Introduction
Nasopharyngeal carcinoma (NPC) is a common
malignancy that develops in the head and neck region, and occurs
primarily in southern China, Southeast Asia and North Africa, while
it is rare in other parts of the world (1). It has been demonstrated that genetic and
infectious factors are associated with NPC. Epithelial cells of NPC
tumors carry Epstein-Barr virus (EBV) genetic material, and EBV
infection has been reported to be associated with 90% of NPC cases
(2). Numerous studies have confirmed
that EBV-encoded proteins, including latent membrane protein 1
(LMP1), BamHI-A rightward frame-1, Epstein-Barr virus
nuclear antigen (EBNA)1 and EBNA2, are essential factors in
EBV-induced NPC cell transformation, and have been demonstrated to
be overexpressed in EBV-associated NPC (3). In particular, the expression of LMP1,
considered to be the principal carcinogenic protein of EBV, is
positively associated with metastasis of NPC and serves as a good
diagnostic marker for NPC (3,4). However, it remains to be elucidated
whether these carcinogenic molecules, including LMP1, may be useful
as therapeutic targets for the treatment of NPC.
MicroRNAs (miRs) are endogenous short non-coding
RNAs of 18–24 nucleotides in length that regulate gene expression
in various biological and metabolic processes (5). miRs have a significant role in the
regulation of gene expression via degradation of target messenger
(m)RNAs or inhibition of translation of target proteins (6). Previous studies focusing on the
regulatory effects of miRs in NPC have recognized several miRs that
are closely associated with tumorigenesis and progression of NPC.
miR-30a (7) and other miRs promote
proliferation, invasiveness and metastasis in vitro and/or
in vivo via various targets and through various mechanisms,
resulting in poor survival of patients with NPC. By contrast, there
are miRs that serve as potential tumor suppressors in NPC,
including miR-9 (8). Furthermore,
certain deregulated miRs in NPC have been reported to be induced by
EBV (5). Oncogenic miRs, including
miR-10b (9) have been recognized to
be induced by or be associated with EBV infection. In addition, EBV
infection induces cellular expression of miR-155 in NPC (10), and upregulated miR-155 during EBV
infection was promoted by expression of EBV-encoded LMP1 (10).
In the present study, in order to confirm the
promotion of miR-155 by LMP1, and to recognize the oncogenic role
of LMP1 and LMP1-promoted miR-155 in NPC, an LMP1-overexpressing
CNE-2 cell line was constructed, and miR-155 upregulation was
examined in this cell line. Subsequently, the regulatory role of
LMP1 and miR-155 on cell proliferation was investigated.
Furthermore, the influence of knockdown of LMP1-induced miR-155 on
the sensitivity of CNE-2 cells to radiation treatment was
assessed.
Materials and methods
Cell culture, LMP1 overexpression and
miR-155 manipulation
NPC CNE-2 cells were purchased from the Cell
Resource Center of the Chinese Academy of Medical Sciences
(Beijing, China). Cells were grown or maintained in RPMI-1640
medium (catalog no., 31800–022; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), which was supplemented with 10% (for growth) or
2% (for maintenance) fetal bovine serum (catalog no., 1009-141-FBS;
Gibco®; Thermo Fisher Scientific, Inc.), 50 µg/ml
penicillin (catalog no., P7794; Sigma-Aldrich, St. Louis, MO, USA)
and 50 µg/ml streptomycin (catalog no., P4333; Sigma-Aldrich).
Cells were incubated at 37°C in an atmosphere of 5% CO2.
For transient LMP1 overexpression, LMP1-pcDNA3.1 recombinant
plasmid (10) was transfected into
CNE-2 cells using Lipofectamine® 2000 (catalog no.,
12566014; Invitrogen®; Thermo Fisher Scientific, Inc.),
at a concentration of 0, 0.2, 0.5 or 1 µg/ml for 12 (for LMP1
messenger (m)RNA assay), 24 (for LMP1 protein, cell viability and
cell proliferation assays), 48 or 72 h (for cell proliferation
assay). For sustained LMP1 overexpression, CNE-2 cells were
transfected with the aforementioned LMP1-pcDNA3.1 recombinant
plasmid, and cultured under Geneticin® (G418; catalog
no., 11811023; Thermo Fisher Scientific, Inc.) pressure (800
µg/ml). The positive cell clones were propagated in RPMI-1640
medium containing 500 µg/ml G418. To manipulate the levels of
miR-155, CNE-2 cells were transfected with miR-155 mimic or miR-155
inhibitor (Qiagen, Inc., Valencia, CA, USA) using Lipofectamine
2000, while miR-Con was utilized as a control miRNA.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total cellular mRNA was isolated and extracted from
CNE-2 cells using RNeasy Mini kit (catalog no., 74104; Qiagen GmbH,
Hilden, Germany.). The sample was purified using the RNase-Free
DNase Set (catalog no., 79254; Qiagen, Inc.) according to the
manufacturer's instructions. RT-qPCR analysis of the mRNA levels of
LMP1 in CNE-2 cells was performed with One-Step SYBR PrimeScript
RT-PCR Kit II (Perfect Real Time) (catalog no., RR086A/B; Takara
Bio, Inc., Otsu, Japan) using a quantitative PCR instrument
(LightCycler® 2.0; Roche Applied Science, Penzberg,
Germany) and the following primers, which were designed by Primer
Express 2 software (Applied Biosystems; Thermo Fisher Scientific,
Inc.) and synthesized by Sangon Biotech Co., Ltd., (Shanghai,
China): Forward, 5′-GCAGCCTCACGCACATCGA-3′ and reverse,
5′-GGGAGGCGCTTGGTGCAAA-3′ for LMP-1; and forward,
5′-TGCACCACCAACTGCTTAG-3′ and reverse, 5′-TCTGGGTGGCAGTGAT-3′ for
glyceraldehyde-3-phosphate dehydrogenase (GAPDH). RT-qPCR was
performed under the following conditions: Denaturation at 42°C for
8 min, 95°C for 30 sec, followed by 40 cycles of 95°C for 10 sec
and at 60°C for 30 sec. The RNA expression levels were normalized
to the levels of GAPDH. miR in CNE-2 cells was extracted using
mirVana™ miRNA Isolation kit (catalog no., Am1561;
Ambion®; Thermo Fisher Scientific, Inc.). Quantitative
analysis of the levels of miR-155 in CNE-2 cells was conducted with
mirVana™ qRT-PCR miRNA Detection kit (catalog no., Am1558;
Ambion; Thermo Fisher Scientific), using the following primers:
5′-UUAAUGCUAAUUGUGAUAGGGGU-3′ for miR-155;
5′-GCAGGGGAACUCAUCAUCUCUG-3′ for U6 spliceosomal RNA; and
5′-UUUCAUCCUUGUGCAGGG-3′ for universal primer. qPCR was performed
under the following conditions: Denaturation at 95°C for 15 min,
followed by 40 cycles of 94°C for 15 sec, 55°C for 15 sec and 70°C
for 20 sec. U6 spliceosomal RNA served as an internal control. The
∆∆Cq method was utilized for relative quantification (11).
Protein sample isolation and western
blot analysis
Protein samples were extracted from CNE-2 cells with
M-PER™ Mammalian Protein Extraction Reagent (catalog no., 78501;
Thermo Fisher Scientific, Inc.). Subsequently, each protein sample
was separated using 8 or 12% sodium dodecyl sulfate-polyacrylamide
gel electrophoresis, prior to being transferred onto a
nitrocellulose membrane. The membranes were blocked at 4°C
overnight with Tris-buffered saline and Tween 20 (50 mM Tris-HCl,
pH 7.5; 150 mM NaCl; 0.05% Tween 20; catalog no., 9005-64-5;
Sigma-Aldrich) containing 5% skimmed milk, prior to be incubated
with mouse monoclonal EBV LMP1 (catalog no., sc-57721; 1:300; Santa
Cruz Biotechnology Inc., Santa Cruz, CA, USA) or rabbit anti-human
polyclonal GAPDH (catalog no., 100242-T40-50; 1:1,000; Sino
Biological, Inc., Beijing, China) primary antibodies for 2 h at
room temperature, followed by incubation with goat anti-rabbit
oligoclonal immunoglobulin G conjugated to horseradish peroxidase
(catalog no., A27036; 1:1,000; Thermo Fisher Scientific, Inc.) at
37°C for 45 min. The target protein band was visualized using the
enhanced chemiluminescence detection system (SuperSignal West Femto
Maximum Sensitivity Substrate; catalog no., 32209;
Pierce®; Thermo Fisher Scientific, Inc.). Protein bands
were quantified using ImageJ 1.45 software (http://imagej.nih.gov/ij/).
Determination of cell viability and
cell proliferation by
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT),
Cell Counting Kit (CCK)-8 and cell colony formation assays
Cell viability was determined via MTT assay (catalog
no., KA1606; Invitrogen; Thermo Fisher Scientific, Inc.). Cells
seeded into 96-well plates (Corning Inc., Tewksbury, MA, USA) were
transfected with 0.00, 0.25, 0.50, 0.75 or 1.00 µg/ml LMP1-pcDNA3.1
or control pcDNA3.1 for 24 h. In another experiment, the seeded
CNE-2 (LMP1 or Con) cells were transfected with 0, 20 or 40 nM
miR-155 inhibitor or miR-Con, respectively, and were irradiated
with 0, 2, 4, 6 or 8 Gy (RS225 X-Ray Research System; Gulmay
Medical Ltd., Surrey, UK). Subsequently, MTT assay was conducted
according to the standard protocol (12). Absorbance was measured at 570 nm with
a reference wavelength of 750 nm using a spectrophotometer (F-2000;
Hitachi, Tokyo, Japan). For the cell proliferation assay, CNE-2
(LMP1), CNE-2 (Con) or CNE-2 cells subjected or not to transfection
with miR-155 inhibitor or miR-155 control, were incubated in the
presence of CCK-8 (catalog no., CK04; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan). The absorbance at 450 nm of
each well was detected following visual color occurrence at 24 h.
All experiments were performed in triplicate. For the cell colony
formation assay, 5×102 cells were incubated in 6-well
plates (Corning Inc.) at 37°C in an atmosphere of 5%
CO2. At 3–6 days post-incubation, the cells were stained
using crystal violet (0.005%; catalog no., C3886; Sigma-Aldrich)
for 20 min, and colony numbers were recorded using ImageJ 1.45
software (http://imagej.nih.gov/ij/).
Statistical evaluations
Results are expressed as the mean ± standard error.
Student's t-tests were performed to compare the differences between
two groups. Statistical analyses were performed using GraphPad
Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-155 expression is upregulated by
overexpression of LMP1 in CNE-2 cells
A previous study by the present group revealed that
expression of miR-155 was upregulated in CNE-2 cells following
transfection with an LMP1-expressing plasmid (10). In order to further confirm the
association between miR-155 expression and overexpression of LMP1,
RT-qPCR was performed in the present study to detect miR-155
expression. LMP1 expression in the CNE-2 (LMP1) group exhibited a
significant difference in terms of mRNA (P<0.01; Fig. 1A) and protein levels (P<0.01;
Fig. 1B), compared with the other
groups. Furthermore, as shown in Fig. 1C
and D, the expression levels of LMP1 in the CNE-2 (LMP1) group
demonstrated no significant difference between the two serial
passages, and was significantly increased, compared with the CNE-2
(Con) group (P<0.01). Overexpression of LMP1 in the CNE-2 cell
line revealed a significant increase in the expression of miR-155,
as demonstrated in Fig. 1E
(P<0.05). Furthermore, the transfection efficiency of
LMP1-pcDNA3.1 plasmid was detected. As shown in Fig. 1F, >0.5 µg of LMP1-pcDNA3.1 was a
concentration that effectively increased the expression levels of
miR-155 in CNE-2 cells (P<0.05). These results suggested that
overexpression of LMP1 was able to upregulate the expression of
miR-155 in CNE-2 cells.
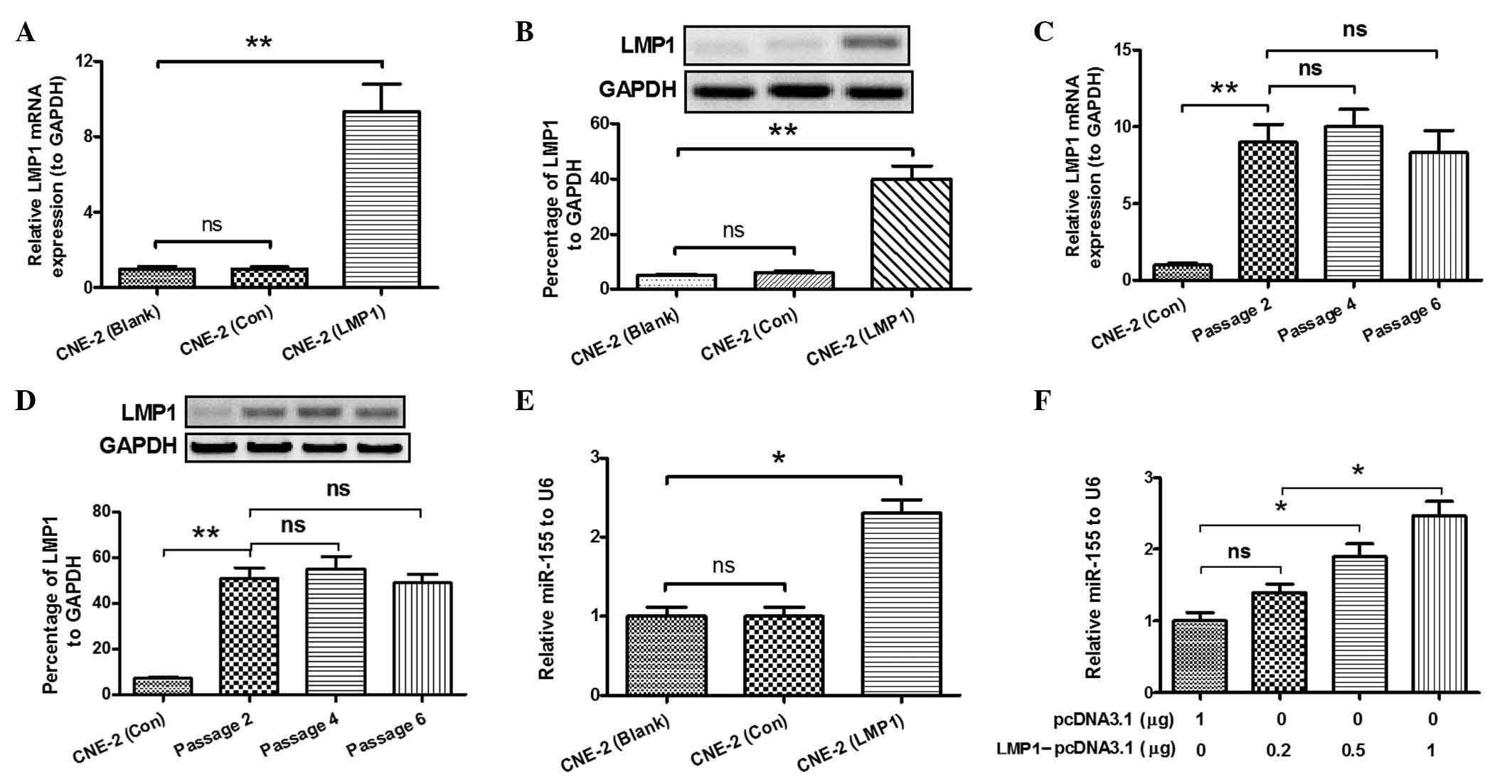 | Figure 1.LMP1 of Epstein-Barr virus promotes
miR-155 expression in CNE-2 cells. CNE-2 cells were separated into
three groups: Non-transfected CNE-2 cells (Blank), CNE-2 cells
transfected with empty pcDNA3.1 as negative control (Con) and
LMP1-pcDNA3.1-transfected CNE-2 cells (LMP1). (A) Relative mRNA
expression levels of LMP1 in the three groups of CNE-2 cells,
compared with the levels of GAPDH. (B) Percentage of LMP1
expression at the protein level in the three CNE-2 cell groups, as
revealed by western blot analysis. (C) Relative mRNA levels of LMP1
vs. GAPDH in the three groups of CNE-2 cells upon a number of
serial passages, as determined by reverse
transcription-quantitative polymerase chain reaction. (D)
Overexpression of LMP1 protein in CNE-2 cells following a number of
serial passages. (E) Relative miR-155 levels in the three groups of
CNE-2 cells, compared with the levels of U6 snRNA. (F) Relative
miR-155 levels in CNE-2 cells transfected with various
concentrations of LMP1-pcDNA3.1, compared with the levels of U6
snRNA. *P<0.05, **P<0.01. LMP1, latent membrane protein 1;
miR, microRNA; mRNA, messenger RNA; GAPDH,
glyceraldehyde-3-phosphate dehydrogenase; ns, not significant; Con,
control; snRNA, spliceosomal RNA. |
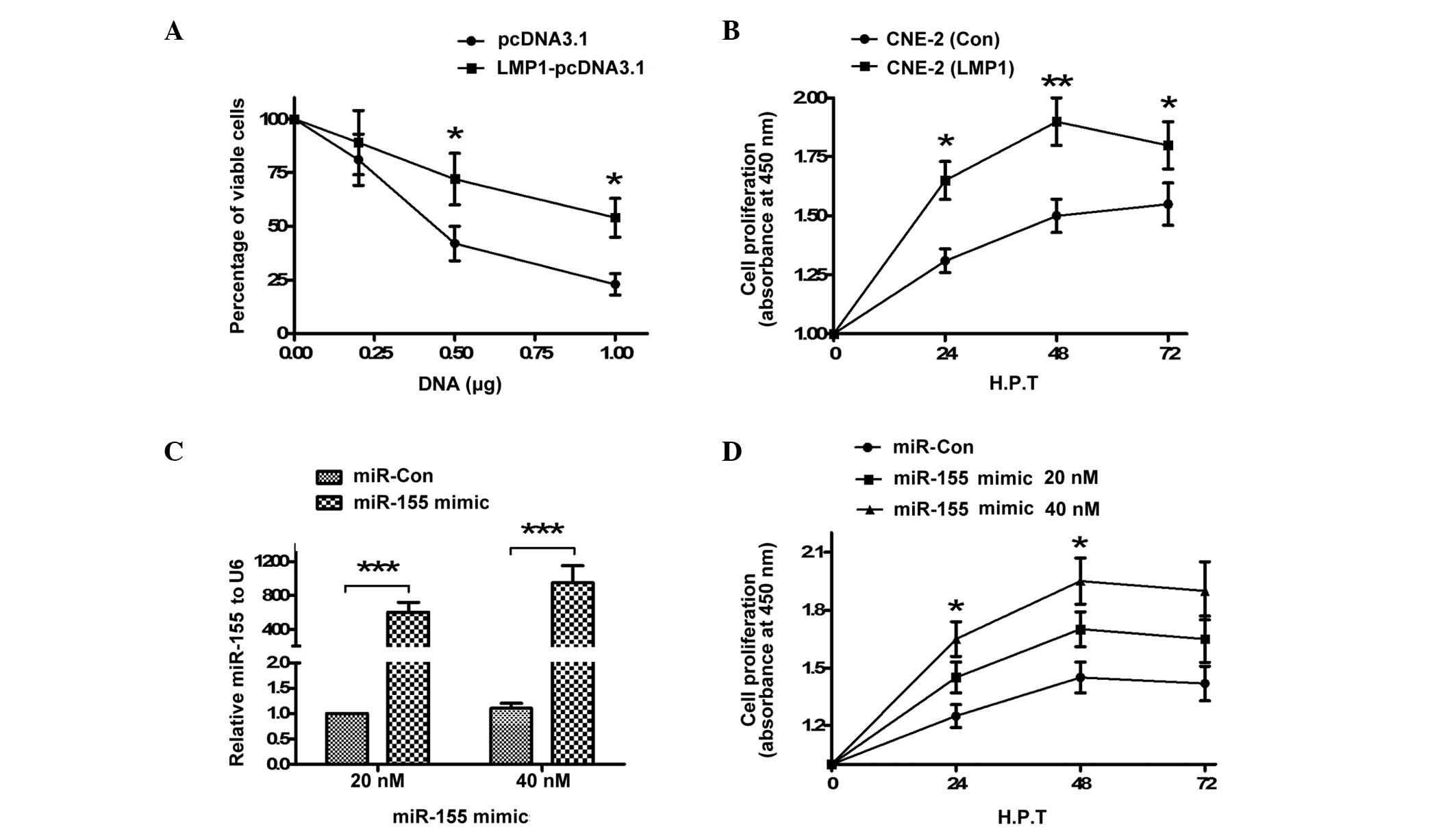
LMP1 overexpression and miR-155 mimic
transfection increase the proliferation of CNE-2 cells
MTT assay was performed in order to assess the
effect of LMP1 overexpression on the viability of the CNE-2 cell
line. For this purpose, cells were transfected with LMP1-pcDNA3.1
or pcDNA3.1 control, according to the manufacturer's protocol,
using a series of concentrations ranging from 0.00 to 1.00 µg. The
viability of LMP1-pcDNA3.1-transfected cells was significantly
increased compared with pcDNA3.1-transfected cells, as shown in
Fig. 2A (P<0.05). Subsequently,
cell proliferation following pcDNA3.1 or LMP1-pcDNA3.1 transfection
was detected by CCK-8 assay in the two groups of CNE-2 cells. As
demonstrated in Fig. 2B, the CNE-2
(LMP1) cells exhibited significantly increased proliferation
compared with the CNE-2 (Con) cells (P<0.05; P<0.01).
Furthermore, the proliferation of CNE-2 cells transfected with
miR-155 mimic or miR-Con was additionally examined. The expression
levels of miR-155 in CNE-2 cells transfected with miR-155 mimic
were significantly increased (P<0.001; Fig. 2C). In addition, CCK-8 assay
demonstrated that transfection with 20 or 40 nM miR-155 mimic
significantly promoted CNE-2 cell proliferation (P<0.05;
Fig. 2D). The results of the present
study confirmed that LMP1 overexpression or miR-155 mimic
transfection promoted the proliferation of CNE-2 cells.
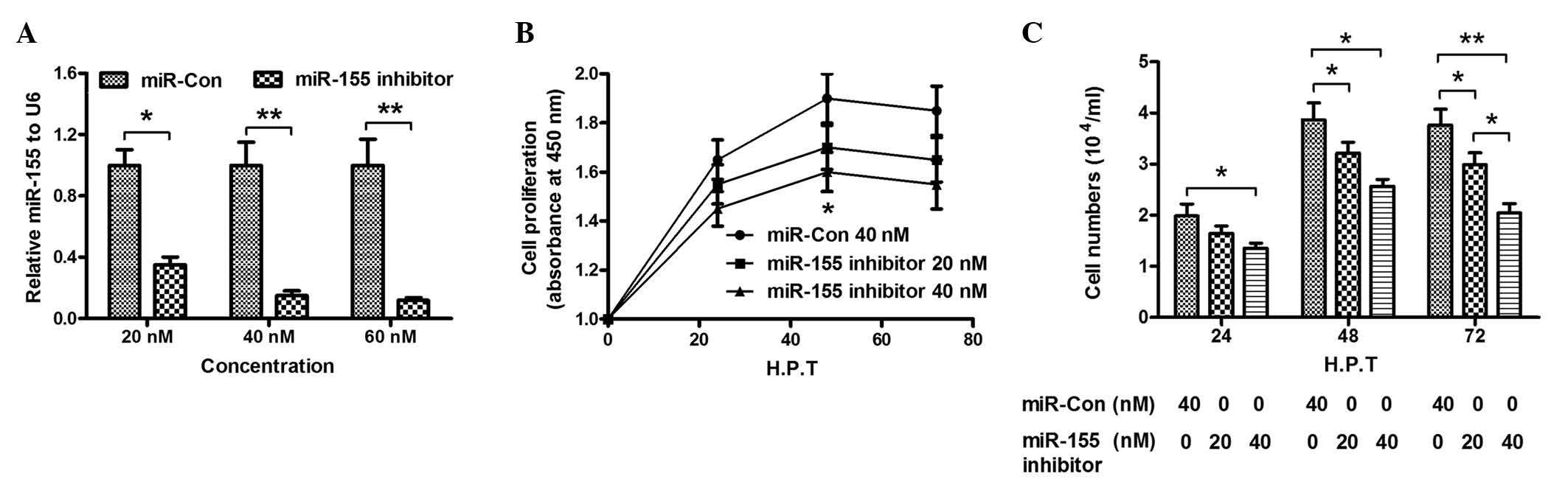
miR-155 knockdown attenuates the
promotion of cell growth in CNE-2 cells caused by overexpression of
LMP1
To further investigate the promotion of CNE-2 cell
growth by miR-155, the expression of miR-155 in CNE-2 cells was
knocked down by addition of miR-155 inhibitor, and the influence of
miR-155 knockdown on the growth of CNE-2 cells was determined. The
results indicated that transfection with 20, 40 or 60 nM miR-155
inhibitor significantly reduced the levels of miR-155 in CNE-2
cells, compared with miR-Con cells (P<0.05; P<0.01; Fig. 3A). Furthermore, cell proliferation
demonstrated a significant decrease upon transfection with miR-155
inhibitor (20 or 40 nM), as shown in Fig.
3B (P<0.05). In addition, the results of CCK-8 assay
indicated that the cell number in the miR-155 inhibitor group was
significantly reduced, compared with the miR-Con group (P<0.05;
P<0.01; Fig. 3C). The results of
the present study confirmed that miR-155 knockdown inhibited the
promotion of proliferation of CNE-2 cells caused by miR-155.
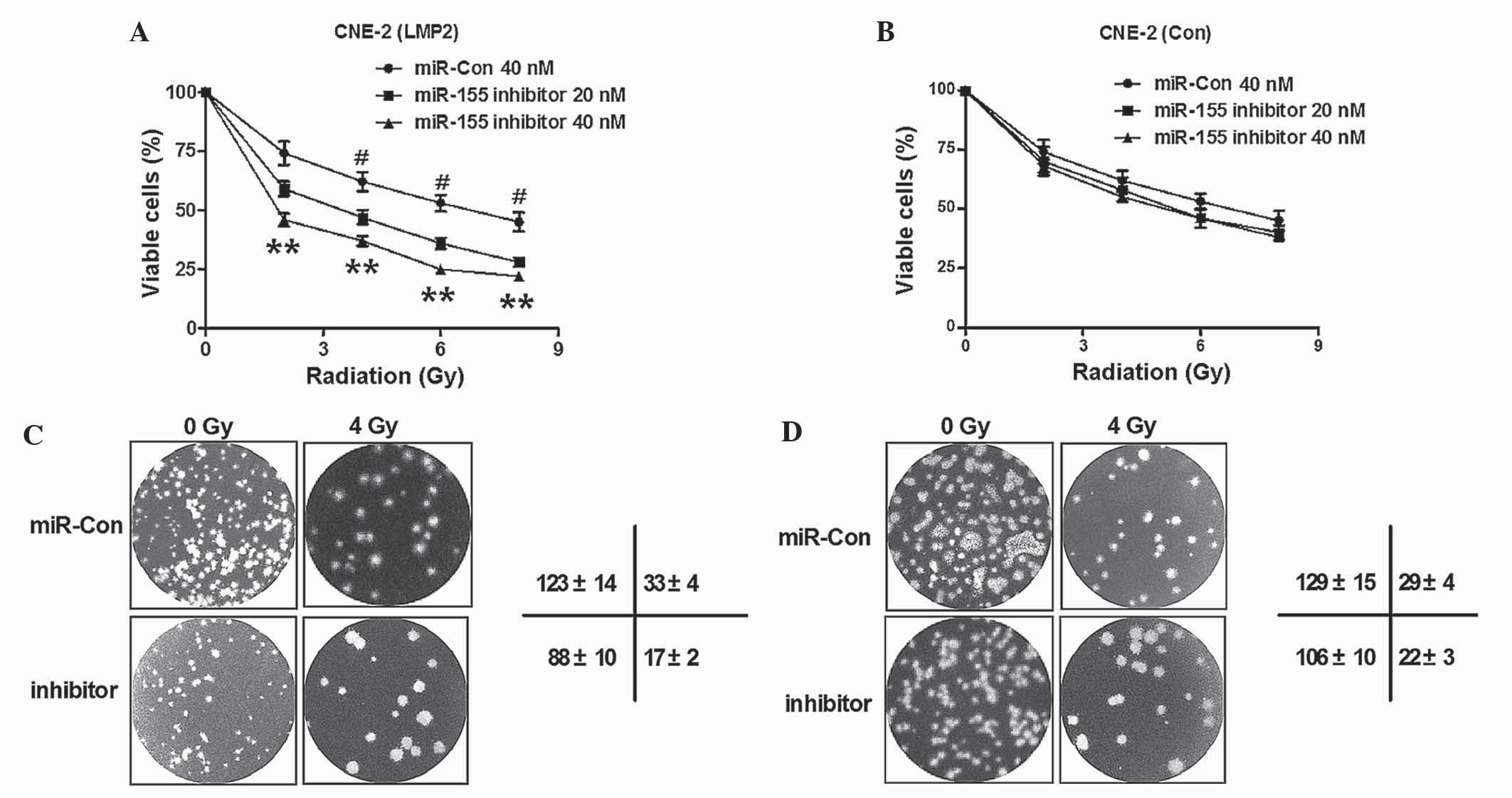
Knockdown of LMP1-induced miR-155
enhances the radiosensitivity of CNE-2 cells
Radiotherapy is widely perceived to be the most
effective treatment for NPC (13,14). In
order to investigate whether miR-155 served as an alternative
target to sensitize NPC cells to radiotherapy, the radiosensitivity
of CNE-2 cells post-miR-155 knockdown was determined. Initially,
the viability of normal CNE-2 cells or CNE-2 (LMP1) cells with
miR-155 knockdown post-radiation was determined. As shown in
Fig. 4A, cell viability was reduced
when miR-155 was blocked in CNE-2 cells overexpressing LMP1
(P<0.05; P<0.01). By contrast, the viability was not
significantly influenced in the CNE-2 (Con) cells post-transfection
with miR-155 inhibitor (P>0.05; Fig.
4B). Furthermore, colony formation assay was performed for
CNE-2 (LMP1) cells and CNE-2 (Con) cells, with or without miR-155
knockdown, following treatment with 0 or 4 Gy radiation. As
demonstrated in Fig. 4C, there were
less colonies formed by CNE-2 (LMP1) cells upon transfection with
miR-155 inhibitor, compared with miR-Con transfected cells (123±14
vs. 88±10 cells, respectively; P<0.01). Furthermore, the colony
number reduction by miR-155 knockdown was more significant in CNE-2
(LMP1) cells transfected with miR-155 inhibitor than in
miR-Con-transfected cells (33±4 vs. 17±2 cells, respectively;
P<0.01). However, the colony number reduction caused by miR-155
knockdown was not significant in CNE-2 (Con) cells, with or without
exposure to 4 Gy radiation (P>0.05; Fig. 4D). Thus, the results of the present
study confirmed that knockdown of LMP1-induced miR-155 decreased
the sensitivity of CNE-2 cells to radiation.
Discussion
EBV infection of primary B cells results in latent
infection, in which a subset of viral genes are expressed (15). There are three types of EBV latent
infection (16); in particular, NPC
correlates with type II latency, in which LMP1 and LMP2 are
overexpressed and contribute to oncogenesis (16). The integral transmembrane protein LMP1
potentiates a variety of signaling pathways, including nuclear
factor-κB, mitogen-activated protein kinase, phosphoinositide
3-kinase/Akt (17) and eukaryotic
translation initiation factor 4E, to promote the proliferation,
migration and invasion of NPC (18).
Previous studies have revealed deregulated miR expression post-EBV
infection in NPC or lymphocytes (9,19,20). Furthermore, a previous study has
recognized the deregulation of miRs by LMP1, including miR-10b
(9). In a previous study by the
present research group, miR-155 was identified to be upregulated by
LMP1 DNA, and contributed to the proliferation and migration of NPC
cells (10).
In the present study, the CNE-2 NPC cell line was
selected to investigate the role of miR-155 induced by LMP1 in the
sensitization of NPC cells to radiotherapy. Initially, the
promotion of miR-155 expression by LMP1 overexpression was
confirmed in CNE-2 cells. Subsequently, it was identified that
overexpression of LMP1 and transfection with miR-155 mimic promoted
the proliferation of CNE-2 cells, according to the results of MTT
and CCK-8 assays. Furthermore, miR-155 knockdown with miR-155
inhibitor attenuated the promotion of CNE-2 cell growth caused by
LMP1, thus confirming the key regulatory role of miR-155 in
LMP1-promoted NPC cell proliferation. Radiotherapy is currently the
most effective treatment for NPC (13,14). The
present study confirmed that miR-155 may serve as a novel target
for the sensitization of NPC cells to radiotherapy. Based on the
results of the cell viability, cell proliferation and colony
formation assays, miR-155 knockdown was confirmed to significantly
enhance the radiosensitivity of CNE-2 cells.
miR-155 is encoded by the MIR155 host gene,
and regulates various physiological and pathological processes
(21), including inflammatory
processes and various signaling pathways in cancer (21). miR-155 has been considered to act as
an oncogene or tumor suppressor, depending on the type of tumor
(22). miR-155 has been observed to
be oncogenic in NPC (10), but
tumor-suppressive in gastric cancer (23). The present study confirmed the
oncogenic role of miR-155 in NPC. Furthermore, the strategy to
knockdown miR-155 has been previously investigated for the control
of cancer progression (24). Previous
studies have demonstrated that knockdown of miR-155 in mice
resulted in rapid regression of lymphadenopathy, in part due to
apoptosis of the malignant lymphocytes (25). In addition, systemic delivery of
antisense peptide nucleic acids against miR-155 that were
encapsulated in unique polymer nanoparticles inhibited miR-155 and
retarded the growth of pre-B-cell tumors in vivo, suggesting
a potential therapeutic option for the treatment of
lymphoma/leukemia (25). In the
present study, it was identified that knockdown of miR-155
inhibited NPC cell growth and sensitized NPC cells to radiotherapy
in vitro. These results suggest that downregulation of
miR-155 may provide a novel approach for the treatment of NPC.
In conclusion, the present study confirmed the
oncogenic role of miR-155 in NPC, and demonstrated that knockdown
of miR-155 inhibited the growth of NPC cells and additionally
sensitized these cells to radiotherapy.
Acknowledgements
The present study was supported by a grant from the
Development and Reform Commission of Jilin Province (Changchun,
China; grant no. 3J113Z363428).
References
|
1
|
Guo X, O'Brien SJ, Zeng Y, Nelson GW and
Winkler CA: GSTM1 and GSTT1 gene deletions and the risk for
nasopharyngeal carcinoma in Han Chinese. Cancer Epidemiol
Biomarkers Prev. 17:1760–1763. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wolf H, zur Hausen H and Becker V: EB
viral genomes in epithelial nasopharyngeal carcinoma cells. Nat New
Biol. 244:245–247. 1973. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu Y, Shi Y, Yuan Q, Liu X, Yan B, Chen L,
Tao Y and Cao Y: Epstein-Barr Virus encoded LMP1 regulates cyclin
D1 promoter activity by nuclear EGFR and STAT3 in CNE1 cells. J Exp
Clin Cancer Res. 32:902013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao Y, Wang Y, Zeng S and Hu X: LMP1
expression is positively associated with metastasis of
nasopharyngeal carcinoma: Evidence from a meta-analysis. J Clin
Pathol. 65:41–45. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Niu Q, Qian M, Liu G, Yang F and Teng Y: A
genome-wide identification and characterization of microRNAs and
their targets in ‘Suli’ pear (Pyrus pyrifolia white pear
group). Planta (Sep). 8:2013.(Epub ahead of print).
|
|
7
|
Wang HY, Li YY, Fu S, Wang XP, Huang MY,
Zhang X, Shao Q, Deng L, Zeng MS, Zeng YX and Shao JY: MicroRNA-30a
promotes invasiveness and metastasis in vitro and in
vivo through epithelial-mesenchymal transition and results in
poor survival of nasopharyngeal carcinoma patients. Exp Biol Med
(Maywood). 239:891–898. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu J, Luo H, Liu X, Peng Y, Zhang B, Wang
L, Xu X, Peng X, Li G, Tian W, et al: miR-9 targets CXCR4 and
functions as a potential tumor suppressor in nasopharyngeal
carcinoma. Carcinogenesis. 35:554–563. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li G, Wu Z, Peng Y, Liu X, Lu J, Wang L,
Pan Q, He ML and Li XP: MicroRNA-10b induced by Epstein-Barr
virus-encoded latent membrane protein-1 promotes the metastasis of
human nasopharyngeal carcinoma cells. Cancer Lett. 299:29–36. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhu X, Wang Y, Sun Y, Zheng J and Zhu D:
MiR-155 up-regulation by LMP1 DNA contributes to increased
nasopharyngeal carcinoma cell proliferation and migration. Eur Arch
Otorhinolaryngol. 271:1939–1945. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real time quantitative PCR and
the 2(Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Freimoser FM, Jakob CA, Aebi M and Tuor U:
The MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide] assay is a fast and reliable method for colorimetric
determination of fungal cell densities. Appl Environ Microbiol.
65:3727–3729. 1999.PubMed/NCBI
|
|
13
|
Tang JM, Ma XM, Hou YL, Dai LY, Cao HB, Ye
M and Bai YR: Analysis of simultaneous modulated accelerated
radiotherapy (SMART) for nasopharyngeal carcinomas. J Radiat Res.
55:794–802. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hua YJ, Han F, Lu LX, Mai HQ, Guo X, Hong
MH, Lu TX and Zhao C: Long-term treatment outcome of recurrent
nasopharyngeal carcinoma treated with salvage intensity modulated
radiotherapy. Eur J Cancer. 48:3422–3428. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kaneda A, Matsusaka K, Aburatani H and
Fukayama M: Epstein-Barr virus infection as an epigenetic driver of
tumorigenesis. Cancer Res. 72:3445–3450. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Murata T, Sato Y and Kimura H: Modes of
infection and oncogenesis by the Epstein-Barr virus. Rev Med Virol.
24:242–253. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dawson CW, Tramountanis G, Eliopoulos AG
and Young LS: Epstein-Barr virus latent membrane protein 1 (LMP1)
activates the phosphatidylinositol 3-kinase/Akt pathway to promote
cell survival and induce actin filament remodeling. J Biol Chem.
278:3694–3704. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao Y, Pang TY, Wang Y, Wang S, Kang HX,
Ding WB, Yong WW, Bie YH, Cheng XG, Zeng C, et al: LMP1 stimulates
the transcription of eIF4E to promote the proliferation, migration
and invasion of human nasopharyngeal carcinoma. FEBS J.
281:3004–3018. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Luo Z, Dai Y, Zhang L, Jiang C, Li Z, Yang
J, McCarthy JB, She X, Zhang W, Ma J, et al: miR-18a promotes
malignant progression by impairing microRNA biogenesis in
nasopharyngeal carcinoma. Carcinogenesis. 34:415–425. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu H, Lu J, Zuo L, Yan Q, Yu Z, Li X,
Huang J, Zhao L, Tang H, Luo Z, et al: Epstein-Barr virus
downregulates microRNA 203 through the oncoprotein latent membrane
protein 1: A contribution to increased tumor incidence in
epithelial cells. J Virol. 86:3088–3099. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
O'Connell RM, Rao DS and Baltimore D:
MicroRNA regulation of inflammatory responses. Annu Rev Immunol.
30:295–312. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen Z, Ma T, Huang C, Hu T and Li J: The
pivotal role of microRNA-155 in the control of cancer. J Cell
Physiol. 229:545–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun S, Sun P, Wang C and Sun T:
Downregulation of microRNA-155 accelerates cell growth and invasion
by targeting c-myc in human gastric carcinoma cells. Oncol Rep.
32:951–956. 2014.PubMed/NCBI
|
|
24
|
Mattiske S, Suetani RJ, Neilsen PM and
Callen DF: The oncogenic role of miR-155 in breast cancer. Cancer
Epidemiol Biomarkers Prev. 21:1236–1243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Babar IA, Cheng CJ, Booth CJ, Liang X,
Weidhaas JB, Saltzman WM and Slack FJ: Nanoparticle-based therapy
in an in vivo microRNA-155 (miR-155)-dependent mouse model
of lymphoma. Proc Natl Acad Sci USA. 109:E1695–E1704. 2012.
View Article : Google Scholar : PubMed/NCBI
|