Introduction
Carcinoma of the exocrine pancreas or pancreatic
cancer (PC) is one of the most aggressive solid tumors (1,2). At
diagnosis, only 10–15% of patients are amenable for surgical
resection, and even for this selected group of patients, prognosis
is poor, with a 5-year survival of 15–40% (1,2). Ampullary
tumors are also aggressive, although to a lesser extent (3). Intraductal papillary mucinous neoplasms
(IPMNs) are a distinct subset of tumors that are increasingly
recognized and may be cured if they are timely detected (3).
Early detection of PC is the best and currently only
option to improve its dismal prognosis (4). The high frequency (75–100%) of
Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations
in PC (5,6), has lead to extensive research aimed at
establishing their role as diagnostic markers for this disease.
However, when assessing pancreatic juice, its clinical utility is
hampered by a lack of specificity (7–9) linked to
the detection of KRAS mutations in a significant proportion
of patients with chronic pancreatitis (CP) (8,10). In
fact, KRAS mutations have been detected in variable
proportion (0–60%) in ductal lesions in CP samples and even in
normal appearing ducts (10).
Pancreatic juice can be considered a good surrogate
of the status of the pancreatic duct epithelium, since it contains
exfoliated cells from all areas of the pancreas (10). In this regard, genetic alterations can
be more easily detected in DNA extracted from pancreatic juice
compared with tissue blocks (10).
This may well reflect a field effect within the pancreas or, when
the tumor is already evident, the possibility that exfoliated cells
only represent part of a tumor that is intrinsically heterogeneous
(11).
Aberrant promoter methylation of tumor suppressor
genes is a frequent and early event in multiple tumors, including
carcinomas of the pancreas and the periampullary area (12,13). The
use of epigenetic abnormalities as biomarkers is based on their
relative high frequency and the development of methodologies that
can sensitively detect methylation even when cancer cells are a
minority of the cells analyzed (14).
Candidate genes for these purposes should ideally have a high
prevalence of hypermethylation in tumors cells not being present in
the absence of disease. The present authors previously reported
that promoter hypermethylation of engrailed-1 (EN1-), histamine
receptor H2 (HRH2), cadherin 13 (CDH13), adenomatous polyposis coli
(APC) and secreted protein acidic and cysteine rich (SPARC)
genes are frequent events in PC, where they may aid the clinical
assessment of fine-needle aspiration (FNA) biopsies of pancreatic
masses (15).
The aim of the present study was to assess the
prevalence of the promoter methylation detection of the above panel
of genes in pancreatic juice using methylation specific-melting
curve analysis (MS-MCA), a sensitive and robust technique for the
analysis of promoter methylation status (15). The present study evaluated the
performance of each gene separately or as a panel, and compared it
with KRAS mutation detection in patients with CP and with
carcinomas of the pancreas and the periampullary area.
Materials and methods
Patients and samples
Between January 2004 and December 2010, a total of
135 patients undergoing surgical resection at the Hospital
Universitari de Bellvitge (L'Hospitalet de LLobregat, Barcelona)
due to pancreatic disease were prospectively included in a study
aimed to identify novel biomarkers in pancreatic diseases. The
diagnosis was as follows: 85 PCs (9.4% of which were well
differentiated, 75.3% moderately differentiated and 15.3% poorly
differentiated), 26 ampullary carcinomas (ACs) (21 of the
pancreaticobiliary subtype and 5 of the intestinal subtype), 10
IPMNs (2 with invasive carcinoma and 1 with carcinoma in
situ) and 14 CPs. Table I
summarizes the main characteristics of the patients included in the
study. One of the inclusion criteria was obtaining a minimum of 200
ml retrograde pancreatic juice from the main pancreatic duct during
the surgical procedure. In a subset of 20 cases (16 PCs and 4 ACs),
paired biopsies were also analyzed. All patients provided written
informed consent to participate in the study and to have their
biological specimens analyzed. The present study was approved by
the Ethical Committee of the University Hospital of Bellvitge
(Barcelona, Spain).
 | Table I.Main characteristics of the study
population. |
Table I.
Main characteristics of the study
population.
|
Characteristics | PC | AC | IPMN | CP |
|---|
| Patients (n) | 85 | 26 | 10 | 14 |
| Age (years) | 62±12 | 71±8 | 66±14 | 47±12 |
| Gender (M/F) | 44/41 | 17/9 | 8/2 | 12/2 |
| Not applicable |
|
| 7 | 14 |
| Not available | 1 |
|
|
|
| TNM stage |
|
|
|
|
|
T1N0 | 2 | 3 |
|
|
|
T2N0 |
| 9 |
|
|
|
T2N1 |
| 6 |
|
|
|
T3N0 | 14 | 3 | 2 |
|
|
T3N1M0 | 60 | 4 |
|
|
|
T3N1M1 | 3 |
|
|
|
|
T4N1M0 | 4 |
|
|
|
|
T4N1M1 | 1 |
|
|
|
| In situ |
| 1 | 1 |
|
Sample processing and DNA
extraction
The intraoperative pure pancreatic juice (PPJ) was
snap frozen immediately after collection. DNA extraction was
directly performed from the PPJ with no further processing. A
modified standard phenol-chloroform method was used to optimize DNA
extraction (16). DNA extraction from
NP9 and NP18 cell lines (previously generated from pancreatic
ductal adenocarcinoma biopsies in the Translational Research
Laboratory, Duran i Reynals Hospital, Barcelona, Spain) used as
controls and biopsies was performed using the saline method
(17), and the DNA was precipitated
with 2-propanol after mechanical disgregation and overnight
digestion with proteinase K at 53°C.
Methylation assessment
MS-MCA was used (15,18) to
assess the methylation status of EN-1, HRH2, SPARC, APC and
CDH13 gene promoters. A total of 800 ng genomic DNA were
treated with sodium bisulfite according to the manufacturer's
protocol (EZ-DNA Methylation-Gold™ kit; Zymo Research Corporation,
Irvine, CA, USA). Quantitative polymerase chain reaction (PCR) with
temperature dissociation or MCA was used to assess the difference
in melting temperatures between methylated and unmethylated
samples. For APC and CDH13, direct amplification
analysis was used, while for the remaining three genes, a nested
PCR approach was selected. PCR was performed using IMMOLASE™ DNA
Polymerase (Bioline, London, UK). For qPCR, either 25 ng
bisulfite-modified DNA or 1 µl pre-amplified DNA was used as a
template, using a LightCycler® 486 (Roche Applied
Science, Penzberg, Germany). Primer sequences, annealing
temperature and CpG residues targeted in each reaction are detailed
in Table II. Whole lymphocyte DNA
amplified with the REPLI-g kit (Qiagen, Inc., Valencia, CA, USA)
was used as unmethylated control. NP18 or NP9 pancreatic cell lines
were used as positive controls. The primers used did not target CpG
residues amenable for methylation, thus resulting in unbiased
amplification.
 | Table II.DNA hypermethylation and
KRAS
mutations analyses. |
Table II.
DNA hypermethylation and
KRAS
mutations analyses.
| A, Primer sequences
and PCR conditions for DNA hypermethylation analysis |
|---|
|
|---|
| Gen | PCR |
| Annealing
temperature (°C) | CpG (n) | Primers sequence
(5′→3′) |
|---|
| EN-1 | External |
| 52 | 28 | F:
ACTATCCTACTTATAAACTC |
|
|
|
|
|
| R:
AGAATAATAAAGATAAGAGAT |
|
| Nested |
| 54 |
| F:
CTACTTATAAACTCAACCAA |
|
|
|
|
|
| R:
GTTTTAGGGATTTAGAGTTT |
| HRH2 | External |
| 56 | 18 | F:
GGGTGGATTTGGAAAGTGT |
|
|
|
|
|
| R:
TTACTCTACTCATCCCACAA |
|
| Nested |
| 62 |
| F:
GGGTGGATTTGGAAAGTGT |
|
|
|
|
|
| R:
TCCAAATATCCCCAACAAAA |
| SPARC | External |
| 54 | 16 | F:
TGGAGGGGAGATAGATTTAGTT |
|
|
|
|
|
| R:
AACCAAAAACAAACACAAAAAA |
|
| Nested |
| 58 |
| F:
TTTTGAGTGGTTTTTTGTTGTT |
|
|
|
|
|
| R:
ATCCACCTTCTAAAAAACAACAA |
| APC |
|
| 64 | 16 | F:
GGTTAGGGTTAGGTAGGTTG |
|
|
|
|
|
| R:
CTACACCAATACAACCAC |
| CDH13 |
|
| 65 | 23 | F:
TGATTTGTGAGGTTGAGTTTTAA |
|
|
|
|
|
| R:
ACCCCTCTTCCCTACCTAAAA |
|
| B, KRAS
mutations analyzed, and probe and primer sequences |
|
| Codona | Aminoacid | WT | Mutation | Control cell
line | Probe sequence
(5′→3′) |
|
| 12 | – | GGT | – | NP18 |
TTGGAGCTGGTGGCGTA |
| 12 | G12C | GGT | TGT | MIA PaCa-2 |
TTGGAGCTTGTGGCGTA |
| 12 | G12V | GGT | GTT | SW480 |
TTGGAGCTGTTGGCGTA |
| 12 | G12D | GGT | GAT | NP9 |
TTGGAGCTGATGGCGTA |
| 12 | G12A | GGT | GCT | SW1116 |
TTGGAGCTGCTGGCGTA |
| 12 | G12S | GGT | AGT | A549 |
TAGTTGGAGCTAGTGGCGTA |
| 12 | G12R | GGT | CGT | CAL-62 |
TTGGAGCTCGTGGCGTA |
| 13 | – | GGC | – | NP18 |
CTTGCCTACGCCACCAG |
| 13 | G13D | GGC | GAC | DLD-1 |
CTTGCCTACGTCACCAG |
The analytical sensitivity and robustness of the
method were assessed using serial dilutions of methylated DNA in
increasing amounts of unmethylated DNA (15). For all selected genes, the technique
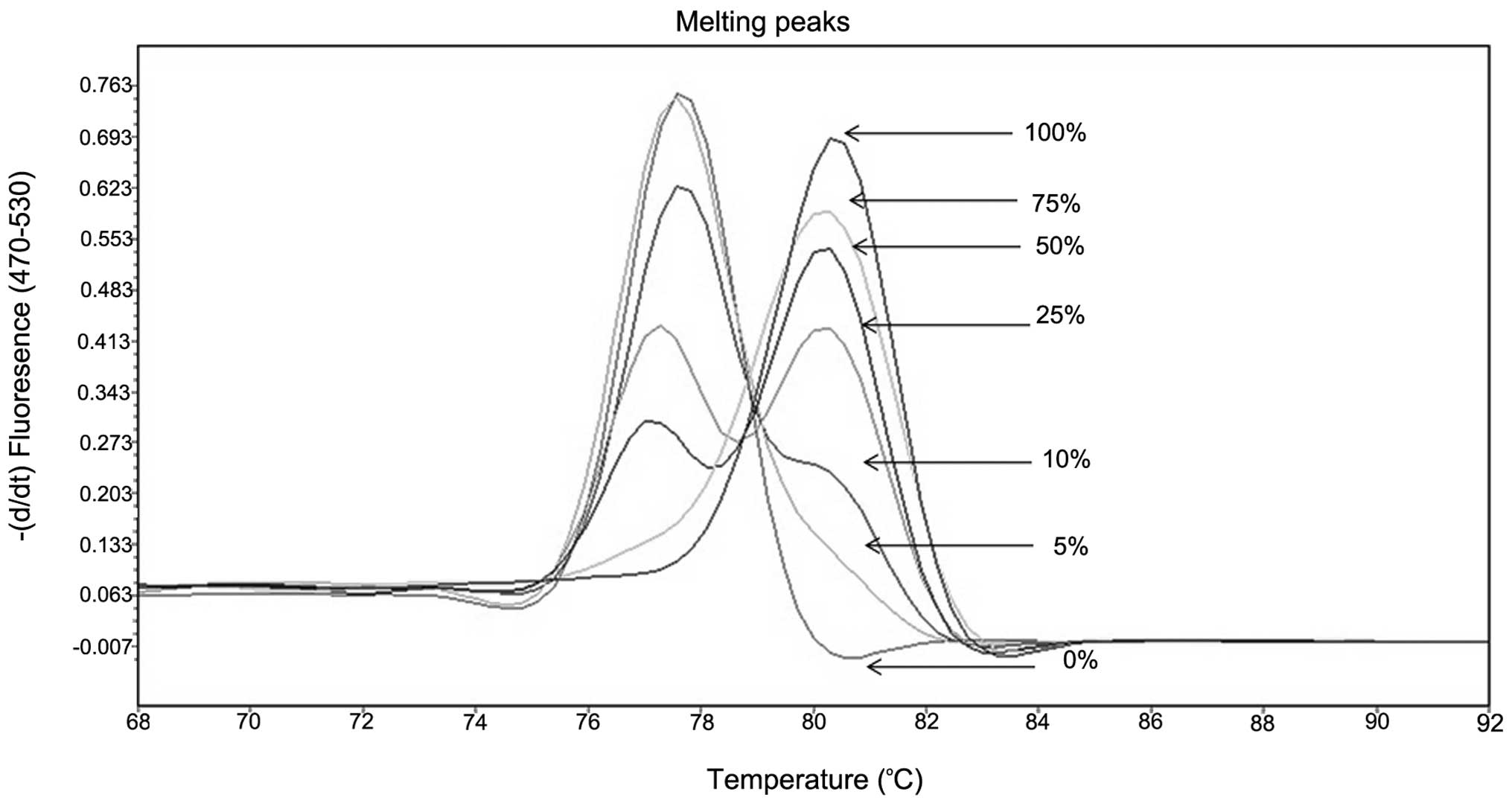
was optimized to have an analytical sensitivity of 5% (Fig. 1). MS-MCA results were compared with
those of direct sequencing of the bisulfite-treated DNA as
previously described (19). All
results were blindly evaluated by M.M.G. and G.C. with 100% of
concordance with the analysis. All analysis depicting the presence
of 5–10% of methylated alleles were repeated, and only when the two
tests yielded the same results, the samples were scored as
methylated.
KRAS mutation detection
KRAS mutation analysis was based on an
allele-specific qPCR assay performed on a Light Cycler®
480 (Roche Applied Science). PCR primers were designed to amplify
the target region, and TaqMan® MGB probes (Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) were
used to screen 7 common mutations in KRAS codons 12 and 13
(Table II). For each allele, one
probe targeted the wild-type variant (tagged with the
VIC® fluorophore; Applied Biosystems; Thermo Fisher
Scientific, Inc.), while the other probe targeted the mutant
variant (tagged with the fluorescein fluorophore). The Light
Cycler® 480 software version 1.5 (Roche Applied Science)
was used to determine the genotype of the sample by measuring the
intensity distribution of the used dyes following PCR
amplification. This technique has an analytical sensitivity of 5%
in a background of wild-type DNA (11).
Statistical analysis
The Fisher's exact test for categorical data was
used to evaluate the association between each methylation marker
and diagnosis. Differences in the prevalence of markers among
groups were assessed by the exact McNemar test. Diagnostic
classifiers from methylation panels were estimated using the random
forests test. Software R version 3.2.0 (https://www.r-project.org/), a language and
environment for statistical computing, was used for all statistical
calculations.
Results
Methylation pattern differs among the
different tumor types
Promoter analysis was informative in the majority of
cases [EN-1, 92% (125/136); HRH2, 95% (129/135);
SPARC 86% (116/135); APC, 98% (132/135); and
CDH13, 90% (122/135)]. Non-informative cases were not
considered. The frequencies of methylation for each marker and
tumor type are detailed in Fig. 2 and
Table III. Out of the 5 markers
analyzed, both APC and HRH2 exhibited the highest
frequency of methylation (71 and 65%, respectively) in PC, while
CDH13, SPARC and EN-1 methylation was detected
at significant albeit lower frequency (57, 49 and 37%,
respectively). In IPMN or AC, APC methylation was identified
at a similar frequency. HRH2 and CDH13 promoters were
less often methylated in AC (P=0.03 and P=0.02, respectively)
(Fig. 2 and Table III).
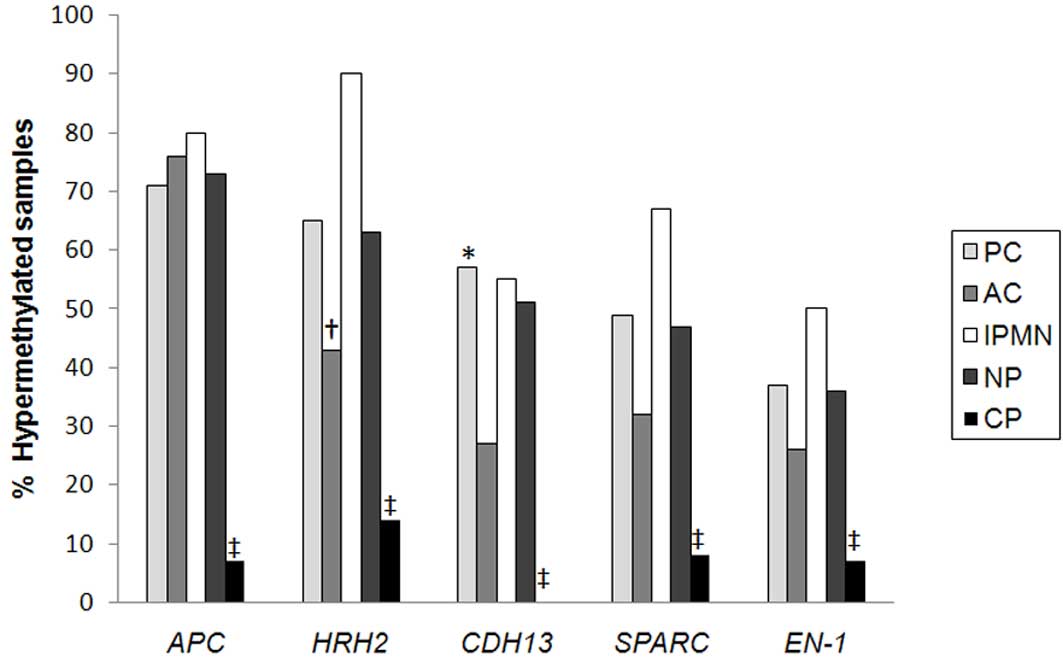 | Figure 2.Proportion of methylated samples
according to sample type. *P<0.05 PC vs. AC;
†P<0.05 AC vs. IPMN; ‡P<0.05 CP vs. PC,
AC and IPMN. PC, pancreatic cancer; AC, ampullary carcinoma; IPMN,
intraductal papillary mucinous neoplasm; NP, neoplasm of pancreatic
area (all types of tumors together); CP, chronic pancreatitis;
APC, adenomatous polyposis coli; HRH2, histamine receptor H2;
CDH13, cadherin 13; EN-1, engrailed-1; SPARC, secreted protein
acidic and cysteine rich. |
 | Table III.Prevalence of all methylation markers
analyzed and KRAS codons 12 and 13 mutations depending on tumor
typea. |
Table III.
Prevalence of all methylation markers
analyzed and KRAS codons 12 and 13 mutations depending on tumor
typea.
| Cancer | Met APC | Met HRH2 | Met
CDH13 | Met
SPARC | Met
EN-1 | ≥2 Met markers | Mut KRAS |
|---|
| PC | 71% | 65% | 57% | 49% | 37% | 72% | 50% |
|
| (59/83) | (53/82) | (44/77) | (35/72) | (29/78) | (61/83) | (41/82) |
| AC | 76% | 43% | 27% | 32% | 26% | 56% | 42% |
|
| (19/25) | (10/23) | (6/22) | (7/22) | (6/23) | (14/25) | (11/26) |
| IPMN | 80% | 90% | 55% | 67% | 50% | 80% | 55% |
|
| (8/10) | (9/10) | (5/9) | (6/9) | (5/10) | (8/10) | (5/9) |
| NP | 73% | 63% | 51% | 47% | 36% | 70% | 47% |
|
| (86/118) | (72/115) | (55/108) | (48/103) | (40/111) | (83/118) | (57/117) |
| CP | 7% | 14% | 0% | 8% | 7% | 7% | 33% |
|
| (1/14) | (2/14) | (0/14) | (1/13) | (1/14) | (1/14) | (4/14) |
The prevalence of methylation in CP was low for all
analyzed markers (0–14%), being only negative for the CDH13
promoter (Fig. 2). In total, 3
samples had 1 marker methylated, while 1 sample had 2 methylated
markers. In spite of the differences observed in methylation
frequencies among the distinct tumor types, it was not possible to
create a classifier that discriminated among them (out-of-bag
estimate of error rate, 34%), indicating that the pattern of
promoter hypermethylation does not predict the site of origin of
the tumor.
As an individual marker, APC yielded the best
performance, with a remarkable sensitivity of 71% and a specificity
of 93% for PC (Table IV). When all
tumors were considered, sensitivity remained high (73%). When all
markers were considered and a threshold of ≥2 markers was
established to score a sample as positive, the panel did not
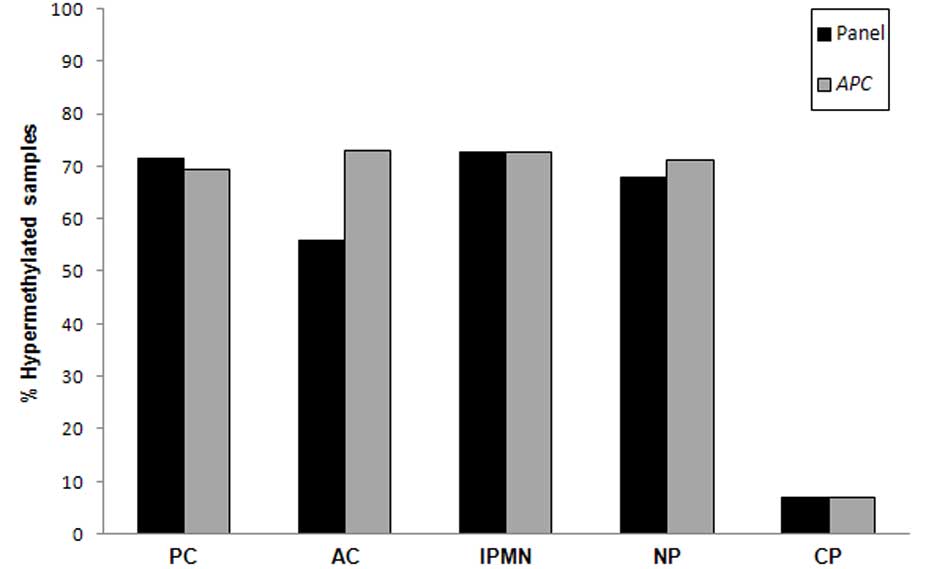
outperform APC hypermethylation as a single marker (Fig. 3) both for PC only or when all tumor
types were considered.
 | Table IV.Sensitivity and specificity of each
methylation marker separately and combined (panel), and sensitivity
and specificity of KRAS mutations detection separately and in
combination with the panel of methylation markers. |
Table IV.
Sensitivity and specificity of each
methylation marker separately and combined (panel), and sensitivity
and specificity of KRAS mutations detection separately and in
combination with the panel of methylation markers.
|
| Sensitivity % (95%
CI) |
|---|
|
|
|
|---|
| Comparison | APC | HRH2 | CDH13 | SPARC | EN-1 | Panel | KRAS |
KRAS+Panel |
|---|
| PC vs. CP | 71% | 65% | 57% | 49% | 37% | 72% | 52% | 78% |
|
| (61–80) | (54–74) | (46–67) | (37–60) | (27–48) | (61–80) |
(41–62)a | (68–85) |
| AC vs. CP | 76% | 43% | 27% | 32% | 26% | 54% | 44% | 69% |
|
| (57–88) | (26–63) | (13–48) | (16–53) | (12–46) | (35–71) | (26–63) | (50-83) |
| IPMN vs. CP | 80% | 90% | 56% | 67% | 50% | 80% | 56% | 80% |
|
| (49–94) | (59–98) | (26–81) | (35–88) | (23–76) | (49–94) | (26–81) | (49–94) |
| NP vs. CP | 73% | 63% | 51% | 47% | 36% | 69% | 50% | 76% |
|
| (64–80) | (53–70) | (41–60) | (37–56) | (27–45) | (60–76) |
(41–59)a |
(68–83)a |
| Specificity | 93% | 86% | 100% | 92% | 93% | 93% | 71% | 79% |
| (95% CI) | (69–99) | (60–96) | (78–100) | (67–99) | (68–98) | (68–99) | (45–88) | (52%-92) |
KRAS
The mutation analysis of KRAS codon 12 was
informative in 134 of 136 (99%) samples, and mutations were
detected in a similar proportion in all tumors (52% PC, 42% AC and
45% IPMN), while the prevalence of mutations in CP was also high
(33%; P>0.05) (Table III). No
mutation was detected at the codon 13 of the KRAS gene. As
expected, the combination of KRAS mutations did not improve
the performance of any individual methylation marker or panel
(Table IV).
APC hypermethylation and KRAS mutation
in paired samples
In order to assess the impact of sampling in the
performance of the test in pancreatic juice, the APC
promoter methylation status was assessed in paired biopsies in a
subset of 20 cases. Regarding the APC methylation status, 11
of 20 (55%) of cases showed concordant results (7 methylated and 4
unmethylated), and in 9 cases, discrepancies were observed. In 6 of
them (30%), the pancreatic juice was methylated, whereas the
corresponding biopsy was not, and in the other 3 cases (15%), the
pancreatic juice was negative in spite of having detected tissue
methylation (data not shown). In all, pancreatic juice analysis
correctly identified 65% of this subset of PCs, which is moderately
higher than the yield of biopsies analyzed (50%). These results
highlight the relevance of sampling when assessing genetic or
epigenetic aberrations in pancreatic juice.
Discussion
The present study has identified APC promoter
hypermethylation as a relevant biomarker for pancreatic juice
assessment when pancreatic and periampullary tumors are suspected.
Previously, APC promoter hypermethylation had been detected at high
frequency in PC biopsies (33–60%) (20–22) or
FNA biopsies (15). In fact,
promoter hypermethylation is likely the most prevalent APC
gene aberration in PC, as loss of heterozygosity and mutations have
been only occasionally detected (23)
[Catalogue of Somatic Mutations in Cancer (COSMIC) database;
http://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=APC].
Of note, APC hypermethylation in pancreatic juice yields a
high sensitivity (71–80%) and specificity (93%) for the
identification of PC in a clinical context where more promising
alterations such as KRAS mutations have failed. The yield is
also good for other periampullary and pancreatic neoplasms, thus
increasing its potential usefulness. In line with the present
observations, APC hypermethylation has been reported in ≤50%
of infiltrative IPMNs (21,24). No previous data are available for
APC promoter methylation in AC, where, in contrast to PC,
APC gene mutations have been detected in ≤42% of cases
(COSMIC database).
Regarding the other markers analyzed in the present
study, HRH2 methylation has also shown a high sensitivity
(65%), which is slightly higher than the 50% observed in the
evaluation of FNA biopsies of pancreatic masses (15). This frequency is similar to that
detected in colorectal carcinoma tissues (25). However, its lower prevalence in AC
diminishes its potential clinical usefulness. According to the
present results, CDH13 was also methylated in ≤50% of PC
juices, in line with the results obtained by Sakai et al
(13). However, while CDH13
methylation was the only marker not being detected in CP, its
relatively low prevalence in AC and IPMN was not high enough to add
diagnostic value. SPARC hypermethylation have been
previously reported at high frequency in PC, IPMN and CP. In the
present study, the prevalence of methylation was still high but
markedly lower (40%) than previously reported (3). Among the markers tested in the present
study, the EN-1 promoter methylation status was the one with
the lowest sensitivity (37%), which is well below the 60%
sensitivity observed in FNA biopsies (15).
EN-1, HRH2, SPARC and CDH13
promoter methylation effectively distinguished between neoplasia
and CP, although, in line with previous observations, 1 of 14
pancreatic juices analyzed exhibited hypermethylation (26). It must be emphasized that
hypermethylation may be detected in normal pancreatic parenchyma
and in non-neoplastic pancreatic juice.
When assessing pancreatic juice, the present panel
did not add information to APC as a single marker. The high
prevalence of APC hypermethylation in all tested tumor
types, together with a low proportion of false positives, account
for the excellent performance of APC. This is in contrast with the
previous study by the present authors evaluating FNA biopsies of
pancreatic masses (15). In that
setting, the panel outperformed any single marker, as previously
described for DNA stool testing for colorectal cancer (27). APC and the other markers analyzed are
methylated not only in PC, but in other tumors arising in the area
(mainly AC or IPMN), thus expanding its potential utility. However,
while certain differences depending on tumor type have been
observed, no evident classifier could be established for a
particular histology.
KRAS mutations at codons 12 and 13 have been
considered good diagnostic markers for PC (5,6) (COSMIC
database; http://cancer.sanger.ac.uk/cosmic/search?q=KRAS).
However, their utility in the assessment of pancreatic juices is
limited due to the relative low prevalence of KRAS mutations
in pancreatic juice and its frequent detection in CP (8–10).
Accordingly, in the present study, KRAS mutations assessment
did not add diagnostic value to APC hypermethylation.
The pancreatic juice provides information from cells
shed from all areas of the pancreatic epithelium, thus yielding a
comprehensive sampling of the target organ (11). However, this may or may not reflect
what occurs in the biopsies of selected areas of the tumor. The
study of paired pancreatic juice-biopsies have revealed a
relatively low degree of concordance between APC methylation
in juice compared with biopsies. Notably, discrepancies go in both
directions (positive in biopsy-negative in juice or vice versa).
Limited representation of pancreatic tumor cells in the juice (as
tumors may not directly shed cells into the juice) is widely
accepted as responsible for false-negative results in pancreatic
juice, which occurred in 3 cases in the present series, whereas 5
cases exhibited reverse results (positive juice-negative biopsy).
Intratumoral heterogeneity of APC status or the presence of
these alterations in non-neoplastic pancreatic ducts may account
for this observation. Irrespective of the cause, in the present
study, APC methylation status still offered a very good
yield in the diagnosis of pancreatic neoplasms. This is in contrast
to the results obtained with highly sensitive KRAS mutation
techniques, where its clinical utility was not obvious (11) due to the high number of KRAS mutations
in CP.
As discussed above, sampling may be a relevant issue
when assessing genetic or epigenetic aberrations in pancreatic
juice. However, the present authors do not consider that the use of
anterograde collection during surgery is worse than retrograde
collection of pancreatic juice during endoscopic retrograde
cholangiopancreatography (ERCP) or collection of duodenal juice
after secretin stimulation. The high yield of APC
hypermethylation and the excellent performance of markers observed
for AC indicate that their relative impact may be less relevant
than anticipated. In fact, the present authors anticipate an even
better performance after secretin stimulation during ERCP, as the
volume collected will certainly increase (26).
Age and chronic inflammation may affect DNA
methylation (28). The well
established association between ageing and hypermeyhylation for
specific genes and tissues makes it necessary to consider age as a
confounder (28). In addition,
persistent inflammation is considered to result in an accelerate
ageing of the affected tissues with increased methylation of
specific markers (29). In the
present limited series, younger cancer patients exhibited higher
methylation levels than those with CP, suggesting that neither
factor is influencing in a significant manner the present
results.
The robustness of the present results may be
partially attributable to the use of MS-MCA, a reliable technique
that assesses the hypermethylation status of all CpG included in
the amplicon analyzed. This technique was selected due to its
simplicity, reproducibility and analytical sensitivity, which
enables the detection of ≤5% of methylated alleles (15). Unlike the often used standard or
quantitative MS-PCR, MS-MCA is dependable (as it uses
methylation-independent primers) and less prone to false positives,
and allows the simultaneous analysis of multiple CpG, thus
providing a more comprehensive picture of the methylation present
in a heterogeneous cell population than MS-PCR.
In conclusion, the present study has observed that
APC promoter hypermethylation is a frequent event in
pancreatic and periampullary neoplasms that may be useful when
assessing pancreatic juice. In the present study, APC
promoter hypermethylation outperformed the other genetic and
epigenetic markers analyzed, based on its high prevalence in all
tumor types evaluated and its great specificity. Differences in the
prevalence of the methylation markers depending on tumor types may
be envisioned, although further studies are required to assess
whether specific methylation profiles may occur in AC and IPMN.
Additionally, the results of the present study highlight the
strengths and weaknesses of pancreatic juice as a surrogate of
tumor biopsies. The present study opens the door to explore whether
APC hypermethylation could be used as a biomarker of
pancreatic and periampullary tumors in less invasive samples such
as duodenal juice or circulating DNA.
Acknowledgements
The present study was supported by grants from the
Carlos III Health Institute, which is seconded to the Ministry of
Economy and Competitiveness (Madrid, Spain) and the European
Regional Development Fund (EDRF) (Brussels, Belgium) (grant nos.
FIS PI060415 and PIE PIE13/00022); Research Foundation in
Gastroenterology (Barcelona, Spain) (grant no. F05-01); the
Ministry of Education and Science (Madrid, Spain) (grant nos.
SAF2011/23638, SAF2015-68016-R and SAF2012-33636); the Thematic
Network of Cooperative Research in Cancer (Madrid, Spain) (grant
no. RD12/0036/0031); the Spanish Association Against Cancer
Scientific Foundation (Madrid, Spain); and the Government of
Catalonia (Barcelona, Spain) (grant nos. 2009 SGR 1356 and 2014 SGR
338). In addition, the current study was co-funded by FEDER
funds/EDRF - a way to build Europe.
Glossary
Abbreviations
Abbreviations:
|
FNA
|
fine-needle aspiration
|
|
qPCR
|
quantitative polymerase chain
reaction
|
|
MCA
|
melting curve analysis
|
|
PC
|
pancreatic cancer
|
|
AC
|
ampullary carcinoma
|
|
IPMN
|
intraductal papillary mucinous
neoplasm
|
|
CP
|
chronic pancreatitis
|
|
PPJ
|
pure pancreatic juice
|
|
APC
|
adenomatous polyposis coli
|
References
|
1
|
Yeo CJ, Cameron JL, Lillemoe KD, Sitzmann
JV, Hruban RH, Goodman SN, Dooley WC, Coleman J and Pitt HA:
Pancreaticoduodenectomy for cancer of the head of the pancreas. 201
patients. Ann Surg. 221:721–731; discussion 731–733. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Busquets J, Fabregat J, Jorba R, Borobia
FG, Valls C, Serrano T, Torras J and Lladó L: Indications and
results of pancreatic surgery preserving the duodenopancreatic
region. Cir Esp. 82:105–111. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hong SM, Kelly D, Griffith M, Omura N, Li
A, Li CP, Hruban RH and Goggins M: Multiple genes are
hypermethylated in intraductal papillary mucinous neoplasms of the
pancreas. Mod Pathol. 21:1499–1507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Majumder S, Chari ST and Ahlquist DA:
Molecular detection of pancreatic neoplasia: Current status and
future promise. World J Gastroenterol. 21:11387–11395. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mu DQ, Peng YS and Xu QJ: Values of
mutations of K-ras oncogene at codon 12 in detection of pancreatic
cancer: 15-year experience. World J Gastroenterol. 10:471–475.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Corbo V, Tortora G and Scarpa A: Molecular
pathology of pancreatic cancer: From bench-to-bedside translation.
Curr Drug Targets. 13:744–752. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mora J, Urgell E, Farré A, Comas L,
Montserrat E and González-Sastre F: Agreement between K-ras
sequence variations detected in plasma and tissue DNA in pancreatic
and colorectal cancer. Clin Chem. 52:1448–1449. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Boadas J, Mora J, Urgell E, Puig P, Roca
M, Cussó X, Capellà G, Lluís F and Farré A: Clinical usefulness of
K-ras gene mutation detection and cytology in pancreatic juice in
the diagnosis and screening of pancreatic cancer. Eur J
Gastroenterol Hepatol. 13:1153–1159. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ha A, Watanabe H, Yamaguchi Y, Ohtsubo K,
Wang Y, Motoo Y, Okai T, Wakabayahi T and Sawabu N: Usefulness of
supernatant of pancreatic juice for genetic analysis of K-ras in
diagnosis of pancreatic carcinoma. Pancreas. 23:356–363. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Löhr M, Müller P, Mora J, Brinkmann B,
Ostwald C, Farré A, Lluis F, Adam U, Stubbe J, Plath F, et al: p53
and K-ras mutations in pancreatic juice samples from patients with
chronic pancreatitis. Gastrointest Endosc. 53:734–743. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Azuara D, Ginesta MM, Gausachs M,
Rodriguez-Moranta F, Fabregat J, Busquets J, Pelaez N, Boadas J,
Galter S, Moreno V, et al: Nanofluidic digital PCR for KRAS
mutation detection and quantification in gastrointestinal cancer.
Clin Chem. 58:1332–1341. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sato N, Fukushima N, Matsubayashi H,
Iacobuzio-Donahue CA, Yeo CJ and Goggins M: Aberrant methylation of
Reprimo correlates with genetic instability and predicts poor
prognosis in pancreatic ductal adenocarcinoma. Cancer. 107:251–257.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sakai M, Hibi K, Koshikawa K, Inoue S,
Takeda S, Kaneko T and Nakao A: Frequent promoter methylation and
gene silencing of CDH13 in pancreatic cancer. Cancer Sci.
95:588–591. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fukushige S and Horii A: Road to early
detection of pancreatic cancer: Attempts to utilize epigenetic
biomarkers. Cancer Lett. 342:231–237. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ginestà MM, Mora J, Mayor R, Farré A,
Peinado MA, Busquets J, Serrano T, Capellá G and Fabregat J:
Genetic and epigenetic markers in the evaluation of pancreatic
masses. J Clin Pathol. 66:192–197. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Müller P, Jesnowski R, Liebe S, Rolfs A
and Löhr M: Simple method for DNA extraction from pancreatic juice
for PCR amplification assays. Int J Pancreatol. 25:39–43. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Miller SA, Dykes DD and Polesky HF: A
simple salting out procedure for extracting DNA from human
nucleated cells. Nucleic Acids Res. 16:12151988. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Azuara D, Rodriguez-Moranta F, de Oca J,
Soriano-Izquierdo A, Mora J, Guardiola J, Biondo S, Blanco I,
Peinado MA, Moreno V, et al: Novel methylation panel for the early
detection of colorectal tumors in stool DNA. Clin Colorectal
Cancer. 9:168–176. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Melki JR, Vincent PC and Clark SJ:
Concurrent DNA hypermethylation of multiple genes in acute myeloid
leukemia. Cancer Res. 59:3730–3740. 1999.PubMed/NCBI
|
|
20
|
Guo M, Jia Y, Yu Z, House MG, Esteller M,
Brock MV and Herman JG: Epigenetic changes associated with
neoplasms of the exocrine and endocrine pancreas. Discov Med.
17:67–73. 2014.PubMed/NCBI
|
|
21
|
Peng DF, Kanai Y, Sawada M, Ushijima S,
Hiraoka N, Kitazawa S and Hirohashi S: DNA methylation of multiple
tumor-related genes in association with overexpression of DNA
methyltransferase 1 (DNMT1) during multistage carcinogenesis of the
pancreas. Carcinogenesis. 27:1160–1168. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Esteller M, Corn PG, Baylin SB and Herman
JG: A gene hypermethylation profile of human cancer. Cancer Res.
61:3225–3229. 2001.PubMed/NCBI
|
|
23
|
McKie AB, Filipe MI and Lemoine NR:
Abnormalities affecting the APC and MCC tumor suppressor gene loci
on chromosome 5q occur frequently in gastric cancer but not in
pancreatic cancer. Int J Cancer. 55:598–603. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
House MG, Guo M, Iacobuzio-Donahue C and
Herman JG: Molecular progression of promoter methylation in
intraductal papillary mucinous neoplasms (IPMN) of the pancreas.
Carcinogenesis. 24:193–198. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rodriguez J, Muñoz M, Vives L, Frangou CG,
Groudine M and Peinado MA: Bivalent domains enforce transcriptional
memory of DNA methylated genes in cancer cells. Proc Natl Acad Sci
USA. 105:19809–19814. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Matsubayashi H, Canto M, Sato N, Klein A,
Abe T, Yamashita K, Yeo CJ, Kalloo A, Hruban R and Goggins M: DNA
methylation alterations in the pancreatic juice of patients with
suspected pancreatic disease. Cancer Res. 66:1208–1217. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Azuara D, Rodriguez-Moranta F, de Oca J,
Sanjuan X, Guardiola J, Lobaton T, Wang A, Boadas J, Piqueras M,
Monfort D, et al: Novel methylation panel for the early detection
of neoplasia in high-risk ulcerative colitis and Crohn's colitis
patients. Inflamm Bowel Dis. 19:165–173. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Matsubayashi H, Sato N, Fukushima N, Yeo
CJ, Walter KM, Brune K, Sahin F, Hruban RH and Goggins M:
Methylation of cyclin D2 is observed frequently in pancreatic
cancer but is also an age-related phenomenon in gastrointestinal
tissues. Clin Cancer Res. 9:1446–1452. 2003.PubMed/NCBI
|
|
29
|
Toyota M and Issa JP: CpG island
methylator phenotypes in aging and cancer. Semin Cancer Biol.
9:349–357. 1999. View Article : Google Scholar : PubMed/NCBI
|