Introduction
Liver cancer is the third most common cause of
cancer-associated mortality worldwide, accounting for an estimated
9.2% of total cancer-associated mortalities in 2008 (1). Surgery is considered the most effective
treatment for patients with hepatocellular carcinoma (HCC)
(2); however, the indications for
surgery are restricted by the size and total number of tumors
(2,3).
Although the 5-year survival rate of patients with HCC has improved
by >30% over the past decade, the recurrence rate following
surgery is estimated to be nearly 50% (4); therefore, systemic chemotherapy is
required for patients with advanced stages of HCC, in order to
prolong their survival.
MicroRNAs (miRNAs) are endogenous non-coding RNAs of
18–22 nucleotides in length (3,5). The
effect of miRNAs on the regulation of the expression of various
genes is so broad that one miRNA controls >200 genes (6). Aberrant expression of miRNAs is a common
feature among various types of human cancer, and has been
reportedly associated with patient survival (7–10).
Regarding the correlation between miRNAs and HCC, several studies
have detected the aberrant expression of specific miRNAs in HCC
tissues when compared with normal tissues (11–14). These
studies indicated that the modulation of non-coding RNAs,
particularly miRNAs, may be a valuable therapeutic target in
HCC.
The aim of the present study was to elucidate the
miRNA profiles that are associated with differentiation and
hepatitis B virus (HBV) infection observed in HCC cell lines. The
characterization of miRNA expression patterns using various
parameters may be a novel approach for the treatment of patients
with HCC.
Materials and methods
Cell lines and culture
The Alex, Hep3B, HepG2, HuH1, HuH7, JHH1, JHH2,
JHH5, JHH6, HLE, HLF and Li-7 HCC cell lines were obtained from the
Japanese Cancer Research Resources Bank (Tokyo, Japan) and
transported to our laboratory. The cell lines were authenticated by
the cell bank using short tandem repeat polymerase chain reaction.
The cells were grown in minimal essential medium (Gibco; Thermo
Fisher Scientific Inc., Waltham, MA, USA) supplemented with 10%
fetal bovine serum (catalog no., 533-69545; Wako Pure Chemical
Industries, Tokyo, Japan) and penicillin (10,000
units/ml)-streptomycin (10,000 µg/ml) (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) in a humidified atmosphere of
5% CO2 at 37°C.
Analysis of microRNA array
Total RNA was extracted from the cancer cell lines
using a miRNeasy Mini kit (Qiagen, Hilden, Germany), according to
the manufacturer's instructions. RNA samples typically showed
A260/280 ratios between 1.9 and 2.1 on an
Agilent 2,100 Bioanalyzer (Agilent Technologies, Santa Clara, CA,
USA).
Following the measurement of the RNA using an RNA
6,000 Nano kit (Agilent Technologies, Tokyo, Japan), the samples
were labeled using a miRCURY Hy3/Hy5 Power Labeling kit (Takara Bio
Inc., Tokyo, Japan) and hybridized onto a human miRNA Oligo chip
(version 19.0; Toray Industries, Inc., Tokyo, Japan). Scanning was
conducted with the 3D-Gene Scanner 3,000 (Toray Industries, Inc.,
Kusatsu, Japan). 3D-Gene extraction software version 1.2 (Toray
Industries, Inc.) was used to read the raw intensity of the image.
To determine the change in miRNA expression between poorly- and
well-differentiated HCC cell lines or HBV-positive and HBV-negative
HCC cell lines, the raw data were analyzed using GeneSpringGX
version 10.0 (Agilent Technologies). Samples were first normalized
to the 28S RNA and the baseline was then corrected to the median of
all samples.
Replicate data were analyzed following their
classification into: i) Poorly- and well-differentiated human HCC
cells, and ii) HBV-positive and -negative human HCC cells, which
were organized by the hierarchical clustering in the GeneSpring
software. For the log2 ratios of the miRNA expression
intensity between two groups, hierarchical clustering was performed
using the furthest neighbor method with the absolute Pearson's
correlation coefficient as a metric. The log2 ratios
were median-centered across each miRNA in a color-coding of the
heat map. The P-value cutoff was set to 0.05. Only changes of
>50% in at least one of the time points for each sample were
considered significant. All of the analyzed data were scaled by
global normalization.
Statistical analysis
All analyses were conducted using the JMP 8.0
software (SAS Institute, Inc., Cary, NC, USA). A paired analysis
between the groups was conducted using a Student's t test.
P<0.05 was used to indicate statistically significant
differences between the groups.
Results
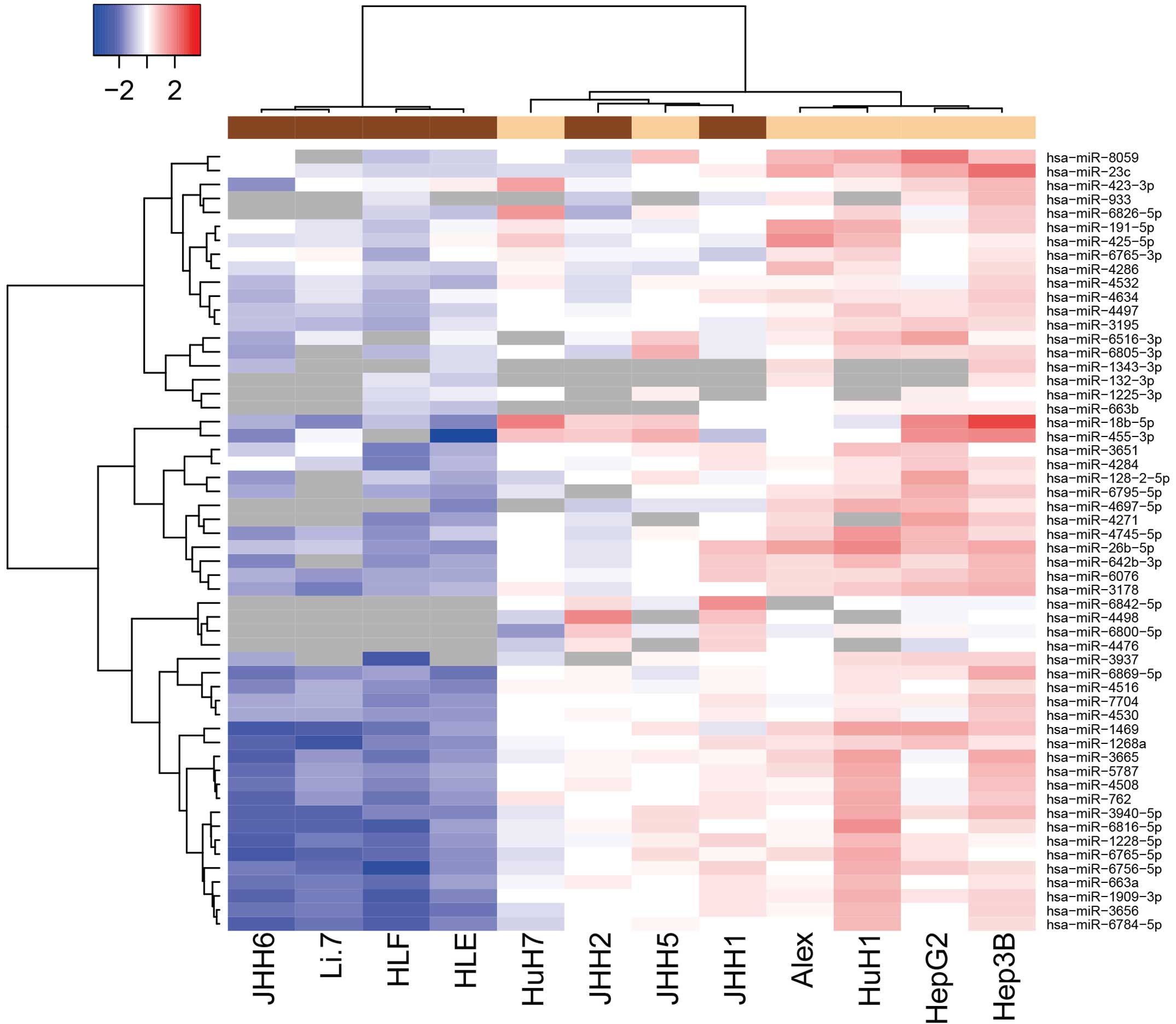
Differences in miRNA expression
between poorly- and well-differentiated human HCC cell lines
Using a custom microarray platform, the expression
levels of 1,719 miRNAs were analyzed in various human HCC cell
lines. As shown in Fig. 1 and
Tables I and II, of the 1,719 miRNAs, 4 were found to be
significantly upregulated and 52 were significantly downregulated
in the poorly-differentiated cells, as compared with the
well-differentiated cells. Unsupervised hierarchical clustering
analysis with Pearson's correlation showed that the
poorly-differentiated HCC cell lines clustered both together and
separately from the well-differentiated HCC cells (Fig. 1).
 | Table I.Upregulated expression of miRNA in
poorly-differentiated HCC cells, as compared with
well-differentiated HCC cells. |
Table I.
Upregulated expression of miRNA in
poorly-differentiated HCC cells, as compared with
well-differentiated HCC cells.
| Upregulated
miRNAs | P-values | PD/WD |
|---|
| hsa-miR-4498 | 0.026931863 | 2.867818244 |
| hsa-miR-6842-5p | 0.025697328 | 2.461117562 |
| hsa-miR-6800-5p | 0.012943201 | 1.941309698 |
| hsa-miR-4476 | 0.030699533 | 1.759074121 |
 | Table II.miRNA downregulation in
poorly-differentiated HCC cells as compared with
well-differentiated HCC cells. |
Table II.
miRNA downregulation in
poorly-differentiated HCC cells as compared with
well-differentiated HCC cells.
| Downregulated
miRNAs | P-value | PD/WD |
|---|
| hsa-miR-3178 | 0.000974376 | 0.348146401 |
| hsa-miR-1469 | 0.001067111 | 0.256078680 |
| hsa-miR-6805-3p | 0.001867476 | 0.415970562 |
| hsa-miR-3195 | 0.001905223 | 0.508164983 |
| hsa-miR-4497 | 0.002391438 | 0.540368285 |
| hsa-miR-4532 | 0.003797834 | 0.570642150 |
| hsa-miR-4745-5p | 0.005612978 | 0.355402751 |
| hsa-miR-6516-3p | 0.008665895 | 0.446770518 |
| hsa-miR-4634 | 0.009685127 | 0.572465267 |
| hsa-miR-8059 | 0.010182045 | 0.344828448 |
| hsa-miR-3940-5p | 0.010700356 | 0.359070586 |
| hsa-miR-1909-3p | 0.011248665 | 0.401354333 |
| hsa-miR-6795-5p | 0.013039021 | 0.381096482 |
| hsa-miR-132-3p* | 0.014002236 | 0.500476019 |
| hsa-miR-26b-5p* | 0.014113042 | 0.357904962 |
| hsa-miR-6765-5p | 0.014456214 | 0.375871481 |
| hsa-miR-4516 | 0.015208421 | 0.529129777 |
| hsa-miR-1268a | 0.015252062 | 0.430113531 |
| hsa-miR-1225-3p | 0.015675303 | 0.690060509 |
| hsa-miR-191-5p | 0.017084147 | 0.504203776 |
| hsa-miR-5787 | 0.019066417 | 0.426932073 |
| hsa-miR-3665 | 0.019620817 | 0.372159995 |
| hsa-miR-6784-5p | 0.021959677 | 0.423492063 |
| hsa-miR-762 | 0.022064969 | 0.428794025 |
| hsa-miR-425-5p | 0.022167838 | 0.469352798 |
| hsa-miR-6076 | 0.02338926 | 0.489615399 |
| hsa-miR-4284 | 0.025395945 | 0.607050985 |
| hsa-miR-6816-5p | 0.025658929 | 0.342345756 |
| hsa-miR-6756-5p | 0.026680935 | 0.418869876 |
| hsa-miR-6765-3p | 0.028318318 | 0.681942211 |
| hsa-miR-1343-3p | 0.029279009 | 0.369035375 |
| hsa-miR-4697-5p | 0.029348892 | 0.356689006 |
| hsa-miR-4286 | 0.029548705 | 0.606212249 |
| hsa-miR-3656 | 0.029629729 | 0.441205506 |
| hsa-miR-6869-5p | 0.030683568 | 0.450864492 |
| hsa-miR-455-3p | 0.031351866 | 0.340391294 |
| hsa-miR-933 | 0.032763996 | 0.446918418 |
| hsa-miR-3937 | 0.032949412 | 0.428511322 |
| hsa-miR-663b | 0.033189114 | 0.645740828 |
| hsa-miR-1228-5p | 0.034737307 | 0.468155726 |
| hsa-miR-4508 | 0.035873986 | 0.465234100 |
| hsa-miR-23c | 0.038312747 | 0.398936550 |
|
hsa-miR-642b-3p | 0.03853801 | 0.474336719 |
| hsa-miR-4530 | 0.03858166 | 0.564198369 |
| hsa-miR-4271 | 0.039079038 | 0.375946014 |
| hsa-miR-18b-5p | 0.046265132 | 0.239201813 |
| hsa-miR-663a | 0.046711854 | 0.497938251 |
| hsa-miR-7704 | 0.046961909 | 0.562374269 |
|
hsa-miR-6826-5p | 0.048141645 | 0.450205273 |
| hsa-miR-3651 | 0.048451237 | 0.591375188 |
| hsa-miR-423-3p | 0.04891406 | 0.585477071 |
|
hsa-miR-128-2-5p | 0.049142734 | 0.487264349 |
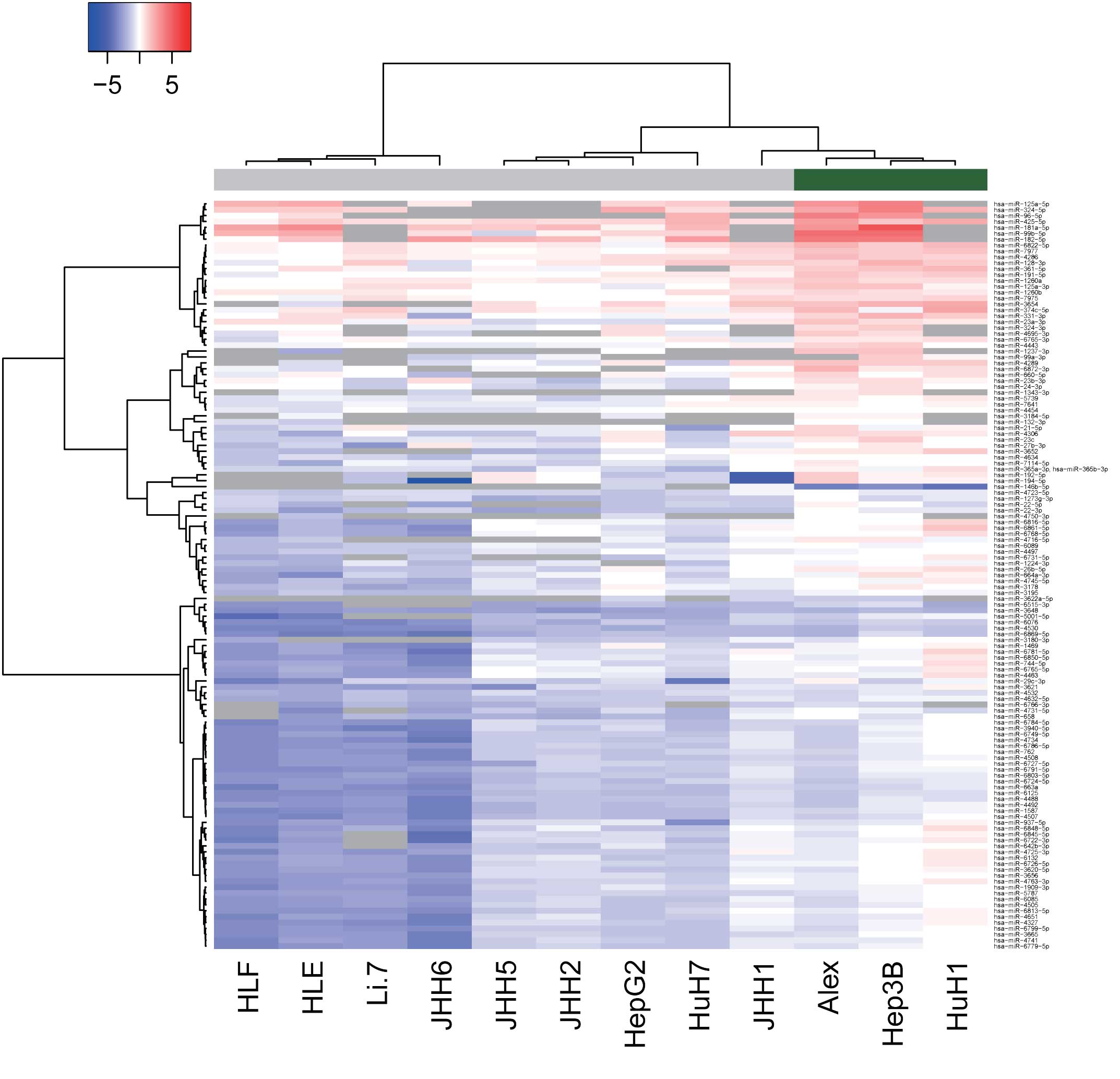
Differences in miRNA expression
between HBV-positive and HBV-negative HCC lines
To examine the effect of HBV infection on
alterations in miRNAs, miRNA profiles were analyzed in HBV-positive
and -negative human HCC cell lines. As shown in Fig. 2 and Tables
III and IV, of the 1,719 miRNAs,
125 miRNAs were found to be significantly upregulated and 2 were
significantly downregulated in the HBV-positive HCC cells, as
compared with the HBV-negative HCC cells. Unsupervised hierarchical
clustering analysis with Pearson's correlation showed that the
HBV-positive HCC cell lines clustered both together and separately
from the HBV-negative HCC cells (Fig.
2).
 | Table III.miRNA upregulation in HBV-positive
HCC cells as compared with HBV-negative HCC cells. |
Table III.
miRNA upregulation in HBV-positive
HCC cells as compared with HBV-negative HCC cells.
| Upregulated
miRNAs | P-value | HBV(+)/HBV(−) |
|---|
| hsa-miR-99b-5p | 0.000000648 | 9.941304892 |
|
hsa-miR-181a-5p | 0.033034307 | 6.386379599 |
| hsa-miR-96-5p | 0.021545231 | 4.758785302 |
|
hsa-miR-1237-3p | 0.037257957 | 4.456964302 |
| hsa-miR-182-5p | 0.000177429 | 4.362818593 |
|
hsa-miR-125a-5p | 0.005613398 | 3.719262548 |
| hsa-miR-99a-3p | 0.045954557 | 3.494856992 |
|
hsa-miR-6861-5p | 0.008595301 | 3.308580171 |
|
hsa-miR-6726-5p | 0.002146572 | 3.247738683 |
|
hsa-miR-4763-3p | 0.003813145 | 3.075732768 |
| hsa-miR-192-5p | 0.018546977 | 3.068893554 |
| hsa-miR-194-5p | 0.027213074 | 3.058433608 |
| hsa-miR-324-5p | 0.047875781 | 3.025508821 |
| hsa-miR-3665 | 0.000663551 | 3.021045838 |
|
hsa-miR-6848-5p | 0.013784248 | 3.003340126 |
| hsa-miR-658 | 0.002645683 | 2.977949153 |
| hsa-miR-3652 | 0.006332146 | 2.969527506 |
| hsa-miR-744-5p | 0.00486586 | 2.967774559 |
| hsa-miR-6132 | 0.00428827 | 2.930151490 |
| hsa-miR-26b-5p | 0.001262254 | 2.924882610 |
|
hsa-miR-6872-3p | 0.013248885 | 2.880715544 |
|
hsa-miR-6813-5p | 0.008803711 | 2.879469305 |
| hsa-miR-4289 | 0.001809926 | 2.859805967 |
|
hsa-miR-3620-5p | 0.004247363 | 2.837639155 |
|
hsa-miR-6779-5p | 0.007205138 | 2.789908382 |
|
hsa-miR-6799-5p | 0.006060554 | 2.738799039 |
| hsa-miR-1587 | 0.005536711 | 2.737246496 |
| hsa-miR-23c | 0.007181466 | 2.716255574 |
|
hsa-miR-4725-3p | 0.026222737 | 2.713962628 |
|
hsa-miR-1343-3p | 0.029279009 | 2.709767325 |
| hsa-miR-937-5p | 0.016562749 | 2.704056512 |
| hsa-miR-22-5p | 0.012966623 | 2.690740441 |
|
hsa-miR-6816-5p | 0.01716898 | 2.690475807 |
|
hsa-miR-6781-5p | 0.043635552 | 2.662786094 |
|
hsa-miR-6768-5p | 0.009273849 | 2.658821540 |
| hsa-miR-331-3p | 0.001849624 | 2.656822543 |
| hsa-miR-4327 | 0.021811096 | 2.620193911 |
|
hsa-miR-6727-5p | 0.006674229 | 2.597922456 |
|
hsa-miR-6722-3p | 0.024746989 | 2.593107400 |
| hsa-miR-5787 | 0.001467763 | 2.579963522 |
| hsa-miR-29c-3p | 0.016910541 | 2.559268566 |
| hsa-miR-22-3p | 0.000110838 | 2.545731351 |
| hsa-miR-3654 | 0.007328507 | 2.536585596 |
| hsa-miR-4507 | 0.013990723 | 2.509463513 |
| hsa-miR-4492 | 0.005952616 | 2.502548594 |
| hsa-miR-4741 | 0.009337902 | 2.486124356 |
| hsa-miR-3621 | 0.015640016 | 2.485247656 |
| hsa-miR-4734 | 0.018933717 | 2.483317331 |
| hsa-miR-6085 | 0.014443652 | 2.479844683 |
| hsa-miR-361-5p | 0.000442472 | 2.474136455 |
|
hsa-miR-5001-5p | 0.017579881 | 2.466921307 |
|
hsa-miR-6845-5p | 0.018960866 | 2.444791599 |
| hsa-miR-4651 | 0.031451007 | 2.433852833 |
|
hsa-miR-664a-3p | 0.022326221 | 2.417433070 |
|
hsa-miR-6850-5p | 0.043000995 | 2.410122248 |
|
hsa-miR-3940-5p | 0.016650066 | 2.406661676 |
|
hsa-miR-4750-3p | 0.034041218 | 2.405803427 |
|
hsa-miR-4716-5p | 0.026554669 | 2.368757616 |
| hsa-miR-365a,
b-3p | 0.002715735 | 2.366494444 |
| hsa-miR-4508 | 0.007102476 | 2.352557384 |
| hsa-miR-191-5p | 0.000120655 | 2.346171925 |
|
hsa-miR-6731-5p | 0.024175847 | 2.333212883 |
|
hsa-miR-6822-5p | 0.001204907 | 2.331567699 |
|
hsa-miR-4745-5p | 0.015938518 | 2.330093060 |
| hsa-miR-1469 | 0.039665847 | 2.329351564 |
| hsa-miR-762 | 0.010944843 | 2.327308371 |
| hsa-miR-4505 | 0.021004934 | 2.322786379 |
| hsa-miR-3656 | 0.012652065 | 2.316840605 |
|
hsa-miR-374c-5p | 0.046201574 | 2.309229277 |
| hsa-miR-4306 | 0.048384309 | 2.306213437 |
| hsa-miR-4463 | 0.043512115 | 2.274070423 |
|
hsa-miR-6749-5p | 0.034695635 | 2.258437039 |
| hsa-miR-425-5p | 0.005354369 | 2.258230321 |
|
hsa-miR-1909-3p | 0.015268122 | 2.256696389 |
| hsa-miR-4443 | 0.002035551 | 2.247501172 |
|
hsa-miR-6784-5p | 0.020580101 | 2.233472310 |
|
hsa-miR-6791-5p | 0.025022751 | 2.227203891 |
|
hsa-miR-6765-5p | 0.03907315 | 2.196934984 |
|
hsa-miR-4695-3p | 0.049035558 | 2.196882473 |
|
hsa-miR-4731-5p | 0.026948571 | 2.153748402 |
| hsa-miR-324-3p | 0.008210293 | 2.148721766 |
| hsa-miR-7977 | 0.000154283 | 2.145089801 |
| hsa-miR-3178 | 0.022352 | 2.127295883 |
|
hsa-miR-642b-3p | 0.020856928 | 2.086744300 |
|
hsa-miR-6786-5p | 0.044762899 | 2.078505858 |
|
hsa-miR-6869-5p | 0.038412235 | 2.071725430 |
| hsa-miR-663a | 0.030021104 | 2.045834444 |
| hsa-miR-4488 | 0.028003788 | 2.038574665 |
|
hsa-miR-7114-5p | 0.007743122 | 2.030825064 |
|
hsa-miR-3180-3p | 0.025916357 | 2.023796991 |
| hsa-miR-6125 | 0.032086556 | 2.008344893 |
| hsa-miR-21-5p | 0.049645958 | 2.003677073 |
|
hsa-miR-125a-3p | 0.011495522 | 1.999755015 |
|
hsa-miR-4632-5p | 0.009562923 | 1.999301680 |
| hsa-miR-132-3p | 0.014002236 | 1.998097736 |
| hsa-miR-23b-3p | 0.022475719 | 1.997567414 |
| hsa-miR-27b-3p | 0.042591151 | 1.976679667 |
|
hsa-miR-6515-3p | 0.043906577 | 1.956257303 |
|
hsa-miR-6803-5p | 0.046392317 | 1.928556399 |
|
hsa-miR-6724-5p | 0.044214734 | 1.926601244 |
| hsa-miR-4286 | 0.000877998 | 1.922172274 |
| hsa-miR-660-5p | 0.029655234 | 1.920831210 |
| hsa-miR-23a-3p | 0.039960215 | 1.913813878 |
|
hsa-miR-1273g-3p | 0.008489419 | 1.887689346 |
| hsa-miR-6076 | 0.046682307 | 1.869956757 |
|
hsa-miR-3184-5p | 0.002114973 | 1.859834906 |
| hsa-miR-24-3p | 0.014737629 | 1.853159621 |
|
hsa-miR-1224-3p | 0.024802116 | 1.852687602 |
| hsa-miR-128-3p | 0.034442661 | 1.852193951 |
| hsa-miR-5739 | 0.015007524 | 1.813507960 |
|
hsa-miR-6766-3p | 0.015357929 | 1.810550072 |
| hsa-miR-6089 | 0.01949948 | 1.777100276 |
| hsa-miR-4530 | 0.045923322 | 1.741259423 |
| hsa-miR-4497 | 0.010048462 | 1.733250168 |
| hsa-miR-4634 | 0.011175767 | 1.733040523 |
| hsa-miR-3195 | 0.048689395 | 1.633334528 |
| hsa-miR-4532 | 0.036380915 | 1.573156813 |
|
hsa-miR-6765-3p | 0.008380175 | 1.570194232 |
| hsa-miR-3648 | 0.018858947 | 1.557642234 |
|
hsa-miR-4723-5p | 0.013307402 | 1.556739349 |
| hsa-miR-1260a | 0.009160577 | 1.550976854 |
| hsa-miR-7641 | 0.040903303 | 1.528537769 |
| hsa-miR-7975 | 0.033849532 | 1.527016010 |
| hsa-miR-1260b | 0.037598062 | 1.399160141 |
| hsa-miR-4454 | 0.044853364 | 1.361601255 |
 | Table IV.miRNA downregulation in HBV-positive
HCC cells as compared with HBV-negative HCC cells. |
Table IV.
miRNA downregulation in HBV-positive
HCC cells as compared with HBV-negative HCC cells.
| Downregulated
miRNAs | P-value | HBV(+)/HBV(−) |
|---|
|
hsa-miR-146b-5p | 0.012478369 | 0.125399188 |
|
hsa-miR-3622a-5p | 0.025065220 | 0.861532186 |
Discussion
The aim of the present study was to elucidate the
targetable miRNAs associated with the etiology, diagnosis and
treatment of HCC. Certain miRNAs, such as miR-26b and miR-132, were
found to be downregulated in poorly-differentiated HCC. It has
recently been reported that dedifferentiation is involved in the
epithelial-mesenchymal transition (EMT), and particularly in the
EMT of cancer (15). In order to
invade and metastasize to different organs, cancer cells shed their
differentiated epithelial phenotype through EMT (15), which suggests that miR-26b or miR-132
may be associated with cancer invasion and metastasis via EMT. In
addition, miR-26b has been shown to directly suppress the
expression of CDK6 and cyclin E1, resulting in reduced
retinoblastoma-associated protein phosphorylation and inhibited
cell proliferation (16). miR-132
also inhibits tumor cell proliferation, invasion and migration by
targeting Sox5 (17). These studies
also indicated that miR-26b and miR-132 may directly inhibit cancer
invasion and metastasis.
In the present study, miR-4476 was upregulated in
poorly-differentiated carcinoma. Recently, it has been demonstrated
that miR-4476 is one of the top 10 validated miRNA markers
differentiating pancreatobiliary cancer from other clinical
conditions, including other types of cancer and healthy controls
(18). Therefore, this result
suggests that advanced stages of HCC, which includes
poorly-differentiated cells, induce cholestasis in a similar
fashion to pancreatobiliary cancers and may increase the miR-4476
upregulation.
Regarding the effect of HBV, miR-99b was found to be
upregulated in HBV-infected HCC cells in the present study. It has
been reported that the expression of miR-99b is associated with the
presence of lymph node metastasis (19). In addition, certain miRNAs are
associated with the oncogenic processes of HBV-related HCC
(3). This data indicates that miRNAs
play an important role in the etiology of HBV-related HCC.
In addition, Wang et al (20) demonstrated that 10 upregulated miRNAs
(miR-217, miR-518b, miR-517c, miR-520g, miR-519a, miR-522,
miR-518e, miR-525-3p, miR-512-3p, and miR-518a-3p) and 11
downregulated miRNAs (miR-138, miR-214, miR-214, miR-199a-5p,
miR-433, miR-511, miR-592, miR-483-5p, miR-483-3p, miRNA-708 and
miRNA-1275) were identified in HBV-associated HCC tissues. In the
present study, the same microRNAs were not detected in HBV-positive
HCC cells; therefore, adjacent normal tissues may be included in
the human HCC tissues. These results indicate that the microRNA
expression patterns are different from cancer cell lines and cancer
tissues. Cell-cell interaction may affect microRNA expression in
the microenvironment of cancer tissues.
In conclusion, changes in the regulation of key
miRNAs due to differentiation and HBV infection were observed in
human HCC cell lines. The present findings suggested that
differences in miRNA expression may serve as a novel marker that
can aid in elucidating the etiology of human HCC and assist in
designing treatments.
Glossary
Abbreviations
Abbreviations:
|
miR/miRNA
|
microRNA
|
|
HBV
|
hepatitis virus B
|
|
HCC
|
hepatacellular carcinoma
|
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Małkowski P, Pacholczyk M, Łagiewska B,
Adadyński L, Wasiak D, Kwiatkowski A, Chmura A and Czerwiński J:
Hepatocellular carcinoma-epidemiology and treatment. Przegl
Epidemiol. 60:731–740. 2006.PubMed/NCBI
|
|
3
|
Belghiti J and Kianmanesh R: Surgical
treatment of hepatocellular carcinoma. HPB (Oxford). 7:42–49. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee PH, Lin WJ, Tsang YM, Hu RH, Sheu JC,
Lai MY, Hsu HC, May W and Lee CS: Clinical management of recurrent
hepatocellular carcinoma. Ann Surg. 222:670–676. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Masaki T: MicroRNA and hepatocellular
carcinoma. Hepatol Res. 39:751–752. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro- RNA genes miR15
and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad
Sci USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Michael MZ, O'Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
9
|
Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner
MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ and Schmittgen
TD: Expression profiling identifies microRNA signature in
pancreatic cancer. Int J Cancer. 120:1046–1054. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
et al: Reduced expression of the let-7 microRNAs in human lung
cancers in association with shortened postoperative survival.
Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gramantieri L, Ferracin M, Fornari F,
Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E,
Grazi GL, et al: Cyclin G1 is a target of miR-122a, a microRNA
frequently down-regulated in human hepatocellular carcinoma. Cancer
Res. 67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wong QW, Lung RW, Law PT, Lai PB, Chan KY,
To KF and Wong N: MicroRNA-223 is commonly repressed in
hepatocellular carcinoma and potentiates expression of Stathmin1.
Gastroenterology. 135:257–269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Varnholt H, Drebber U, Schulze F,
Wedemeyer I, Schirmacher P, Dienes HP and Odenthal M: MicroRNA gene
expression profile of hepatitis C virus-associated hepatocellular
carcinoma. Hepatology. 47:1223–1232. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li L and Li W: Epithelial-mesenchymal
transition in human cancer: Comprehensive reprogramming of
metabolism, epigenetics, and differentiation. Pharmacol Ther.
150:33–46. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu Y, Lu Y, Zhang Q, Liu JJ, Li TJ, Yang
JR, Zeng C and Zhuang SM: MicroRNA-26a/b and their host genes
cooperate to inhibit the G1/S transition by activating the pRb
protein. Nucleic Acids Res. 40:4615–4625. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Renjie W and Haiqian L: MiR-132, miR-15a
and miR-16 synergistically inhibit pituitary tumor cell
proliferation, invasion and migration by targeting Sox5. Cancer
Lett. 356:568–578. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kojima M, Sudo H, Kawauchi J, Takizawa S,
Kondou S, Nobumasa H and Ochiai A: MicroRNA markers for the
diagnosis of pancreatic and biliary-tract cancers. PLoS One.
10:e01182202015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Feber A, Xi L, Pennathur A, Gooding WE,
Bandla S, Wu M, Luketich JD, Godfrey TE and Litle VR: MicroRNA
prognostic signature for nodal metastases and survival in
esophageal adenocarcinoma. Ann Thorac Surg. 91:1523–1530. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang W, Zhao LJ, Tan YX, Ren H and Qi ZT:
Identification of deregulated miRNAs and their targets in hepatitis
B virus-associated hepatocellular carcinoma. World J Gastroenterol.
18:5442–5453. 2012. View Article : Google Scholar : PubMed/NCBI
|