction
Chronic lymphocytic leukemia (CLL) is the most
common type of leukemia observed in people aged >50 years in
Western countries. CLL is characterized by a heterogeneous clinical
course, with a time to progression ranging from months to decades
(1). The presence of cytogenetic
abnormalities is a hallmark of CLL. The most common recurrent
aberrations in CLL affect chromosomes 11q, 13q, 14q, 17p and the
whole of chromosome 12. Certain abnormalities, including deletions
in 11q22.3, the ataxia telangiectasia mutated (ATM) gene
(10–20% of all CLL cases), and 17p13.1, the tumor protein 53
(TP53) gene (5–10% of all CLL cases), are associated with a
poor clinical outcome. Therefore, the detection of these
aberrations is important for identifying high-risk patients, who
suffer from rapid disease progression and a decreased overall
survival time (2). Other frequent
chromosomal aberrations in CLL are associated with a good
(deletions in 13q14 or 14q32.33) or intermediate (trisomy 12)
prognosis (1,3–5).
CLL is considered to be an insidious disease.
Certain CLL patients, particularly patients with a good prognosis,
survive for several years without requiring treatment; however,
another subgroup of patients experience an aggressive disease
course and have a short life expectancy, despite aggressive
treatment (6). The latter group tends
to exhibit a particular lack of response to fludarabine-based
regimens, which are generally considered to be the first line of
treatment for CLL (6). In a large
fraction of these patients, the molecular basis of the aggressive
clinical course remains unclear; however, in ~40% of patients, the
molecular basis is hypothesized to be due to TP53 disruption
(7). In addition, activation of the
nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB)
pathway is considered to be a mechanism of resistance to disease
eradication (7).
From a clinical perspective, CLL cases may be
divided into three major clinical phases: i) newly diagnosed CLL;
ii) progressive CLL; and iii) relapsed or fludarabine-refractory
CLL. TP53 abnormalities are observed in 40–50% of relapsed
and fludarabine-refractory CLL cases and the deletion of 11q22–23
occurs in 25–30% of relapsed or fludarabine-refractory CLL patients
(8). In a large previous study, 637
patients were classified into four risk groups according to a
multivariate analysis of overall survival, which was based on
genomic abnormalities and the mutational status of TP53,
baculoviral IAP repeat-containing 3 (BIRC3),
translocation-associated notch homolog 1 and splicing factor 3B
subunit 1. Notably, the high-risk group was composed of patients
that exhibited disruption to TP53 and/or BIRC3
(9).
In CLL, deletions within the long arm of chromosome
11 may be highly variable in size. The deletion may be
distinguished as the more common ‘classical or large deletion’ or
an ‘atypical or small deletion’, which is uncommon and more
frequently associated with ATM mutations. This variation
indicates that other genes may contribute to the pathobiology of
11q deletions in CLL, and one of the genes that is hypothesized to
be involved is BIRC3 (10).
BIRC3 disruption, mutations or deletions are rarely detected
in CLL at diagnosis (4% of patients), but are detected in 24% of
fludarabine-refractory CLL patients. In a previous study,
fludarabine-sensitive patients did not exhibit BIRC3
mutations initially, which suggests that BIRC3 disruption
may be specifically associated with a chemo-refractory CLL subtype
(7). Therefore, BIRC3
disruption may be added to the panel of cytogenetic abnormalities,
as it may be helpful in the early identification of relapsed and
fludarabine-refractory CLL patients. Affected patients should be
considered for other treatment regimens, including cyclin-dependent
kinase inhibitor, Bruton's tyrosine-kinase inhibitor, B-cell
lymphoma 2 inhibitor or and alemtuzumab/corticosteroids (8,10).
BIRC3 abnormalities provide a molecular rationale for using
NF-κB inhibitors, which remain under development (7).
Materials and methods
Patients and sample preparation
The present study included 117 CLL patients, and 45
B-cell acute lymphocytic leukemia (B-ALL) patients that were
diagnosed according to standard criteria (11). The samples were obtained with the
informed consent from the corresponding patients and according to
the institutional Ethical Committee guidelines. For CLL cases, DNA
was extracted from lymphocytes using the Gentra®
Puregene® Blood kit (Qiagen, Hilden, Germany), according
to the manufacturer's protocol. For B-ALL cases, DNA was derived
from cytogenetically prepared cells, as previously described
(2), which were fixed in
methanol/acetic acid (dilution, 3:1) (Table I).
 | Table I.Gender, age and cytogenetic results
of the B-ALL and CLL cases used in the present study. |
Table I.
Gender, age and cytogenetic results
of the B-ALL and CLL cases used in the present study.
| Case no. | Gender | Age, years | DNA extracted
from | Cytogenetic
results |
|---|
| A-1 | Male | 84 | BM | 46,XY,
−9,t(9;22)(q34;q11),del(11)(q),+mar[cp3]/46,XY[5] |
| A-2 | Male | 23 | BM |
Hyperdiploid/46,XY |
| A-3 | Male | 34 | BM | 46,XY |
| A-4 | Male | 19 | BM | 46,XY |
| A-5 | Female | 76 | BM |
45,X,-X[14]/46,XX[2] |
| C-1 | Male | 73 | BM |
46–47,XY,del(11)(q22q2?3),add(17)t(17;?)(p11.2;?)[cp5]/
45–46,XY,del(11)(q22q2?3),del(17)(p11.2)[cp4]/ 43–46,XY,del(11)(q22q2?3)[cp2]/ 46,XY[7] |
| C-2 | Female | 50 | B | n.a. |
| C-3 | Female | 39 | BM |
43–46,XY,del(11)(q2?2q2?4)[cp5]/ 45–46,XY,del(11)(q2?2q2?4),del(15)(q1?1q2?3)[cp11]/ 46,XY[1] |
| C-4 | Male | 64 | BM | 46,XY |
| C-5 | Male | 43 | BM | 46,XY |
| C-6 | Male | 67 | BM | 46,XY |
| C-7 | Male | 77 | BM | 46,XY,del(11)(q?21),add(20)(p13)[7]/ 45,X,-Y[10]/ 46,XY[3] |
| C-8 | Male | 53 | BM | 46,XY |
| C-9 | Male | 59 | BM | n.a. |
| C-10 | Male | 73 | BM | 45,XY,der(2)t(2;13)(q?37;q?14),?del(6)(p?23), del(11)(q?21)der(12)t(12;13)(q?24;q?22),-13[cp4]/
46,XY[19] |
| C-11 | Male | 72 | B | n.a. |
| C-12 | Female | 73 | BM | 46,XX,add(11)(q?22)[3]/ 46,XX[12] |
| C-13 | Male | 54 | B | 46,XY |
| C-14 | Male | 68 | BM | 46,XY |
| C-15 | Male | 53 | BM | 46,XY |
| C-16 | Male | 75 | BM | n.a. |
| C-17 | Female | 67 | BM | 46,XX[18]
45,X,-X[1] |
| C-18 | Male | 74 | BM | n.a. |
| C-19 | Male | 65 | BM | 46,XY |
| C-20 | Male | 77 | B |
45–46,XY,del(11)(q?22q?23)[cp14] 46,XY[5] |
| C-21 | Male | 83 | BM |
47,XY,-11,+12,+mar[cp3]/
47,XY,del(5)(p1?3),-11,+12,-17,+mar1,+mar2[cp6]/
46,XY[9] |
| C-22 | Male | 72 | BM | 46,XY |
| C-23 | Male | 59 | B | 46,XY |
| C-24 | Female | 66 | B | n.a. |
| C-25 | Female | 71 | B | 46,XX |
| C-26 | Male | 65 | BM |
46,XY,?t(3;?)(p21;?),add(17)(p?12)ort(17;?),-8,+mar[cp7]
46,XY[9] |
| C-27 | Female | 74 | B | 46,XX |
| C-28 | Female | 74 | BM | 46,XX,i(17)(q10)[1]/ 46,XX,+12,i(17)(q10),-21[9]/
46,XX,t(3;?)(q2?9;?)[4],-7[4],+12[4],i(17)(q10)[4][cp4]/ 46,XX[4] |
| C-29 | Female | 90 | B | n.a. |
| C-30 | Male | 56 | BM | n.a. |
| C-31 | Female | 65 | BM | 46,XX |
Interphase fluorescence in situ
hybridization (iFISH) analysis
iFISH analyses were performed as previously
described (2), using the following
commercially available probes: LSI p53/LSI ATM (in 17p13.1 and
11q22.3), CEP 3 (D3Z1 in 3p11.1-q11.1), CEP 4 (D4Z1 in 4p11-q11),
CEP 7 (D7Z1 in 7p11.1-q11.1), CEP 11 (D11Z1 in 11p11.11-q11), CEP
16 (D16Z2 in 16p11.1-q11.1), CEP 17 (D17Z1 in 17p11.1-q11.1) and
CEP 18 (D18Z1 in 18p11.1-q11.1), all from Vysis (Abbott GmbH &
Company, KG, Wiesbaden, Germany); and ZytoLight® SPEC
BIRC3/MALT1 DualColor Dual Fusion probe (in 11q22.2
and 18q21.32) from ZytoVision GmbH (Bremerhaven, Germany). For each
iFISH analysis, 100–200 interphase nuclei were examined per patient
and probe.
Multiplex ligation-dependent probe
amplification (MLPA) analysis
MLPA was performed using the SALSA MLPA probemix
P377-A1 for Hematological Malignancies kit (MRC-Holland, Amsterdam,
Netherlands). The P377-A1 probemix kit contains 52 probes for 37
genes. The TP53 and ATM genes were assessed by four
probes each; however, probes for the BIRC3 gene were not
included in the kit (2). MLPA was
successfully performed on 85/117 CLL samples and 32/45 B-ALL
samples. MLPA was not successful for the remaining samples due to
fragmentation of DNA.
Array-comparative genomic
hybridization (aCGH)
aCGH was performed using the Agilent SurePrint G3
Human Genome Microarray 180 K (Agilent Technologies, Santa Clara,
CA, USA), as previously described (12). aCGH was applied in a total of 3 CLL
patients that carried a BIRC3 duplication and in 1 B-ALL
patient that possessed a BIRC3 deletion.
Results
Gene copy numbers
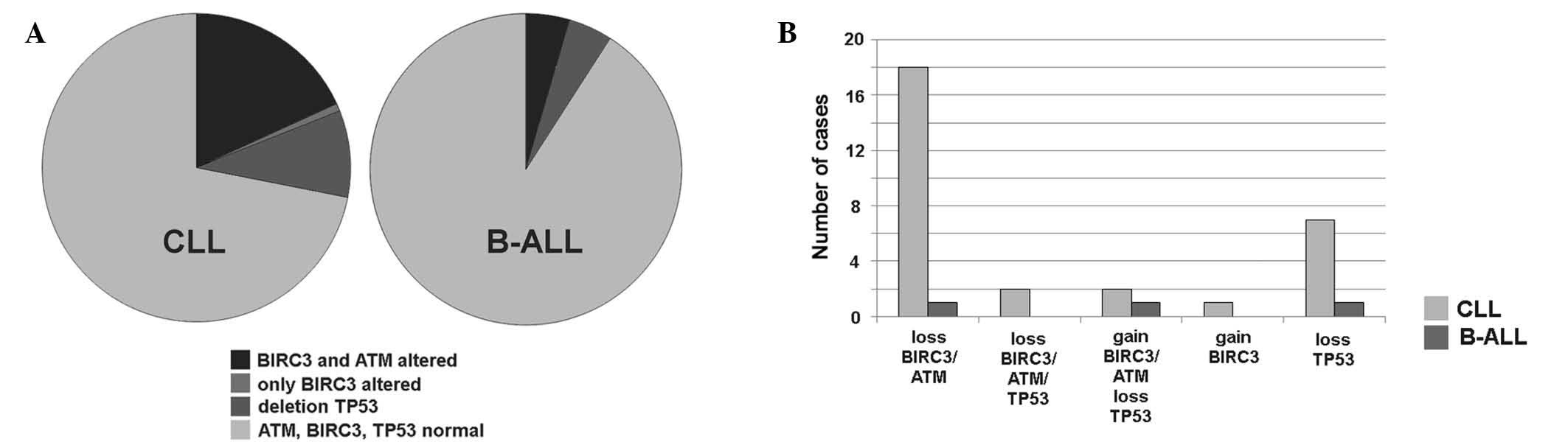
BIRC3 gene copy number variations were
detected in 23/117 (~20%) of CLL and 2/45 (~4%) of B-ALL cases, as
summarized and detailed in Fig. 1.
BIRC3 deletions were identified in 20 cases of CLL (cases
C-1 to C-20) and in 1 case of B-ALL (case A-1). ATM deletion
was detected in the identical 20 CLL and 1 B-ALL cases. Therefore,
all patients with a BIRC3 deletion also possessed an
ATM deletion. However, in cases C-1, C-8, C-10, C-13 and
C-20 the detected clone sizes with deletions in BIRC3 and
ATM were extremely varied from one another. In case C-1, the
clone with the ATM deletion was 8× smaller compared with
that with the BIRC3 deletion, whereas in cases C-8, C-10,
C-13 and C-20, the clone with the BIRC3 deletion was 2–3×
smaller than that with the ATM deletion. A BIRC3
duplication was identified in 1 case of B-ALL (case A-2) and in 3
CLL patients (cases C-21 to C-23) (Table
II).
 | Table II.Summary of MLPA and iFISH results of
TP53, ATM, BIRC3 and MALT1 in all
studied cases. |
Table II.
Summary of MLPA and iFISH results of
TP53, ATM, BIRC3 and MALT1 in all
studied cases.
|
| TP53
(%) | ATM (%) | BIRC3
(%) | MALT1
(%) |
|---|
|
|
|
|
|
|
|---|
| Case no. | MLPA | iFISH | MLPA | iFISH | iFISH | iFISH |
|---|
| A-1 | N | N | D | D (76.5) | D (75.0) | N |
| A-2 | n.a. | A (100.0) | n.a. | A (100.0) | A (100.0) | A (100.0) |
| A-3 | D | D (8.5) | N | N | N | N |
| A-4 | D | D (10.0) | N | N | N | N |
| A-5 | D | D (10.0) | N | N | N | N |
| A-6 to A-33 | N | N | N | N | N | N |
| A-34 to A-45 | n.a. | N | n.a. | N | N | N |
| C-1 | D | D (86.0) | D | D (11.0) | D (80.0) | N |
| C-2 | D | D (21.0) | N | D (23.0) | D (22.0) | N |
| C-3 | N | N | D | D (98.0) | D (90.0) | N |
| C-4 | N | N | D | D (23.5) | D (30.0) | N |
| C-5 | N | N | D | D (24.0) | D (25.0) | N |
| C-6 | N | N | D | D (88.0) | D (85.0) | N |
| C-7 | N | N | D | D (90.0) | D (80.0) | N |
| C-8 | N | N | D | D (77.0) | D (50.0) | N |
| C-9 | N | N | D | D (98.0) | D (75.0) | N |
| C-10 | N | N | D | D (87.0) | D (60.0) | N |
| C-11 | N | N | D | D (95.0) | D (90.0) | N |
| C-12 | N | N | D | D (83.0) | D (80.0) | N |
| C-13 | N | N | D | D (93.0) | D (25.0) | N |
| C-14 | N | N | N | D (33.0) | D (15.0) | N |
| C-15 | N | N | N | D (12.0) | D (13.0) | N |
| C-16 | n.a. | N | n.a. | D (80.0) | D (78.0) | N |
| C-17 | n.a. | N | n.a. | D (10.0) | D (9.0) | N |
| C-18 | n.a. | N | n.a. | D (73.0) | D (64.0) | N |
| C-19 | n.a. | N | n.a. | D (9.0) | D (10.0) | N |
| C-20 | n.a. | N | n.a. | D (96.0) | D (42.0) | N |
| C-21 | D | D (16.0) | N | A (50.0) | A (50.0) | A (50.0) |
| C-22 | D | D (40.0) | N | A (40.0) | A (40.0) | A (40.0) |
| C-23 | N | N | N | N | A (36.0) | N |
| C-24 | D | D (19.0) | N | N | N | N |
| C-25 | D | D (36.0) | N | N | N | N |
| C-26 | D | D (89.0) | N | N | N | N |
| C-27 | D | D (77.0) | N | N | N | N |
| C-28 | D | D (95.0) | N | N | N | N |
| C-29 | N | D (11.5) | N | N | N | N |
| C-30 | n.a. | D (86.5) | n.a. | N | N | N |
| C-31 | N | N | N | N | N | A (75.0) |
| C-32 to C-91 | N | N | N | N | N | N |
| C-92 to C-117 | n.a. | N | n.a. | N | N | N |
With regard to TP53 abnormalities, 3 patients
with B-ALL possessed a TP53 deletion in the absence of any
aberrations in BIRC3. TP53 deletions were present in
11 CLL patients, 7 of which possessed no associated BIRC3
aberrations and, notably, 2 of which were accompanied by
BIRC3 and ATM amplification.
BIRC3 duplication
In total, 3 CLL patients harbored a BIRC3
duplication (cases C-21 to C-23), 2 of which (C-21 and C-22) were
accompanied by ATM and MALT1 duplications in addition
to TP53 deletion. To study these cases in greater depth,
iFISH was performed using centrometric probes for chromosomes (CEP)
3, 4, 7, 11, 16 and 18. For these chromosomes, 3 signals were
detected in 11% (case C-21) and 25% (case C-22) of the cells, and 4
signals were detected in 29% (case C-21) and 25% (C-22) of the
cells, respectively (Fig. 2).
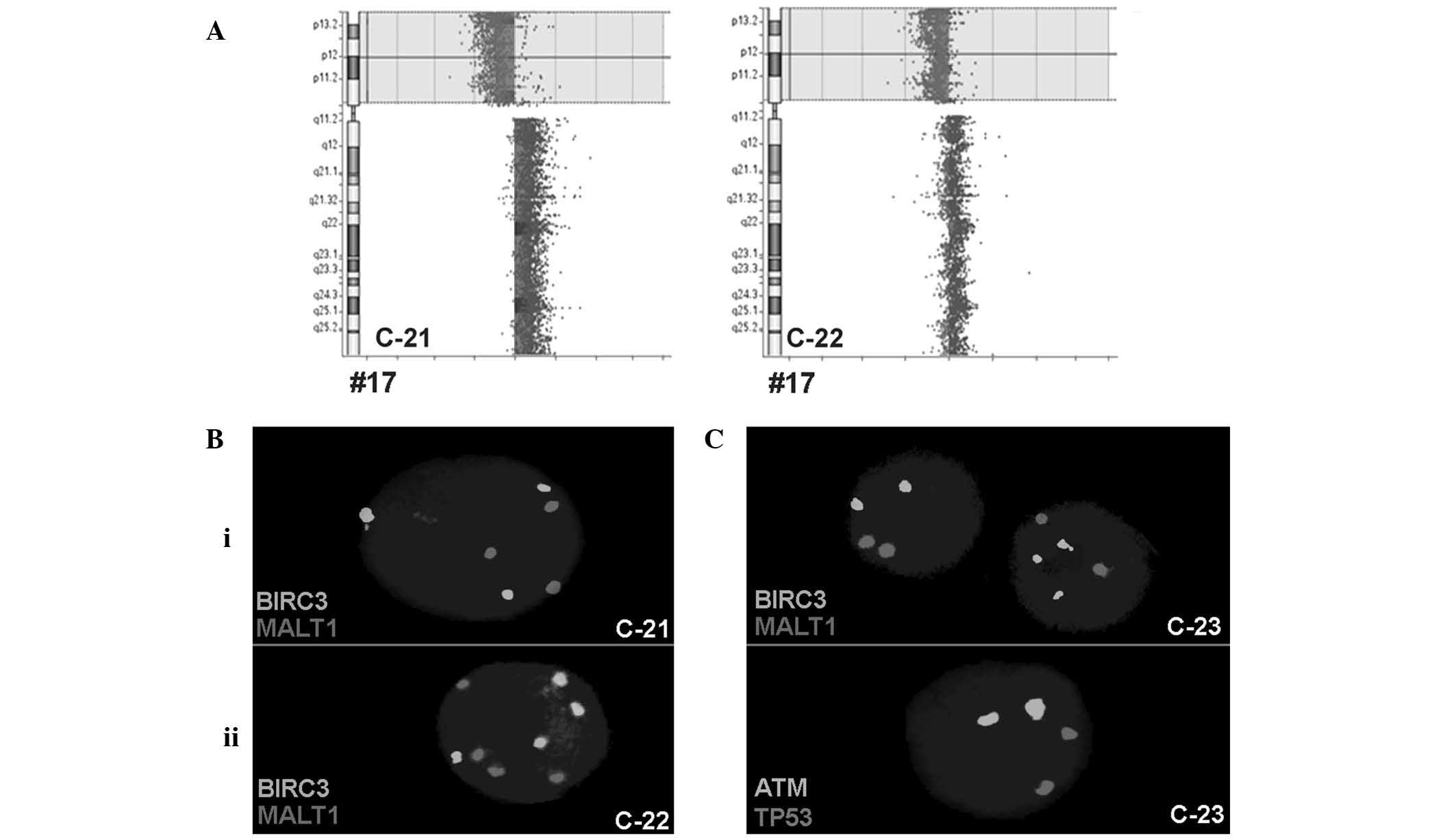 | Figure 2.(A) Array comparative genomic
hybridization confirmed the deletion in TP53, which was
detected initially using iFISH and multiplex ligation dependent
probe amplification for CLL cases C-21 and C-22. The whole short
arm was deleted and the long arm was possibly duplicated due to an
isochromosome 17 formation, at least in case C-21. (B) Examples for
gain of copy numbers for BIRC3 and MALT1 in the 2
cases by iFISH: i) C-21, an example of 3 copies and ii) C-22, an
example of 4 copies. (C) iFISH results of the CLL case C-23.
BIRC3 had 3 copies in certain cells; however ATM,
MALT1 and TP53 exhibited only 2 copies each, in all
cells. TP53, tumor protein 53; CLL, chronic lymphocytic
leukemia; iFISH, interphase fluorescence in situ
hybridization; BIRC3, baculoviral IAP repeat-containing 3;
MALT1, mucosa-associated lymphoid tissue lymphoma
translocation gene 1; ATM, ataxia telangiectasia
mutated. |
The third CLL case (C-23) with BIRC3
duplication was associated with normal copy numbers of TP53
and ATM; the centromeric probes for chromosomes 11 and 17
only revealed 2 signals each (Fig.
2).
Based on the aCGH results for cases C-21, C-22 and
C-23, the TP53 deletion in C-21 and C-22 was confirmed;
however, BIRC3 was normal in all 3 patients (Fig. 2). Therefore, cases C-21 and C-22 had a
mixture of a malignant triploid and tetraploid cell clones and a
deletion in TP53. Case C-23 demonstrated the selective gain
of copy numbers for BIRC3, without ATM involvement,
in 36% of the cells; however, this finding was not detectable using
aCGH.
MALT1 duplication
The patient with a MALT1 duplication (case
C-31) possessed an acquired trisomy of chromosome 18, which was
confirmed using MLPA and iFISH. The probes for the deleted in
colorectal cancer gene on 18q21.2 and RNA
(guanine-7-)methyltransferase gene on 18p11.22 revealed a
duplication by MLPA, which was confirmed by iFISH in 75% of the
cells.
B-ALL patients
Regarding B-ALL patients, 1 case revealed a deletion
in BIRC3 along with ATM (case A-1), and another case
was identified as possessing a triploid/hyperdiploid karyotype in
the iFISH analysis using CEP 11, 17 and 18 (case A-2, result not
shown), as was observed in the cases C-21 and C-22.
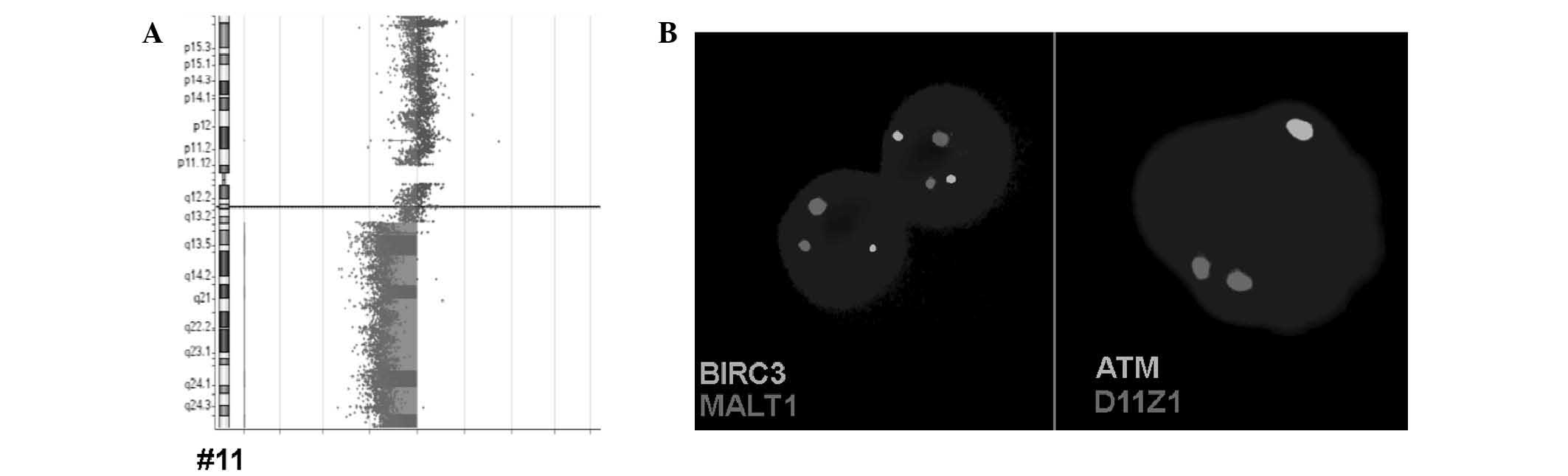
The BIRC3 deletion in B-ALL case A-1 was
confirmed by aCGH, which reveled that the deletion in the long arm
of chromosome 11 covered between chr11:67,773,863 and 134,945,165
(GRCh37/hg19) (Fig. 3). The
ATM and BIRC3 genes are located between positions
102,188,181 and 108,239,826.
Discussion
The BIRC3 gene, located on 11q22.2, is ~6 Mb
centromeric to the ATM gene locus and is considered to be a
negative regulator of the non-canonical NF-κB signaling pathway
(13,14). BIRC3 cooperates with tumor
necrosis factor receptor-associated factors 2 and 3, in the same
protein complex that negatively regulates the mitogen-activated
protein kinase 14, a serine-threonine kinase and central activator
of non-canonical NF-κB signaling (7).
In addition, a frequent aberration associated with BIRC3 is
the recurrent t(11;18)(q21;q21) translocation, which involves the
mucosa-associated lymphoid tissue lymphoma translocation gene 1
(MALT1), located on 18q21.32. This type of alteration
appears in mucosa-associated lymphoid tissue (MALT) lymphoma
(15).
The present study regarding BIRC3 copy number
variations in 117 CLL and 45 B-ALL patients has revealed several
major findings that, to the best of our knowledge, have not been
previously reported. Firstly, BIRC3 duplications were
detected in 3 cases of CLL, and 2 of these were associated with
ATM and MALT1 duplications, in addition to
TP53 deletions; BIRC3 amplification was detected in these 2
cases as a result of a hyperdiploid cell clone and has been
previously reported in cases of CLL, but is not a frequent event
(16,17). In addition, 1 B-ALL patient possessed
a duplication of BIRC3 due to partial hyperdiploidy, which
is more common in B-ALL compared with CLL, and is associated with
good prognosis in pediatric patients (18). A CLL-case with a BIRC3
duplication possessed normal ATM and TP53 copy
numbers; however, the duplication was not detected by aCGH, most
likely due to the low sensitivity of aCGH for mosaic detection,
despite being present in 36% of the cells. Previous studies on the
interaction of BIRC3 with the NF-κB pathway indicate that
BIRC3 duplication may lead to the inactivation of tumor
suppressor activity (19–21).
As the predominant morphological feature of CLL is
the accumulation of small B lymphocytes (1), B-ALL patients were chosen to be the
second group to be tested for BIRC3-alterations in the
present study. Therefore the second important finding of the
present study is the detection of a BIRC3 deletion in 1 of
the 45 studied B-ALL cases. The aCGH for case A-1 revealed the
deletion of almost all of the long arm of chromosome 11, and the
most frequent aberrations associated with chromosome 11 in B-ALL
patients are structural abnormalities in band 11q23, which harbors
the myeloid/lymphoid leukemia gene (22).
Chromosomal deletions involving 11q have been
reported in certain subtypes of hematological malignancies,
including B-cell CLL, and are associated with a poor prognosis in
mantle cell lymphomas or T-cell prolymphocytic leukemia (23). Therefore, the prognosis for the B-ALL
patient in the present study may be poor or extremely poor.
Additional studies are required to determine the role of
BIRC3 in the prognosis of B-ALL patients.
The disruption of BIRC3 is specifically
restricted to chemo-refractory cases in progressive CLL patients,
and may selectively associate with fludarabine-refractory patients
with normal TP53 (7).
Therefore, another notable finding of the present study is that
BIRC3 abnormalities were associated with TP53
deletion in only 4/117 CLL cases. According to previous studies,
the frequency of BIRC3 disruption is low at diagnosis;
however, BIRC3 disruptions tend to accumulate among
refractory CLL and emerge over time. Patients harboring a
BIRC3 disruption typically experience an aggressive disease
course, even compared with other clinically aggressive groups
(14,24). This aspect of the disease was not the
focus of the present study. However, examining BIRC3
duplication cases for the presence of mutations may be interesting
for future study.
The application of a BIRC3 probe in iFISH
increased the diagnostic yield in CLL and ALL cases. As in MLPA or
aCGH by BIRC3-directed iFISH, only gain or loss of copy
numbers can be registered. However, to the best of our knowledge,
BIRC3 activation by a translocation has not previously been
reported. Therefore, routine iFISH and MLPA testing in CLL and ALL
should be enhanced by a probe specific for BIRC3. iFISH is
superior to MLPA as it is able to detect low level mosaics in the
probe sample, however, MLPA is more cost-effective (2). A combination of the two approaches, by
performing MLPA followed by iFISH in selected cases may be the best
compromise if corresponding MLPA sets are available (2).
In conclusion, the hypothesis by Rose-Zerilli et
al (25) that BIRC3
deletions are always associated with ATM deletions is
questioned at least for a small percentage of cases. As screening
of the BIRC3 gene is not routinely undertaken for CLL
patients (2,26), the results of the present study
suggest that screening may be considered as necessary in the
future, particularly to aid in making the correct treatment
decisions; particularly, whether to treat with or without
fludarabine.
Acknowledgements
The present study was supported by Katholischer
Akademischer Ausländer-Dienst (fellowship to Mr. Eyad Alhourani)
and Deutscher Akademischer Austauschdienst (fellowship to Mr.
Moneeb A.K. Othman; PROBRAL (grant no., 57054562 to Dr Thomas
Liehr; University Partnership Program of Friedrich Schiller
University Jena to Dr Thomas Liehr).
References
|
1
|
Rodríguez-Vicente AE, Díaz MG and
Hernández-Rivas JM: Chronic lymphocytic leukemia: A clinical and
molecular heterogenous disease. Cancer Genet. 206:49–62. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alhourani E, Rincic M, Othman MA, Pohle B,
Schlie C, Glaser A and Liehr T: Comprehensive chronic lymphocytic
leukemia diagnostics by combined multiplex ligation dependent probe
amplification (MLPA) and interphase fluorescence in situ
hybridization (iFISH). Mol Cytogenet. 7:792014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Döhner H, Stilgenbauer S, Benner A,
Leupolt E, Kröber A, Bullinger L, Döhner K, Bentz M and Lichter P:
Genomic aberrations and survival in chronic lymphocytic leukemia. N
Engl J Med. 343:1910–1916. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gunn SR, Hibbard MK, Ismail SH,
Lowery-Nordberg M, Mellink CH, Bahler DW, Abruzzo LV, Enriquez EL,
Gorre ME, Mohammed MS and Robetorye RS: Atypical 11q deletions
identified by array CGH may be missed by FISH panels for prognostic
markers in chronic lymphocytic leukemia. Leukemia. 23:1011–1017.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Greipp PT, Smoley SA, Viswanatha DS,
Frederick LS, Rabe KG, Sharma RG, Slager SL, Van Dyke DL, Shanafelt
TD, Tschumper RC and Zent CS: Patients with chronic lymphocytic
leukaemia and clonal deletion of both 17p13.1 and 11q22.3 have a
very poor prognosis. Br J Haematol. 163:326–333. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Campregher PV and Hamerschlak N: Novel
prognostic gene mutations identified in chronic lymphocytic
leukemia and their impact on clinical practice. Clin Lymphoma
Myeloma Leuk. 14:271–276. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rossi D, Fangazio M, Rasi S, Vaisitti T,
Monti S, Cresta S, Chiaretti S, Del Giudice I, Fabbri G, Bruscaggin
A, et al: Disruption of BIRC3 associates with fludarabine
chemorefractoriness in TP53 wild-type chronic lymphocytic leukemia.
Blood. 119:2854–2862. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rossi D, Fangazio M and Gaidano G: The
spectrum of genetic defects in chronic lymphocytic leukemia.
Mediterr J Hematol Infect Dis. 4:e20120762012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rossi D, Rasi S, Spina V, Bruscaggin A,
Monti S, Ciardullo C, Deambrogi C, Khiabanian H, Serra R, Bertoni
F, et al: Integrated mutational and cytogenetic analysis identifies
new prognostic subgroups in chronic lymphocytic leukemia. Blood.
121:1403–1412. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Puiggros A, Blanco G and Espinet B:
Genetic abnormalities in chronic lymphocytic leukemia: Where we are
and where we go. BioMed Res Int. 2014:4359832014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hallek M, Cheson BD, Catovsky D,
Caligaris-Cappio F, Dighiero G, Döhner H, Hillmen P, Keating MJ,
Montserrat E, Rai KR and Kipps TJ: International Workshop on
Chronic Lymphocytic Leukemia: Guidelines for the diagnosis and
treatment of chronic lymphocytic leukemia: A report from the
International Workshop on Chronic Lymphocytic Leukemia updating the
National Cancer Institute-Working Group 1996 guidelines. Blood.
111:5446–5456. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Othman MA, Rincic M, Melo JB, Carreira IM,
Alhourani E, Hunstig F, Glaser A and Liehr T: Novel cryptic
three-way translocation t(2;9;18)(p23.2;p21.3;q21.33) with deletion
of tumor suppressor genes in 9p21.3 and 13q14 in a T-cell acute
lymphoblastic leukemia. Leukemia Res Treat. 2014:3571232014.
|
|
13
|
Rossi D, Ciardullo C, Spina V and Gaidano
G: Molecular bases of chronic lymphocytic leukemia in light of new
treatments. Immunol Lett. 155:51–55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cortese D, Sutton LA, Cahill N, Smedby KE,
Geisler C, Gunnarsson R, Juliusson G, Mansouri L and Rosenquist R:
On the way towards a ‘CLL prognostic index’: Focus on TP53, BIRC3,
SF3B1, NOTCH1 and MYD88 in a population-based cohort. Leukemia.
28:710–713. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Morgan JA, Yin Y, Borowsky AD, Kuo F,
Nourmand N, Koontz JI, Reynolds C, Soreng L, Griffin CA,
Graeme-Cook F, et al: Breakpoints of the t(11;18)(q21;q21) in
mucosa-associated lymphoid tissue (MALT) lymphoma lie within or
near the previously undescribed gene MALT1 in chromosome 18. Cancer
Res. 59:6205–6213. 1999.PubMed/NCBI
|
|
16
|
Shao L, Kang SH, Li J, Hixson P, Taylor J,
Yatsenko SA, Shaw CA, Milosavljevic A, Chang CC, Cheung SW and
Patel A: Array comparative genomic hybridization detects
chromosomal abnormalities in hematological cancers that are not
detected by conventional cytogenetics. J Mol Diagn. 12:670–679.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Specchia G, Albano F, Anelli L, Storlazzi
CT, Monaco M, Capalbo S, Rocchi M and Liso V: Concomitant tetrasomy
3q and trisomy 18 in CD5(−), CD13(+) chronic lymphocytic leukemia.
Cancer Genet Cytogenet. 133:160–163. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kebriaei P, Anastasi J and Larson RA:
Acute lymphoblastic leukaemia: Diagnosis and classification. Best
Pract Res Clin Haematol. 15:597–621. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ryan KM, Ernst MK, Rice NR and Vousden KH:
Role of NF-kappaB in p53-mediated programmed cell death. Nature.
404:892–897. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Keller U, Huber J, Nilsson JA, Fallahi M,
Hall MA, Peschel C and Cleveland JL: Myc suppression of Nfkb2
accelerates lymphomagenesis. BMC Cancer. 10:348–358. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu F, Bardhan K, Yang D, Thangaraju M,
Ganapathy V, Waller JL, Liles GB, Lee JR and Liu K: NF-κB directly
regulates Fas transcription to modulate Fas-mediated apoptosis and
tumor suppression. J Biol Chem. 287:25530–25540. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cox MC, Panetta P, Lo-Coco F, Del Poeta G,
Venditti A, Maurillo L, Del Principe MI, Mauriello A, Anemona L,
Bruno A, et al: Chromosomal aberration of the 11q23 locus in acute
leukemia and frequency of MLL gene translocation: Results in 378
adult patients. Am J Clin Pathol. 122:298–306. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Monni O and Knuutila S: 11q deletions in
hematological malignancies. Leuk Lymphoma. 40:259–266. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Strefford JC, Sutton LA, Baliakas P,
Agathangelidis A, Malčíková J, Plevova K, Scarfó L, Davis Z,
Stalika E, Cortese D, et al: Distinct patterns of novel gene
mutations in poor-prognostic stereotyped subsets of chronic
lymphocytic leukemia: The case of SF3B1 and subset #2. Leukemia.
27:2196–2199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rose-Zerilli MJ, Forster J, Parker H,
Parker A, Rodríguez AE, Chaplin T, Gardiner A, Steele AJ, Collins
A, Young BD, et al: ATM mutation rather than BIRC3 deletion and/or
mutation predicts reduced survival in 11q-deleted chronic
lymphocytic leukemia: Data from the UK LRF CLL4 trial.
Haematologica. 99:736–742. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Goorha S, Glenn MJ, Drozd-Borysiuk E and
Chen Z: A set of commercially available fluorescent in-situ
hybridization probes efficiently detects cytogenetic abnormalities
in patients with chronic lymphocytic leukemia. Genet Med. 6:48–53.
2004. View Article : Google Scholar : PubMed/NCBI
|