Introduction
Bladder cancer is one of the most common types of
cancer in the Western male population (1). It is mainly classified into two types:
Superficial bladder cancer (pTis, pTa and pT1), which does not
invade into the muscle layer; and invasive bladder cancer (pT2, pT3
and pT4), which invades through the muscle layer. These two cancer
types exhibit different clinical behaviors, with superficial
bladder cancers being generally low-grade (grade 1 or 2) tumors,
the majority of which have a good prognosis; however, in 10–20% of
cases, cancer cells become more malignant. By contrast, invasive
bladder cancers are high-grade (grade 3) tumors, which are very
aggressive, as they develop and progress rapidly and metastasize in
an early stage (2).
Almost half of all the bladder cancer cases
diagnosed to date have been attributed to environmental carcinogens
such as tobacco smoking. Genetic and epigenetic changes that drive
cells into carcinogenesis have been linked to environmental and
occupational exposures in non-smokers, and these constitute a
significant proportion among non-smoker bladder cancer cases.
Molecular studies are being currently conducted to measure the
effects of inheritance that may be important in the epidemiology of
bladder cancer (3).
The majority of bladder cancers are transitional
cell carcinomas (TCCs), which originate from the cells lining the
inside of the bladder. TCC of the bladder is the second most common
malignancy of the genitourinary tract, and the second most common
cause of mortality among all genitourinary tumors (4). TCC provides a good model for
understanding the genetic basis of tumors, since the histological
progression of this tumor is correlated with p53 gene
mutations (5). p53 is a human
tumor suppressor gene that codes for the p53 protein. p53 is a
nuclear phosphoprotein that acts as a transcription factor and
regulates cell cycle events such as cell arrest or apoptosis under
different circumstances, including hypoxia, DNA damage and
metabolic changes that alter cell activity (6). Regardless the type and stage of cancer,
p53 has come to the forefront of research because it is
commonly mutated in multiple human cancers (7), including bladder cancer (8). Mutations in the p53 gene are
usually located in functionally important regions that have been
highly conserved over the evolution of species. These regions are
located in exons 5–8 (codons 126–306) of the p53 gene (8). In addition to p53 mutations,
mitochondrial DNA (mtDNA) is prone to mutations due to the absence
of protective histone backbones and efficient repair mechanisms, as
emerged in cancer research. Although several reports suggested the
important role of mtDNA in different cancer types, very few
publications have reported the detection of mtDNA mutations
in bladder cancer (9–13). In addition, alterations in the
p53 gene and in mtDNA genes have been examined in a
variety of solid tumors, but the number of studies focussing on
bladder cancer are insufficient (14). Therefore, the aim of the present study
is to identify the association between p53 and mtDNA
mutations in TTC of superficial bladder cancer.
Materials and methods
Clinical samples
Tissue specimens (n=30) were collected by
transurethral resection from patients diagnosed with TCC of the
bladder who were treated at the Department of Urology, Faculty of
Medicine, Marmara University (Istanbul, Turkey) from 1997 to 2010.
All patients provided written informed consent prior to inclusion
in the study. All tumors were staged and graded by pathology at
Marmara University Hospital (Istanbul, Turkey). Following
pathological evaluation for histological grade and stage, DNA
extraction was performed from the snap-frozen samples. In addition,
samples of normal bladder tissues (n=27) were obtained during
transurethral resection of the prostate and radical prostatectomy
operations. The specimens were stored at −80°C until DNA
extraction. The demographic, tumoral and progression
characteristics of the patients are summarized in Table I. The experimental protocol was
approved by the ethical committee of Marmara University
(MAR-AEK-09-2010-0055).
 | Table I.Demographic, tumoral and progression
characteristics of patients. |
Table I.
Demographic, tumoral and progression
characteristics of patients.
| Patient number | Gender | Stage | Grade | Recurrence | Progression |
|---|
| 1 | Male | T1 | G2 | Absent | Absent |
| 2 | Male | T1 | G3 | Absent | Absent |
| 3 | Male | T1 | G2 | Present | Absent |
| 4 | Female | T1 | G2 | Present | Absent |
| 5 | Female | T1 | G2 | Present | Present |
| 6 | Male | T1 | G2 | Present | Absent |
| 7 | Female | Ta | G1 | Present | Absent |
| 8 | Female | T1 | G1 | Present | Absent |
| 9 | Male | T1 | G3 | Present | Present |
| 10 | Male | Ta | G2 | Present | Present |
| 11 | Female | T1 | G2 | Present | Absent |
| 12 | Male | T1 | G2 | Absent | Absent |
| 13 | Male | T1 | G2 | Present | Absent |
| 14 | Female | T1 | G3 | Absent | Absent |
| 15 | Male | Ta | G2 | Present | Absent |
| 16 | Male | Ta | G1 | Present | Absent |
| 17 | Male | T1 | G1 | Present | Absent |
| 18 | Male | T1 | G2 | Present | Present |
| 19 | Female | T1 | G1 | Present | Absent |
| 20 | Male | T1 | G2 | Present | Present |
| 21 | Female | T1 | G2 | Present | Absent |
| 22 | Male | Ta | G2 | Present | Absent |
| 23 | Female | T1 | G1 | Present | Present |
| 24 | Male | Ta | G1 | Absent | Absent |
| 25 | Male | Ta | G1 | Absent | Absent |
| 26 | Male | T1 | G2 | Present | Absent |
| 27 | Male | T1 | G2 | Present | Absent |
| 28 | Male | Ta | G1 | Present | Absent |
| 29 | Female | Ta | G1 | Present | Absent |
| 30 | Male | Ta | G1 | Present | Present |
DNA extraction
Genomic DNA was extracted from tissues using a
commercial kit (InViTek, Berlin, Germany), according to the
manufacturer's protocol. The final DNA pellets were dissolved in
double distilled water, and the DNA concentrations were measured by
spectrophotometry. DNA samples were kept at −20°C until use.
Polymerase chain reaction (PCR)
amplification of mtDNA
For detection of mtDNA mutations, the reduced
nicotinamide adenine dinucleotide dehydrogenase 1
(ND1),adenosinetriphosphatase 6 (ATPase6) and
cytochrome b (Cytb) genes, and the D310 region, were
amplified by PCR using 10–20 ng total DNA. Table II lists the sequences of primers
specific for human mtDNA genes and the D310 region.
 | Table II.Sequences of the primers used for
polymerase chain reaction analysis of mutations in human mtDNA
genes and D310 region and in the human p53 gene (exons 5–8). |
Table II.
Sequences of the primers used for
polymerase chain reaction analysis of mutations in human mtDNA
genes and D310 region and in the human p53 gene (exons 5–8).
| Genes | Primer
sequence |
|---|
| mtDNA |
|
|
ND1 | Forward,
5′-CCAACCTCCTACTCCTCATTGT-3′ |
|
| Reverse,
5′-TGATCAGGGTGAGCATCAAA-3′ |
|
ATPase6 | Forward,
5′-AACGAAAATCTGTTCGCTTCAT-3′ |
|
| Reverse,
5′-ATGTGTTGTCGTGCAGGTAGAG-3′ |
|
Cytb | Forward,
5′-TATCCGCCATCCCATACATT-3′ |
|
| Reverse,
5′-GGTGATTCCTAGGGGGTTGT-3′ |
|
D310 | Forward,
5′-ACAATTGAATGTCTGCACAGCCACTT-3′ |
|
| Reverse,
5′-GGCAGAGATGTGTTTAAGTGCTG-3′ |
| p53,
exon |
|
| 5 | Forward,
5′-TCTTCCTACAGTACTCCCCT-3′ |
|
| Reverse,
5′-AGCTGCTCACCATCGCTATC-3′ |
| 6 | Forward,
5′-CCTCTGATTCCTCACTGATTGC-3′ |
|
| Reverse,
5′-CTCCTCCCAGAGACCCCAG-3′ |
| 7 | Forward,
5′-TCTCCTAGGTTGGCTCTGAC-3′ |
|
| Reverse,
5′-CCAGTGTGCAGGGTGGCAAG-3′ |
| 8 | Forward,
5′-TCCTGAGTAGTGGTAATCTA-3′ |
|
| Reverse,
5′-GCTTGCTTACCTCGCTTAGT-3′ |
PCR amplifications of the ND1, ATPase6
and Cytb genes were performed in a total volume of 50 µl
containing 50–100 ng DNA template in 10 mM Tris-HCl (pH 8.0), 50 mM
KCl, 1.5 mM MgCl2, deoxynucleotides (dNTPs; 100 mM
each), 1.0 U Taq DNA polymerase and primers (12.5 pmol each). The
conditions of PCR amplification were as follows: A denaturation
step at 94°C for 5 min, followed by 35 cycles at 94°C for 1 min,
annealing at 59°C for 1 min, extension at 72°C for 1 min, a final
extension at 72°C for 5 min and stop at 4°C. PCR amplifications for
the D310 region were performed in a total volume of 50 µl
containing 50–100 ng DNA template in 10 mM Tris-HCl (pH 8.0), 50 mM
KCl, 1.25 mM MgCl2, 100 mM each of the different dNTPs,
1.0 U Taq DNA polymerase and 10 pmol each primer. The conditions of
PCR amplification of the D310 region were as follows: A
denaturation step at 95°C for 2 min, followed by 35 cycles at 95°C
for 30 sec, annealing at 60°C for 30 sec, extension at 72°C for 1
min, a final extension at 72°C for 5 min and stop at 4°C. All PCR
products were fractionated by electrophoresis on a 2% agarose gel.
The sizes of the fragments of the ND1, ATPase6 and
Cytb genes following amplification were 934, 675 and 1,064
bp respectively. The size of the fragment containing the D310
region was 109 bp.
PCR amplification of the p53 gene
Tumors were screened for mutations in the p53
gene between exons 5 and 8, which encompass the DNA-binding domain
of the encoded protein. Table II
lists the primer sequences for the human p53 gene between
exons 5 and 8. Primers used for the amplification of exon 6 also
amplified the end of intron 5. The sizes of the fragments obtained
upon amplification were 205, 173, 149 and 157 bp for exons 5, 6, 7
and 8 of the p53 gene, respectively. PCR amplifications were
performed in a total volume of 50 µl containing 50–100 ng DNA
template in 10 mM Tris-HCl (pH 8.0), 50 mM KCl, 1.5 mM
MgCl2, dNTPs (100 mM each), 0.5 U Taq DNA polymerase and
15 pmol each primer. For all exons, the PCR procedures were the
same, with the exception of the annealing temperature. The
conditions of PCR amplification were as follows: A denaturation
step at 94°C for 5 min, followed by 35 cycles at 94°C for 1 min,
extension at 72°C for 1 min, a final extension at 72°C for 5 min
and stop at 4°C. Annealing temperatures and durations were 57°C for
1 min, 61°C for 1 min, 60°C for 1 min and 55°C for 1 min, for exons
5, 6, 7 and 8, respectively. All PCR products were fractionated by
electrophoresis on a 2% agarose gel, and products exhibiting
appropriate sizes were purified using a commercial kit (Roche
Applied Science, Penzberg, Germany), according to the
manufacturer's protocol.
Purification of PCR products and
direct sequencing of mtDNA and p53 gene regions
The purified PCR products were sequenced with an ABI
PRISM 310 genetic analyzer (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA).
Statistical analysis
SPSS version 21.0 (IBM SPSS, Armonk, NY, USA) was
used for statistical analysis. The χ2 and Fisher's exact
tests were used to determine the association between patient and
control groups for mtDNA and p53 mutations. The
Bonferroni-corrected P-value was used to evaluate the statistical
significancy. Additionally, the Spearman's rank correlation
coefficient (r) test was used to evaluate the correlation between
mtDNA and p53 mutations.
Results
DNA sequencing
The results of DNA sequencing demonstrated various
point mutations in the mitochondrial genome of the human tumor
samples. A total of 34 polymorphisms were identified in
mtDNA genes, 15 of which cause amino acid substitutions. In
addition, 3 novel polymorphisms were detected in the p53
gene. The distribution of mutations/polymorphisms is summarized in
Table III.
 | Table III.mtDNA mutations identified in the
D310 region and in the ATPase6, Cytb and ND1
genes. |
Table III.
mtDNA mutations identified in the
D310 region and in the ATPase6, Cytb and ND1
genes.
| Nucleotide
position | Nucleotide
change | Amino acid
change | Mutation frequency
(% of cases) | Mutation frequency
(% of cases) | P-value | Reported in
Mitomapb and other
tumors |
|---|
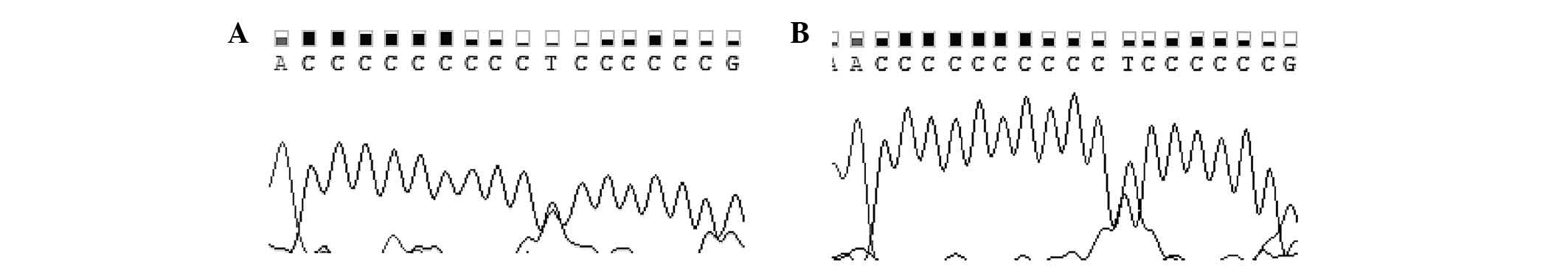
| D310-ND1;
P<0.05 was considered to indicate a statistically significant
difference |
|
| – | 7C→9C/10C | – | 1/30 (3) | 0/27 (0) | 1.000 | x |
| – | 7C→10C/11C | – | 1/30 (3) | 0/27 (0) | 1.000 | x |
|
| ATPase6-ND1;
P<0.0011 was considered to indicate a statistically significant
difference |
|
| 8557 | G→A | Ala→Thr | 1/30 (3) | 0/27 (0) | 1.000 | x |
| 8573 | G→A | Gly→Asp | 1/30 (3) | 0/27 (0) | 1.000 | x |
| 8584 | G→A | Ala→Thr | 1/30 (3) | 1/27 (4) | 1.000 | x |
| 8684 | C→T | Thr→Ile | 2/30 (7) | 1/27 (4) | 1.000 | x |
| 8697 | G→A | Silent | 8/30 (27) | 2/27 (7) | 0.083 | x |
| 8701 | A→G | Thr→Ala | 2/30 (7) | 1/27 (4) | 1.000 | x |
| 8730 | A→G | Silent | 1/30 (3) | 0/27 (0) | 1.000 | x |
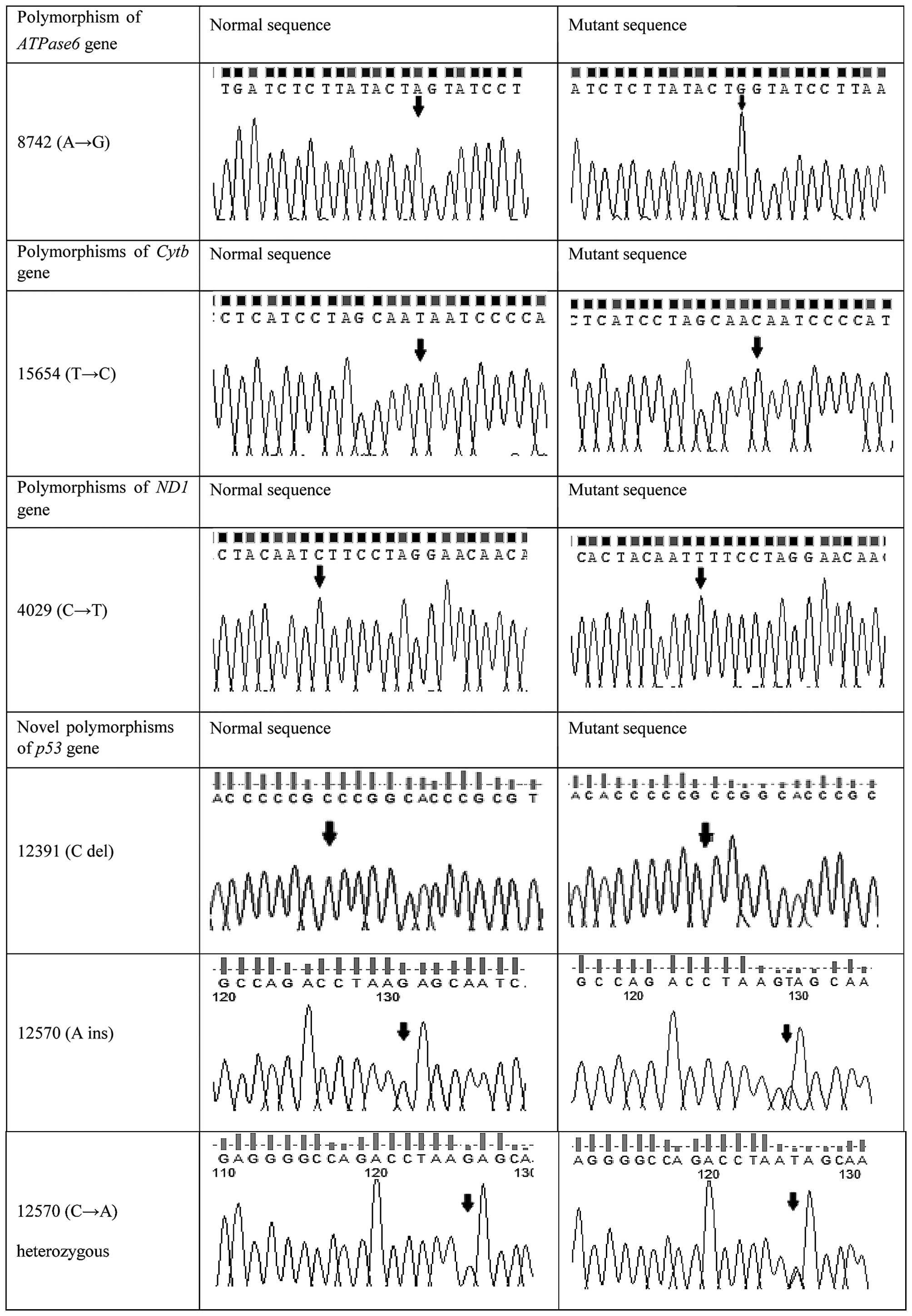
| 8742 | A→G | Silent | 1/30 (3) | 0/27 (0) | 1.000 | x |
| 8950 | G→A | Val→Ile | 1/30 (3) | 1/27 (4) | 1.000 | x |
| 9055 | G→A | Ala→Thr | 6/30 (20) | 1/27 (4) | 0.105 | x |
|
| Cytb-ND1;
P<0.0006 was considered to indicate a statistically significant
difference |
|
| 14783 | T→C | Leu→Ile | 2/30 (7) | 1/27 (4) | 1.000 | x |
| 14798 | T→C | Phe→Leu | 7/30 (23) | 3/27 (11) | 0.304 | x |
| 14905 | G→A | Silent | 8/30 (27) | 2/27 (7) | 0.083 | x |
| 15043 | G→A | Silent | 4/30 (13) | 2/27 (7) | 0.673 | x |
| 15218 | A→G | Thr→Ala | 2/30 (7) | 1/27 (4) | 1.000 | x |
| 15301 | G→A | Silent | 2/30 (7) | 1/27 (4) | 1.000 | x |
| 15452 | C→A | Leu→Ile | 12/30 (40) | 6/27 (22) | 0.149 | x |
| 15454 | T→C | Silent | 1/30 (3) | 1/27 (4) | 1.000 | x |
| 15498 | G→A | Gly→Asp | 3/30 (10) | 0/27 (0) | 0.239 | x |
| 15607 | A→G | Silent | 8/30 (27) | 1/27 (4) | 0.029 | x |
| 15622 | T→C | Silent | 1/30 (3) | 0/27 (0) | 1.000 | x |
| 15654 | T→C | Met→Thr | 1/30 (3) | 0/27 (0) | 1.000 | x |
| 15783 | T→C | Silent | 1/30 (3) | 0/27 (0) | 1.000 | x |
|
| ND1;
P<0.0013 was considered to indicate a statistically significant
difference |
|
| 3480 | A→G | Silent | 6/30 (20) | 1/27 (4) | 0.105 | x |
| 3507 | C→T | Silent | 2/30 (7) | 0/27 (0) | 0.492 | x |
| 3546 | C→A | Silent | 1/30 (3) | 0/27 (0) | 1.000 | x |
| 3703 | C→T | Leu→Trp | 2/30 (7) | 0/27 (0) | 0.492 | x |
| 3741 | C→T | Silent | 2/30 (7) | 1/27 (4) | 1.000 | x |
| 3930 | C→T | Silent | 1/30 (3) | 1/27 (4) | 1.000 | x |
| 4029 | C→T | Silent | 1/30 (3) | 1/27 (4) | 1.000 | x |
| 4188 | A→G | Silent | 2/30 (7) | 1/27 (4) | 1.000 | x |
| 4216 | T→C | Tyr→His | 10/30 (26) | 5/27 (19) | 0.241 | x |
|
| p53 |
|
| 12391 | C del | Premature stop
codon | 1/30 (3) | 0/27 (0) | 1.000 | Novel |
| 12570 | A ins | – | 20/30 (67) | 0/27 (0) | 0.001a | Novel |
| 12570 | C→A | – | 4/30 (13) | 17/27 (63) | 0.001a | Novel |
Comparision of mutations/polymorphisms
between patient and control groups
The A15607G polymorphism in the Cytb gene and
the 12570-A insertion in the p53 gene were significantly
higher in patients than in controls (P<0.05), whereas the
C12570A heterozygote mutation was significantly higher in controls
than in patients (P<0.05). Representative DNA sequencing
chromatograms of various polymorphisms in the ATPase6,
Cytb and ND1 genes, and of novel polymorphisms in the
p53 gene, are shown in Fig. 1,
while representative chromatograms of the D310 region are shown in
Fig. 2. The numbers and percentages
of mutations in different stages, grades, progression and
recurrence are shown in Table
IV.
 | Table IV.Numbers and percentages of mutations
in different stages, grades, progression and recurrence. |
Table IV.
Numbers and percentages of mutations
in different stages, grades, progression and recurrence.
|
| Gene mutations, n
(%) |
|---|
|
|
|
|---|
|
|
|
|
|
|
| p53 |
|---|
|
|
|
|
|
|
|
|---|
| Classification of
tumors | ATPase6
(n=22) | Cytb
(n=25) | ND1
(n=23) | D310 (n=11) | A ins (n=20) | C/A het (n=4) | C del (n=1) |
|---|
| Stage |
|
|
|
|
|
|
|
| Ta
(10) | 7
(31.8) | 9
(36.0) | 8
(34.8) | 1
(9.1) | 4
(20.0) | 3 (75.0) | 0 (0.0) |
| T1
(20) | 15 (68.2) | 16 (64.0) | 15 (65.2) | 10 (90.9) | 16 (80.0) | 1 (25.0) | 1 (100.0) |
| Grade |
|
|
|
|
|
|
|
| G1
(11) | 8
(36.4) | 11 (44.0) | 8
(34.8) | 1
(9.1) | 5
(25.0) | 3 (75.0) | 0 (0.0) |
| G2
(16) | 12 (54.5) | 12 (48.0) | 14 (60.9) | 9
(81.8) | 12 (60.0) | 1 (25.0) | 1 (100.0) |
| G3
(3) | 2
(9.1) | 2
(8.0) | 1
(4.3) | 1
(9.1) | 3
(15.0) | 0 (0.0) | 0 (0.0) |
| Progression |
|
|
|
|
|
|
|
| Yes
(7) | 6
(27.3) | 6
(24.0) | 4
(17.4) | 2
(18.2) | 5
(25.0) | 0 (0.0) | 0 (0.0) |
| No
(23) | 16 (72.7) | 19 (76.0) | 19 (82.6) | 9
(81.8) | 15 (75.0) | 4 (100.0) | 1 (100.0) |
| Recurrence |
|
|
|
|
|
|
|
| Yes
(24) | 18 (81.8) | 20 (80.0) | 19 (82.6) | 8
(72.7) | 16 (80.0) | 3 (75.0) | 1 (100.0) |
| No
(6) | 4
(18.2) | 5
(20.0) | 4
(17.4) | 3
(27.3) | 4
(20.0) | 1 (25.0) | 0 (0.0) |
Comparision of mutations/polymorphisms
according to tumor stage
A15607G polymorphism and 12570-A insertion were
significant in patients at T1 stage (P<0.05).
Correlation results
P<0.05 and r=0.25-0.49 indicate a weak positive
correlation between two mutations/polymorphisms, while r> 0.5
and r<0.5 indicate a moderate positive and negative correlation,
respectively, between two mutations/polymorphisms. In the patient
group, 5 different positive moderate correlations were observed
between A3480G (ND1)-G9055A (ATPase6), C3741T
(ND1)-C8684T (ATPase6), G8573A
(ATPase6)-T15622C (Cytb), G8697A
(ATPase6)-G14905A (Cytb) and A8701G
(ATPase6)-T14783C (Cytb), and 1 negative moderate
correlation was noticed between 12570-A insertion
(p53)-C12570A heterozygous (p53) (r>0.5,
P<0.05). Additionally, 3 different positive correlations were
detected between 12570-A insertion (p53)-G8697A
(ATPase6), 12570-A insertion (p53)-G14905A
(Cytb) and 12570-A insertion (p53)-A15607G
(Cytb) (r>0.5, P<0.05).
Discussion
The human mitochondrial genome consists of a
circular double-stranded DNA of 16,569 bp, which includes genes
encoding for the electron transport chain (complexes I–IV), ATP
synthase (or complex V in oxidative phosphorylation), a
displacement loop (D-loop) region, 2 ribosomal RNAs (16 and 23) and
22 transfer RNAs (15). In comparison
with nuclear DNA, mtDNA has a higher mutation rate, thus being more
susceptible to damage, primarily due to the lack of a histone
backbone, absence of an efficient repair mechanism and constant
exposure to reactive oxygen species (ROS) (16). mtDNA mutations are also
commonly observed in various tumors (17), both in the non-coding and in the
coding regions of mtDNA in cancer cells (12,13).
Therefore, in the present study, mtDNA mutations were
analyzed in the coding and non-coding regions of mtDNA.
The human tumour suppressor gene p53 is located in
17p13.1, and consists of 11 exons spanning over 20 kb of DNA. The
open reading frame of the p53 gene codes for a 53 kDa protein (p53
protein) with 393 amino acids. The protein is involved in multiple
biological functions, including cell cycle regulation, apoptosis,
DNA replication and repair, and maintenance of genomic stability.
Genetic changes in the p53 gene are observed in various
types of human cancer (6). Previous
studies on p53 mutations in urinary bladder tumors have
reported mutation frequencies between 6 and 61% (18,19). The
majority of p53 mutations are missense point mutations, and
~80% are present in the evolutionarily highly conserved and
functionally important exons 5–8 of the gene (20). Therefore, in the present study,
p53 gene mutations were analyzed in exons 5–8 and intron
5.
The role of the tumor suppressor gene p53 in
bladder TCC has been extensively studied, and it is known that
mutations in this gene correlate with the grade of cellular
dedifferentiation, stage of local infiltration, recurrence and
tumor progression (21,22). Therefore, mutations in the p53
gene are associated with the grade and stage of bladder cancer, and
may be important in the multistep progression of bladder cancer.
Lorenzo Romero et al (8)
observed that mutations in p53 did not appear in healthy
bladder mucosa, while they were significantly more frequent in pT1
and high-grade (grade 2 and 3) tumors. In the present study,
12570-A insertion on the p53 gene was significant in
patients at T1 stage (P<0.05). In another study, 2 different
mutations were identified in exon 5, 1 of which was detected on
codon 153 (C>T) (6). In the
present study, a novel frameshift mutation (12391-C deletion) was
noticed on the same codon, which causes a premature stop codon. In
addition, 2 novel mutations were identified in intron 5, of which,
the 12570-A insertion mutation was only detected in patients (67%).
At the same location, the C12570A heterozygous mutation was
significantly higher in controls (63%) than in patients (13%)
(P<0.05). Therefore, the 12570-A insertion mutation may
contribute to the development of the disease, whereas the C12570A
heterozygous mutation may have a protective effect in this
process.
All the mitochondrial protein coding genes encode
subunits of the oxidative phosphorylation enzymes that are
responsible for the energy generation pathway (23). The oxidative phosphorylation system is
composed of 5 complexes (I–V) that are assembled from multiple
polypeptides, a number of which are encoded by mtDNA and others by
nuclear DNA (24). Among the above 5
complexes, complex III is a membrane-bound enzyme that catalyzes
the transfer of electrons from ubiquinol to Cytc (25). In this pathway, Cytb is fundamental
for the assembly and function of complex III (11). It is known that compromised
mitochondrial function due to mtDNA mutations enhances the
generation of ROS. It has been widely demonstrated that ROS are
also involved in numerous proliferating signaling pathways
associated with tumor promotion (16). Another complex, complex V, is
important in adenosine triphosphate (ATP) production and apoptosis
(26,27). The contribution of mtDNA complex V
variants in cell transformation, elevated ROS production and tumor
progression has been described (28).
Additionally, efficient programmed cell death requires the
molecular machinery of the ATP synthase (29). The ATPase6 gene, one of the complex V
genes, contributes to mtDNA maintenance (15). Furthermore, ND1 (or complex I) has the
most number of subunits encoded by mtDNA. The production of ROS
probably occurs when complex I activity is interrupted (23). In addition, the majority of mutations
in cancer occur in the mitochondrial D-loop region, which functions
as a promoter for both the heavy and light strands of mtDNA
(13). Therefore, in the present
study, the mtDNA genes ATPase6, Cytb and
ND1, and the D-loop region of mtDNA, were analyzed in 30 TCC
patients and 27 healthy individuals.
The various mutations frequencies were determined
due to the variations in the tumor stage and grade, and a much
higher number of mutations were observed in tumors of high stage
and grade than in tumors of low stage and grade (2). Thus, the present study also investigated
the association between gene mutations and tumor stage and
grade.
The A15607G polymorphism on the Cytb gene was
significant in patients at T1 stage (P<0.05). A total of 34
mtDNA mutations were identified in TCC patients, of which,
15 cause an amino acid change. The present results are consistent
with a previous study by the present authors, in which mtDNA
mutations were examined in TCC patients and healthy individuals. In
that study, a total of 68 mutations were identified, a number of
which are similar to the ones reported in the present study
(30). The D-loop region, while being
non-coding, has been demonstrated to have a higher mutation rate
than coding mtDNA within cancer patients. Previous studies
suggested that mutations in the D-loop region could alter mtDNA
transcription and further lead to respiratory chain alteration, and
thereby disrupt mitochondrial-induced apoptosis (31,32).
Therefore, mutations in the D-loop region may play an indirect role
in the tumorigenic process, since this region is responsible for
the control of mtDNA proliferation (16). Chang et al (13) observed that of 88 tumors with
p53 mutations, 34 (38.6%) had D-loop mutations, and its
frequency was significantly higher in tumors with p53
mutations than in tumors without p53 mutations (23/106;
21.7%) (13). Although the present
study identified numerous mutations in the D-loop region, no
correlation was observed between mutations in the D-loop region and
in the p53 gene in bladder cancer patients.
Analyses of the mutational spectra for the
mtDNA and p53 genes may provide clues about cancer
etiology, mechanisms of mutations and the role of p53 and
mtDNA genes in the development of urinary bladder cancer.
mtDNA and p53 mutations have been previously described in
different tumors, whereas similar studies focusing on bladder
cancer are scarce (14,16). In the present study, the p53
and mtDNA mutational spectra for urinary bladder tumors were
analyzed by studying the mutation frequency in exons 5–8 as well as
in intron 5 of the p53 gene, and in the ND1,
ATPase6 and Cytb genes, in addition to the D-loop
D310 region of mtDNA, in patients with TCC at various stages and
grades. p53 gene mutations were analyzed in patients who had
mtDNA mutations, and the association between mtDNA
and p53 mutations in TCC of the bladder was investigated.
Achanta et al (33)
demonstrated an association between the expression of cytoplasmic
p53 and the vulnerability of mtDNA to exogenous damage. The
authors also revealed that p53 has a role in maintaining
mitochondrial genetic stability through its ability to translocate
to mitochondria and interact with DNA polymerase γ, thus enhancing
its DNA replication function, in response to mtDNA damage. In
addition, the authors also noticed that loss of p53 resulted
in a significant increase in mtDNA vulnerability to damage, leading
to increased frequency of in vivo mtDNA mutations. Gochhait
et al (14) suggested that the
concomitant presence of somatic alterations in mtDNA and the DNA
binding domain of the p53 gene facilitates cell survival and
tumorigenesis. To the best of our knowledge, the present is the
first study that attempts to correlate mtDNA and p53
mutations in TCC. In the present study, the r test was used to
investigate the association between mtDNA and p53
mutations in TCC of the bladder. Positive correlations were
observed between 12570-A insertion (p53 mutation) and G8697A
(ATPase6 mutation), G14905A and A15607G (Cytb
mutation) (r>0.3, P<0.05).
The present results, as well as the published data
regarding the sequence analysis of the mutated p53 gene and
mtDNA, revealed that there were no consistent patterns of
p53 and mtDNA mutations in bladder cancer. Further
studies are required to determine whether p53 and
mtDNA mutations could serve as an important predictor for
tumor progression or as a useful marker for selection of a more
suitable treatment, and to explain whether there are consistent
patterns of mutation in bladder cancer. Therefore, future analyses
of the progression of bladder cancer patients with mutated
p53 and mtDNA genes will help clarify this
aspect.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global Cancer Statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fujimoto K, Yamada Y, Okajima E, Kakizoe
T, Sasaki H, Sugimura T and Terada M: Frequent association of p53
gene mutation in invasive bladder cancer. Cancer Res. 52:1393–1398.
1992.PubMed/NCBI
|
|
3
|
Kiriluk KJ, Prasad SM, Patel AR, Steinberg
GD and Smith ND: Bladder cancer risk from occupational and
environmental exposures. Urol Oncol. 30:199–211. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Williams SG and Stein JP: Molecular
pathways in bladder cancer. Urol Res. 32:373–385. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin HY, Huang CH, Wu WJ, Chou YH, Fan PL
and Lung FW: Mutation of the p53 tumor suppressor gene in
transitional cell carcinoma of the urinary tract in Taiwan.
Kaohsiung J Med Sci. 21:57–64. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Berggren P, Steineck G, Adolfsson J,
Hansson J, Jansson O, Larsson P, Sandstedt B, Wijkström H and
Hemminki K: p53 mutations in urinary bladder cancer. Br J Cancer.
84:1505–1511. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Greenblatt MS, Bennett WP, Hollstein M and
Harris CC: Mutations in the p53 tumor suppressor gene: Clues to
cancer etiology and molecular pathogenesis. Cancer Res.
54:4855–4878. 1994.PubMed/NCBI
|
|
8
|
Romero JG Lorenzo, Sánchez AS Salinas,
Bachs JM Giménez, Sánchez F Sánchez, Martínez J Escribano, Millán
IR Hernández, Martín M Segura and Rodríguez JA Virseda: p53 Gene
mutations in superficial bladder cancer. Urol Int. 73:212–218.
2004. View Article : Google Scholar
|
|
9
|
Chen GF, Chan FL, Hong BF, Chan LW and
Chan PS: Mitochondrial DNA mutations in chemical carcinogen-induced
rat bladder and human bladder cancer. Oncol Rep. 12:463–472.
2004.PubMed/NCBI
|
|
10
|
Wada T, Tanji N, Ozawa A, Wang J,
Shimamoto K, Sakayama K and Yokoyama M: Mitochondrial DNA mutations
and 8-hydroxy-2′-deoxyguanosine Content in Japanese patients with
urinary bladder and renal cancers. Anticancer Res. 26:3403–3408.
2006.PubMed/NCBI
|
|
11
|
Dasgupta S, Hoque MO, Upadhyay S and
Sidransky D: Mitochondrial cytochrome B gene mutation promotes
tumor growth in bladder cancer. Cancer Res. 68:700–706. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Carew JS and Huang P: Mitochondrial
defects in cancer. Mol Cancer. 1:92002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chang SC, Lin PC, Yang SH, Wang HS, Liang
WY and Lin JK: Mitochondrial D-loop mutation is a common event in
colorectal cancers with p53 mutations. Int J Colorectal Dis.
24:623–628. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gochhait S, Bhatt A, Sharma S, Singh YP,
Gupta P and Bamezai RN: Concomitant presence of mutations in
mitochondrial genome and p53 in cancer development-a study in north
Indian sporadic breast and esophageal cancer patients. Int J
Cancer. 123:2580–2586. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ghaffarpour M, Mahdian R, Fereidooni F,
Kamalidehghan B, Moazami N and Houshmand M: The mitochondrial
ATPase6 gene is more susceptible to mutation than the ATPase8 gene
in breast cancer patients. Cancer Cell Int. 14:212014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Prior SL, Griffiths AP and Lewis PD: A
study of mitochondrial DNA D-loop mutations and p53 status in
nonmelanoma skin cancer. Br J Dermatol. 161:1067–1071. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou S, KAchap S and Singh KK:
Mitochondrial impairment in p53-deficient human cancer cells.
Mutagenesis. 18:287–292. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shipman R, Schraml P, Moch H, Colombi M,
Sauter G, Mihatsch M and Ludwig C: p53 protein accumulation and p53
gene alterations (RFLP, VNTR and p53 gene mutations) in
non-invasive versus invasive human transitional bladder cancer. Int
J Oncol. 10:801–806. 1997.PubMed/NCBI
|
|
19
|
Sidransky D, Von Eschenbach A, Tsai YC,
Jones P, Summerhayes I, Marshall F, Paul M, Green P, Hamilton SR,
Frost P, et al: Identification of p53 gene mutations in bladder
cancers and urine samples. Science. 252:706–709. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Phillips HA, Howard GC and Miller WR: p53
mutations as a marker of malignancy in bladder washing samples from
patients with bladder cancer. Br J Cancer. 82:136–141.
2000.PubMed/NCBI
|
|
21
|
Kindelán J Álvarez, López-Beltrán A and
Tapia MJ Requena: Molecular biology in bladder cancer. Actas Urol
Esp. 24:604–625. 2000.(In Spanish). View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lianes P: Biología Molecular del Cáncer de
VejigaTumores Vesicales Superficiales. Vicente J, Chéchile G and
Salvador J: Acción Médica; Madrid: pp. 47–60. 2000, (In
Spanish).
|
|
23
|
Akouchekian M, Houshmand M, Akbari MH,
Kamalidehghan B and Dehghan M: Analysis of mitochondrial ND1 gene
in human colorectal cancer. J Res Med Sci. 16:50–55.
2011.PubMed/NCBI
|
|
24
|
Petros JA, Baumann AK, Ruiz-Pesini E, Amin
MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, et
al: mtDNA mutations increase tumorigenicity in prostate cancer.
Proc Natl Acad Sci USA. 102:719–724. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Blakely EL, Mitchell AL, Fisher N, Meunier
B, Nijtmans LG, Schaefer AM, Jackson MJ, Turnbull DM and Taylor RW:
A mitochondrial Cytochrome b mutation causing severe respiratory
chain enzyme deficiency in human and yeast. FEBS J. 272:3583–3592.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jonckheere AII, Smeitink JA and Rodenburg
RJ: Mitochondrial ATP synthase: architecture, function and
pathology. J Inherit Metab Dis. 35:211–225. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Czarnecka AM, Kukwa W, Krawczyk T, Scinska
A, Kukwa A and Cappello F: Mitochondrial DNA mutations in cancer -
from bench to bedside. Front Biosci (Landmark Ed). 15:437–460.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Amuthan G, Biswas G, Zhang SY,
Klein-Szanto A, Vijayasarathy C and Avadhani NG:
Mitochondria-to-nucleus stress signaling induces phenotypic
changes, tumor progression and cell invasion. Embo J. 20:1910–1920.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Matsuyama S, Xu Q, Velours J and Reed JC:
The Mitochondrial F0F1-ATPase proton pump is required for function
of the proapoptotic protein Bax in yeast and mammalian cells. Mol
Cell. 1:327–336. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guney AI, Ergec DS, Tavukcu HH, Koc G,
Kirac D, Ulucan K, Javadova D and Turkeri L: Detection of
mitochondrial DNA mutations in nonmuscle invasive bladder cancer.
Genet Test Mol Biomarkers. 16:672–678. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Simonnet H, Alazard N, Pfeiffer K, Gallou
C, Béroud C, Demont J, Bouvier R, Schägger H and Godinot C: Low
mitochondrial respiratory chain content correlates with tumor
aggressiveness in renal cell carcinoma. Carcinogenesis. 23:759–768.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Meierhofer D, Mayr JA, Foetschl U, Berger
A, Fink K, Schmeller N, Hacker GW, Hauser-Kronberger C, Kofler B
and Sperl W: Decrease of mitochondrial DNA content and energy
metabolism in renal cell carcinoma. Carcinogenesis. 25:1005–1010.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Achanta G, Sasaki R, Feng L, Carew JS, Lu
W, Pelicano H, Keating MJ and Huang P: Novel role of p53 in
maintaining mitochondrial genetic stability through interaction
with DNA Pol gamma. EMBO J. 24:3482–3492. 2005. View Article : Google Scholar : PubMed/NCBI
|