Introduction
Colorectal cancer (CRC) is a global health challenge
with >1.2 million novel cases and 600,000 mortalities every year
(1). Survival rates of CRC greatly
depend on the stage at primary diagnosis, and the 5-year survival
rate is 93 and 8% for stage I and IV, respectively (2). Systematic methods for diagnosis,
including flexible sigmoidoscopy and implementation of the fecal
occult blood test, may contribute to a high detection rate of
patients in the early stages of CRC, thus reducing mortality rates
(3). There are numerous screening
methods, drugs and chemotherapy regimens, including irinotecan and
oxaliplatin, available; however, the majority of these have low
sensitivity and treatment-limiting side effects (4–6).
Therefore, the development of highly sensitive and CRC-specific
diagnostic and prognostic biomarkers is required for the diagnosis
and treatment of early stage CRC.
MicroRNAs (miRs) have emerged as important molecules
involved in tumorigenesis and disease progression and metastasis in
various cancers, including CRC (7).
miRs are endogenous, short non-coding RNAs that effectively inhibit
target gene activity by binding to target mRNAs and hindering their
translation (8). Each miR achieves
functional specificity by targeting a core network of genes and has
potential effects on almost every genetic pathway (9). Increasing evidence suggests that
aberrant miR expression is clearly associated with the initiation
and progression of certain cancers, and miRs may act as oncogenes
and/or tumor suppressor genes (10).
miR-21 downregulates tumor suppressor programmed cell death 4 at a
post-transcriptional level and promotes the invasion, intravasation
and metastasis of CRC (11). miR-335
exhibits anti-tumoral activity in various tumors, including gastric
cancer (12), ovarian cancer
(13) and CRC (14). Therefore, pivotal miRs may be
informative biomarkers for detection, diagnosis and prognosis of
various types of cancer. Furthermore, miRs have emerged as highly
tissue-specific biomarkers (15,16), and
previous studies have paved the way for miR-based cancer tissue
classification (17,18). However, miRs associated with CRC and
their roles in the molecular pathogenesis of CRC remain
unclear.
To date, one of the most interesting findings is
that secreted miRs potentially influence target cell function via
exosomes (19). Exosomes are small
(30–90 nm) membrane vesicles that carry mRNAs, proteins, lipids and
miRs, depending on the origin of the secreting cells (20,21).
Previous studies have demonstrated that exosomes are the newest
family members of ‘bioactive vesicles’ that promote intercellular
communication and immunoregulatory processes via shuttling
molecules between cells (22,23). miRs carried in exosomes are secreted
from cancer cells into body fluids, such as blood, urine, breast
milk and saliva (21). Therefore,
exosomal miRs in body fluids may be useful diagnostic and
prognostic biomarkers of cancers (24). However, research on the association
between exosomal miR profiles in blood and the pathological
condition of cancer patients are limited.
In a previous study, the exosomal miR profile
(GSE39833) (25) has been used to
highlight the most abundant miRs in the blood of patients with CRC
using an arbitrary boundary. In silico analysis of these
miRs was performed to explore the function of miRs as tumor
surveillance mechanisms exerting continuous inhibition on tumor
formation. There was no systematic and comprehensive analysis for
the miR functions in CRC. Therefore, the present study used
microarray analysis to screen the differentially expressed miRs in
the sera of patients with CRC. Additionally, in order to improve
the understanding of miR functions, the predicted target genes of
miRs were identified. Comprehensive bioinformatics methods were
used to investigate the function and pathways of target genes of
miRs and construct a protein-protein interaction (PPI) network to
identify the hub target genes and miRs associated with CRC. The
present study aimed to investigate the key role of potentially
important miRs and their target genes in the initiation and
progression of CRC. The present study may provide a basis for
screening novel biomarkers of CRC for therapeutic
interventions.
Materials and methods
Microarray data of miRs
The miR expression profile was downloaded from the
Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/) using the series accession
number GSE39833, which was deposited by Ogata-Kawata et al
(3). The GSE39833 dataset consisted
of 88 CRC patients with various tumor-necrosis-metastasis stages
(stage I, 20 patients; II, 20 patients; IIIa, 20 patients; IIIb, 16
patients; IV, 12 patients) between the ages of 45–65 years and 11
healthy controls. An Agilent Human miRNA Microarray platform was
used (product no., G4470C; design ID, 021827; Agilent Technologies,
Inc., Santa Clara, CA, USA).
Data preprocessing and screening of
differentially expressed miRs
Limma (26) (version
2.7.10; bioconductor.org/packages/release/bioc/html/limma.html)
package in R (version 2.3.1; www.r-project.org/) was used for data preprocessing
and normalization. A T-test (26) in
Limma package was used to identify the significantly differentially
expressed miRs in the CRC group compared with the healthy control
group. P<0.05 and |log2 fold change|>1.5 were considered to
indicate a statistically significant difference.
Prediction of the target genes for
miRs
Identification of the predicted target genes of miRs
is conducive to improve the understanding of miR functions.
miRecords (version 3; c1.accurascience.com/miRecords/) and miRWalk (verison
2.0; zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/) were used to
retrieve the target genes of the differentially expressed miRs.
miRecords (27), an integrated
database for miR-target interactions (MTIs), hosts a large,
high-quality, manually curated database of experimentally validated
MTIs. miRWalk (28), a comprehensive
database on miRs, hosts predicted and validated miR binding sites
and information on all known genes of human, mouse and rat. Only
miR-target interactions present in the two databases were used,
since target genes shared by the two databases were considered to
be more reliable.
Function and pathway enrichment
analysis
Functional analysis of large gene lists from
high-throughput genomic, proteomic and bioinformatics scanning
approaches increases the likelihood for researchers to identify
biological processes most associated with their study (29). Gene Ontology (GO) analysis is
increasingly applied for functional studies of microarray data
(30). Kyoto Encyclopedia of Genes
and Genomes (KEGG) is the major public pathway-associated database
(31), and Database for Annotation
Visualization and Integrated Discovery (DAVID) is a tool for
providing functional annotation behind large-scale genomic or
transcriptomic data (32). DAVID
online analytical tool (version 6.7; david.ncifcrf.gov/) was utilized to analyze the
significantly enriched GO terms (geneontology.org/) and KEGG pathways (www.genome.jp/kegg/) for target genes of miRs. Each GO
term or KEGG pathway included at least two genes, and P<0.05 was
considered to indicate a statistically significant difference.
Construction of a PPI network
Search Tool for the Retrieval of Interacting Genes
(STRING) (33) is a database
providing the predicted protein interaction information in a given
cell context. PPI networks are increasingly becoming the focus of
research in the identification of the cellular function of proteins
in various organisms (34). Based on
the information of the STRING database (version 8.3; string-db.org/), a protein interaction network was
constructed by mapping the target genes of miRs. The interaction
pairs with a combined score of 0.4 were selected in this
network.
Results
Screening of differentially expressed
miRs
In the present study, the expression levels of 851
miRs were acquired following normalization. Following a comparison
of the expression between the CRC and healthy control samples using
a t-test, 18 differentially expressed miRs were identified
(Table I). Among them, 8 miRs were
upregulated (Table IA) and 10 were
downregulated (Table IB).
 | Table I.Differentially expressed miRs between
colorectal cancer and healthy control samples. |
Table I.
Differentially expressed miRs between
colorectal cancer and healthy control samples.
| A, Upregulated
differentially expressed miRs |
|---|
|
|---|
| miR | Log2, FC | P-value |
|---|
| hsa-let-7b | 0.58796649 |
2.89×10−3 |
| hsa-miR-1246 | 2.59391919 |
7.14×10−10 |
| hsa-miR-126 | 3.16477901 |
6.27×10−9 |
| hsa-miR-1290 | 1.32767795 |
3.49×10−5 |
| hsa-miR-181b | 1.08748377 |
3.88×10−2 |
| hsa-miR-181d | 1.41294860 |
2.45×10−2 |
| hsa-miR-23a | 1.87225376 |
6.94×10−8 |
| hsa-miR-654-5p | 1.35980450 |
1.25×10−3 |
|
| B, Downregulated
differentially expressed miRs |
|
| miR | Log2, FC | P-value |
|
|
hsa-miR-1225-5p | −0.81943754 |
9.73×10−4 |
| hsa-miR-1287 | −0.72720715 |
2.84×10−3 |
| hsa-miR-1295 | −1.44266151 |
5.07×10−5 |
| hsa-miR-1299 | −1.22393055 |
4.15×10−3 |
| hsa-miR-144 | −2.48669140 |
1.47×10−9 |
| hsa-miR-16 | −1.35722402 |
2.42×10−5 |
|
hsa-miR-513a-5p | −0.90185107 |
3.77×10−4 |
| hsa-miR-575 | −0.86229919 |
5.94×10−10 |
| hsa-miR-652 | −1.77364130 |
8.49×10−12 |
| hsa-miR-760 | −0.91368658 |
1.36×10−2 |
Prediction of the target genes for
miRs
Based on the information of miRecords and miRTarBase
database, 27 target genes corresponding to the 8 upregulated miRs
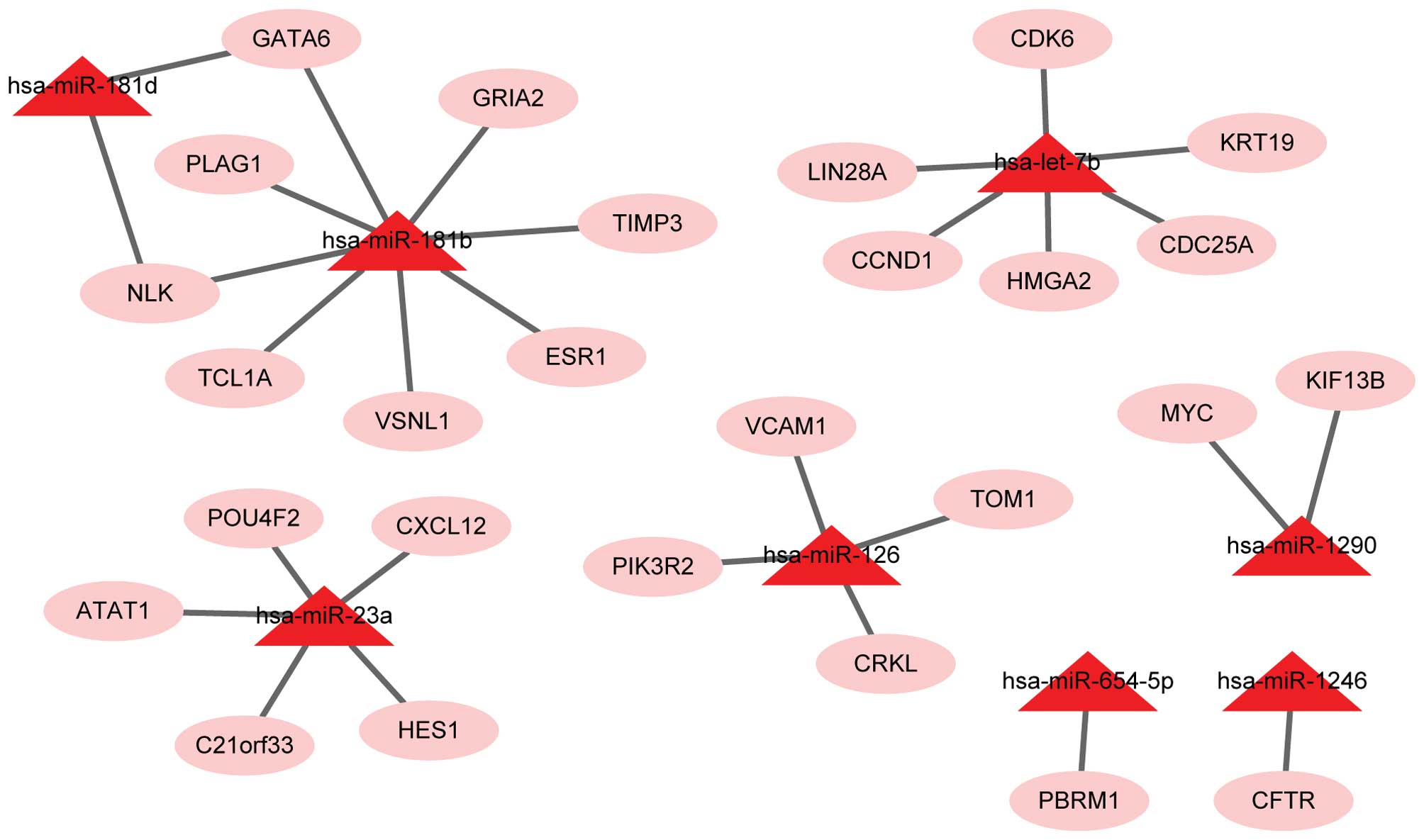
(Fig. 1) and 79 target genes
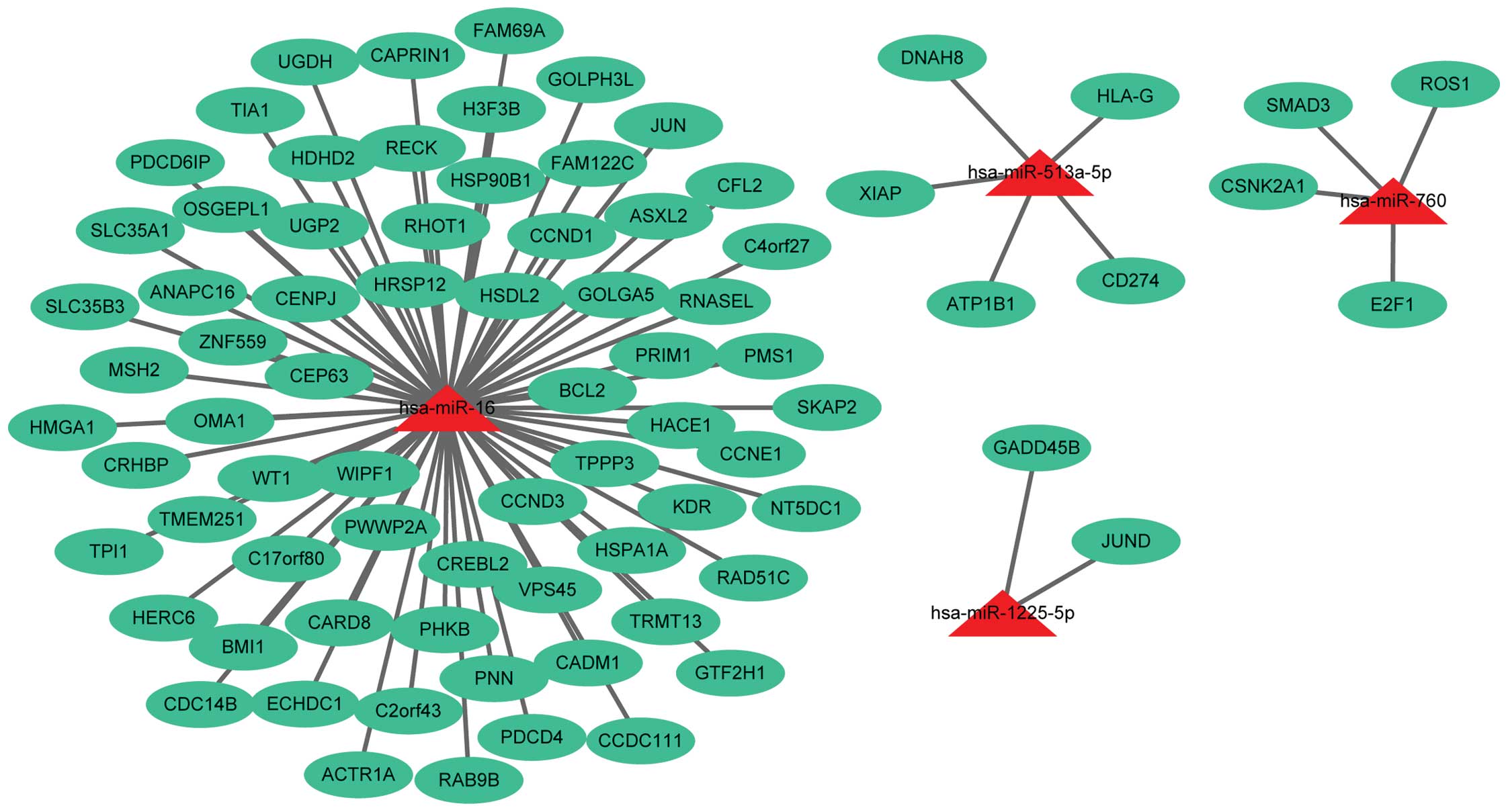
corresponding to 4 out of 10 downregulated miRs (Fig. 2) were obtained. In total, 6 out of 10
downregulated miRs, miR-1287, miR-1295, miR-1299, miR-144, miR-575
and miR-652, did not have their target genes identified.
Function and pathway enrichment
analysis
In the present study, 15 KEGG pathways corresponding
to the upregulated miRs and 9 KEGG pathways corresponding to the
downregulated miRs were enriched, respectively, using a cutoff
criteria of P<0.05 (Table II).
The present results revealed that the pathways enriched by
differentially expressed miRs were not only associated with various
types of cancer, including chronic myeloid leukemia, small cell
lung cancer, endometrial cancer and CRC, but also with important
signaling pathways, including the ErbB, Wnt, Jak-STAT and p53
signaling pathways (Table II).
Notably, the present results demonstrated that the target genes of
upregulated miRs, such as cyclin D1 (CCND1), v-myc avian
myelocytomatosis viral oncogene homolog (MYC) and
phosphoinositide-3-kinase, regulatory subunit 2 (beta) (PIK3R2)
(Table IIA), and target genes of
downregulated miRs, such as CCND1, mutS homolog 2 (MSH2), jun
proto-oncogene (JUN), B-cell CLL/lymphoma 2 (BCL2) and SMAD family
member 3 (SMAD3) (Table IIB), were
all enriched in CRC pathways; therefore, these target genes may be
closely associated with CRC. Additionally, the upregulated miRs of
these target genes were let-7b (CCND1), miR-1290 (MYC) and miR-126
(PIK3R2). The downregulated miRs of these target genes were miR-16
(CCND1, MSH2, JUN, BCL2) and miR-760 (SMAD3).
 | Table II.Enriched KEGG pathways of target
genes in colorectal cancer samples. |
Table II.
Enriched KEGG pathways of target
genes in colorectal cancer samples.
| A, Enriched KEGG
pathways of upregulated target genes |
|---|
|
|---|
| KEGG ID | Description | Count | P-value | Genes |
|---|
| hsa05220 | Chronic myeloid
leukemia | 5 |
1.33×10−5 | CCND1, CRKL, CDK6,
MYC, PIK3R2 |
| hsa05200 | Pathways in
cancer | 5 |
3.90×10−3 | CCND1, CRKL, CDK6,
MYC, PIK3R2 |
| hsa05222 | Small cell lung
cancer | 4 |
6.52×10−4 | CCND1, CDK6, MYC,
PIK3R2 |
| hsa04110 | Cell cycle | 4 |
2.07×10−3 | CCND1, CDK6, MYC,
CDC25A |
| hsa05213 | Endometrial
cancer | 3 |
5.32×10−3 | CCND1, MYC,
PIK3R2 |
| hsa05223 | Non-small cell lung
cancer | 3 |
5.73×10−3 | CCND1, CDK6,
PIK3R2 |
| hsa05221 | Acute myeloid
leukemia | 3 |
6.58×10−3 | CCND1, MYC,
PIK3R2 |
| hsa05214 | Glioma | 3 |
7.73×10−3 | CCND1, CDK6,
PIK3R2 |
| hsa05218 | Melanoma | 3 |
9.75×10−3 | CCND1, CDK6,
PIK3R2 |
| hsa05212 | Pancreatic
cancer | 3 |
1.00×10−2 | CCND1, CDK6,
PIK3R2 |
| hsa05210 | Colorectal
cancer | 3 |
1.35×10−2 | CCND1, MYC,
PIK3R2 |
| hsa04012 | ErbB signaling
pathway | 3 |
1.44×10−2 | CRKL, MYC,
PIK3R2 |
| hsa04670 | Leukocyte
transendothelial migration | 3 |
2.56×10−2 | VCAM1, CXCL12,
PIK3R2 |
| hsa04310 | Wnt signaling
pathway | 3 |
4.04×10−2 | CCND1, NLK,
MYC |
| hsa04630 | Jak-STAT signaling
pathway | 3 |
4.24×10−2 | CCND1, MYC,
PIK3R2 |
|
| B, Enriched KEGG
pathways of downregulated target genes |
|
| KEGG ID | Description | Count | P-value | Genes |
|
| hsa05200 | Pathways in
cancer | 9 |
5.85×10−4 | E2F1, CCNE1, CCND1,
HSP90B1, XIAP, MSH2, JUN, BCL2, SMAD3 |
| hsa04110 | Cell cycle | 7 |
8.70×10−5 | E2F1, CCNE1, CCND1,
CCND3, CDC14B, SMAD3, GADD45B |
| hsa04510 | Focal adhesion | 6 |
6.74×10−3 | CCND1, CCND3, XIAP,
JUN, BCL2, KDR |
| hsa05210 | Colorectal
cancer | 5 |
1.55×10−3 | CCND1, MSH2, JUN,
BCL2, SMAD3 |
| hsa05222 | Small cell lung
cancer | 5 |
1.55×10−3 | E2F1, CCNE1, CCND1,
XIAP, BCL2 |
| hsa05215 | Prostate
cancer | 5 |
1.92×10−3 | E2F1, CCNE1, CCND1,
HSP90B1, BCL2 |
| hsa04310 | Wnt signaling
pathway | 5 |
1.26×10−2 | CCND1, CSNK2A1,
CCND3, JUN, SMAD3 |
| hsa04144 | Endocytosis | 5 |
2.43×10−2 | VPS45, HSPA1A,
PDCD6IP, HLA-G, KDR |
| hsa04115 | p53 signaling
pathway | 4 |
7.87×10−3 | CCNE1, CCND1,
CCND3, GADD45B |
Furthermore, based on GO enrichment analysis
associated with the biological processes of target genes, 35 and 96
GO terms were identified, which were enriched corresponding to the
up/downregulated miRs, respectively (Table III). The results revealed that the
target genes of differentially expressed miRs were significantly
associated with cell cycle processes and response to hormone
stimulus.
 | Table III.Ten of the most enriched GO terms
associated with the biological process of target genes in
colorectal cancer samples. |
Table III.
Ten of the most enriched GO terms
associated with the biological process of target genes in
colorectal cancer samples.
| A, Enriched GO
terms of upregulated target genes |
|---|
|
|---|
| GO ID | Description | Count | P-value |
|---|
| GO:0045449 | Regulation of
transcription | 10 |
2.14×10−2 |
| GO:0010033 | Response to organic
substance | 7 |
1.04×10−3 |
| GO:0022402 | Cell cycle | 6 |
2.25×10−3 |
| GO:0007049 | Cell cycle | 6 |
8.69×10−3 |
| GO:0010604 | Positive regulation
of macromolecule metabolic process | 6 |
1.30×10−2 |
| GO:0043627 | Response to
estrogen stimulus | 5 |
2.71×10−5 |
| GO:0048545 | Response to steroid
hormone stimulus | 5 |
2.82×10−4 |
| GO:0045596 | Negative regulation
of cell differentiation | 5 |
4.41×10−4 |
| GO:0009725 | Response to hormone
stimulus | 5 |
3.14×10−3 |
| GO:0043627 | Response to
estrogen stimulus | 5 |
3.23×10−3 |
|
| B, Enriched GO
terms of downregulated target genes |
|
| GO ID | Description | Count | P-value |
|
| GO:0006915 | Apoptosis | 13 |
1.80×10−5 |
| GO:0012501 | Programmed cell
death | 13 |
2.08×10−5 |
| GO:0008219 | Cell death | 13 |
1.01×10−4 |
| GO:0016265 | Cell death | 13 |
1.08×10−4 |
| GO:0007049 | Cell cycle | 12 |
8.20×10−4 |
| GO:0051726 | Regulation of cell
cycle | 11 |
2.71×10−6 |
| GO:0006793 | Phosphorus
metabolic process | 11 |
1.38×10−2 |
| GO:0006796 | Phosphate metabolic
process | 11 |
1.38×10−2 |
| GO:0016310 |
Phosphorylation | 10 |
1.13×10−2 |
| GO:0042981 | Regulation of
apoptosis | 10 |
1.17×10−2 |
PPI network construction of
differentially expressed miR target genes
According to the information from STRING database, a
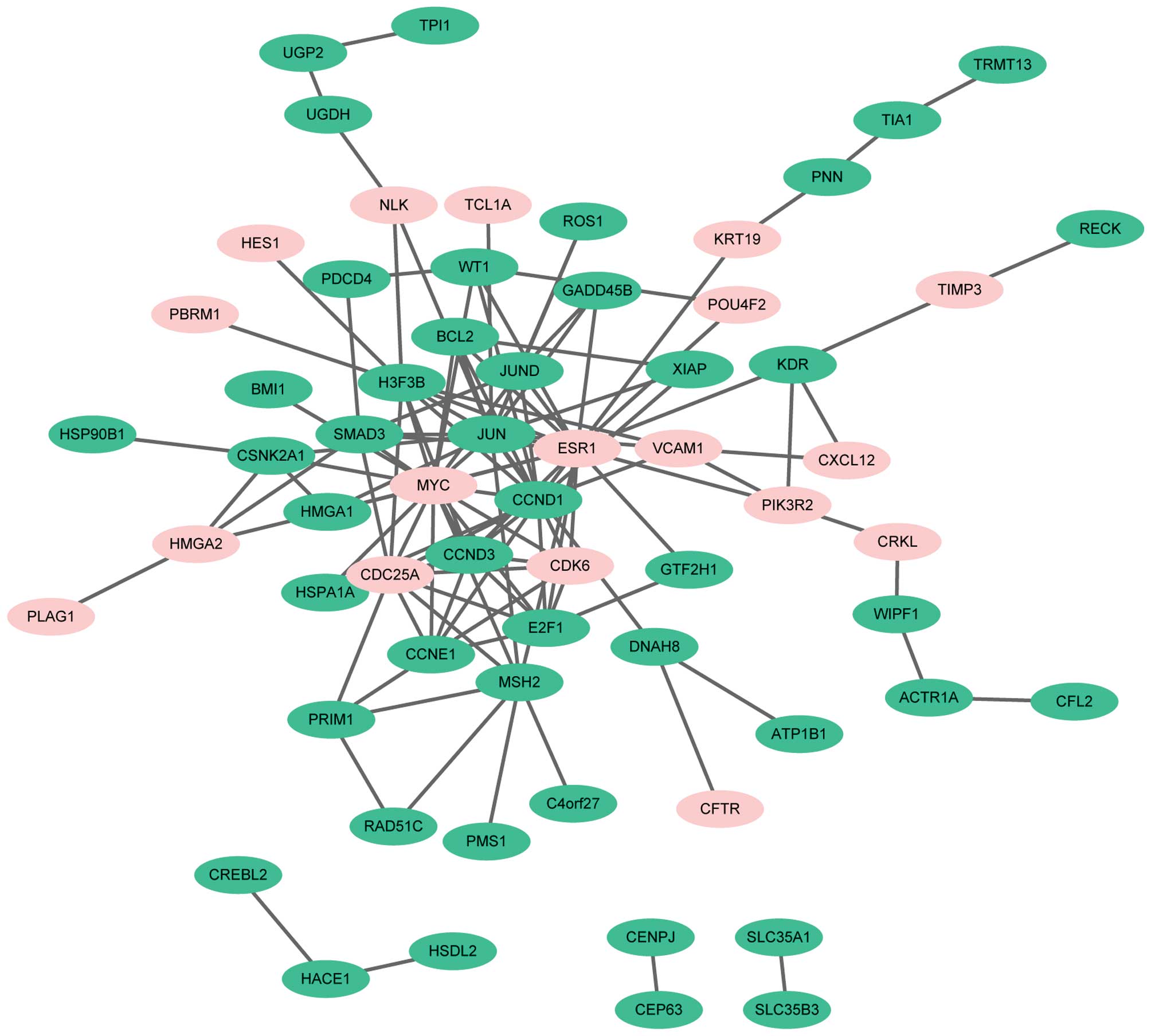
PPI network of differentially expressed miR target genes was
constructed (Fig. 3). The results
demonstrated that the top 6 target genes, from high to low
expression based on node degree, were estrogen receptor 1
(degree=19), MYC (degree=18), JUN (degree=17), CCND1 (degree=15),
cell division cycle 25A (degree=10) and H3 histone, family 3B
(degree=10). Notably, all the target genes enriched in the CRC
pathway, including CCND1, MYC, PIK3R2, MSH2, JUN, BCL2 and SMAD3,
had higher node degrees in the PPI network and the node degrees of
these genes was >4.
Discussion
CRC is a complex disease with genetic and epigenetic
alterations in numerous key oncogenes and tumor suppressor genes
(35). However, the precise molecular
mechanisms leading to anticancer activities in CRC remain unclear.
miR biology has increasingly become a focus in the field of cancer
research, and secreted miRs embedded in exosomes have emerged as
diagnostic biomarkers for cancer detection (24). The present study analyzed the exosomal
miR profile in the sera of CRC patients, derived from a GEO
database, to screen differentially expressed miRs between CRC
patients and healthy controls, and their target genes for
therapeutic intervention.
A total of 18 differentially expressed miRs were
identified in the sera of CRC patients compared to healthy
controls. Furthermore, the present results revealed that 3
upregulated (let-7b, miR-1290 and miR-126) and 2 downregulated
(miR-16 and miR-760) differentially expressed miRs and their target
genes, including CCND1, MYC, PIK3R2 and SMAD3, were significantly
enriched in the CRC pathway. All these target genes had higher node
degrees in a PPI network constructed in the present study.
CCND1 was the identified crucial target gene of
let-7b and miR-16 in the network of miRs and target genes. let-7b
is a member of the let-7 family, which may inhibit cancer cell
growth and promote cell apoptosis via post-transcriptional
repression of cytochrome P450 epoxygenase 2J2 (CYP2J2) (36). CYP2J2 has been observed to have a
tumor-promotion function in human cancer (36). In addition, miR-16 is a regulator of
microtubule-associated protein 7 in modulating the proliferation in
several types of cancer cell (37).
In vitro experiments reveal that overexpression of miR-16
represses the proliferation and induces apoptosis of CRC cells by
regulating the p53/survivin signaling pathway (38). Furthermore, CCND1 is widely recognized
to be involved in cancer cell cycle progression and is vital in
controlling G1/S cell cycle transition (39). CCND1 regulates the switch between
proliferation and growth arrest, and the CCND1 G870A polymorphism
contributes to CRC susceptibility (40). Additionally, CCND1 is regulated by the
miR-16 family, and the miR-16 family induces G1 arrest by
regulating the expression of CCND1 (41). let-7b inhibits cell cycle progression
and anchorage-independent growth of melanoma cells via the
downregulation of CCND1 (42). The
results of the present study are similar to those of previous
studies, as let-7b and miR-16 could directly target CCND1. The
present results additionally suggest that let7b and miR-16 inhibit
CRC progression by targeting CCND1.
miR-1290 was also demonstrated to be upregulated in
the present study. miR-1290 and its target gene MYC are considered
to be crucial in the regulation of CRC. Upregulation of miR-1290 in
colon cancer cells impairs cytokinesis, which is the final step of
cell division (43). Furthermore,
overexpression of miR-1290 activates the Wnt pathway (43), which has been demonstrated in several
types of cancer, including CRC (44),
and is important in tumorigenesis (45). The MYC proto-oncogene encodes a
prototypical oncogenic transcription factor that is crucial in the
genesis of various cancers (46). MYC
is part of the Mad-Max network that regulates the cell cycle, and
cell differentiation and apoptosis (47,48).
Previous experimental data indicates that even a slight inhibition
of c-MYC expression may permanently inhibit tumor growth and
enhance the regression of tumors (49). A previous study by Wu et al
(43) demonstrated that miR-1290 was
involved in the reprogramming of colon cancer cells by increasing
the expression of c-MYC. Thus, miR-1290 may be a putative oncogenic
miR in promoting CRC progression.
Similarly, the upregulated miR-126 and target gene
PIK3R2 were also identified in the network of miRs and target
genes. miR-126 is upregulated in primary CRC tissues and cell
lines; a previous study verified that epigenetic silencing of
miR-126 in CRC cells effectively leads to cell growth, migration
and invasion (50,51). PIK3R2 is a key regulator in the
phosphoinositide 3-kinase (PI3K) pathway, which is a key signal
transduction system that associates oncogenes with numerous
cellular functions and has a role in human cancer (52). In addition, miR-126 may be important
in tumorigenesis and metastasis by targeting PIK3R2 to regulate the
vascular endothelial growth factor/PI3K/protein kinase B signaling
pathway in human breast cancer (53).
On the basis of the present results, the present authors
hypothesise that miR-126 may be an important molecule in the
pathogenesis of CRC via the regulation of PIK3R2.
In the present study miR-760 was downregulated, and
was demonstrated to interact with SMAD3. SMAD3 is a downstream
mediator of the transforming growth factor-β (TGF-β) signaling
pathway (54). TGF-β signaling
regulates tumorigenesis, and its signaling pathway is crucial in
tumor progression in human cancer (55). Additionally, SMAD3 expression
decreases susceptibility to tumorigenicity in the early stages of
gastric carcinogenesis (56). In the
present study, miR-760 was downregulated, which was similar to a
previous finding that miR-760 had a lower expression in CRC tissues
and might be a biomarker for the early detection of CRC (57). Therefore, miR-760 may indirectly
inhibit tumorigenesis and progression of CRC by targeting SMAD3.
However, few functional studies have been performed on miR-760;
therefore, additional experimental validation and high throughput
data analysis are required.
In conclusion, the present study has identified
differentially expressed miRs in the sera of CRC patients compared
with healthy controls. let-7b, miR-16, miR-1290, miR-126 and
miR-760, and their target genes, CCND1, MYC, PIK3R2 and SMAD3, may
be important in the initiation and progression of CRC. let-7b and
miR-16 may regulate cell cycle processes by interacting with CCND1,
thus inhibiting cancer cell proliferation and growth and induce
cell apoptosis. miR-1290 and miR-126 may function in promoting the
tumorigenesis and metastasis of CRC cells via the regulation of MYC
and PIK3R2, respectively, while miR-760 may indirectly prevent the
progression of CRC by targeting SMAD3. Therefore, these 5 miRs and
their target genes may be candidate biomarkers for the diagnosis
and treatment of CRC. In addition, the present study provides
intriguing clues as to the identification of potential candidate
biomarkers. The target genes of 6 downregulated miRs, including
miR-1287, miR-1295, miR-1299, miR-144, miR-575 and miR-652, were
not identified in the present study; therefore, whether they are
associated with CRC requires addition study. Overall, the present
study may aid additional clinical molecular target therapy
experiments concerning CRC.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Løvf M, Nome T, Bruun J, Eknaes M, Bakken
AC, Mpindi JP, Kilpinen S, Rognum TO, Nesbakken A, Kallioniemi O,
et al: A novel transcript, VNN1-AB, as a biomarker for colorectal
cancer. Int J Cancer. 135:2077–2084. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ogata-Kawata H, Izumiya M, Kurioka D,
Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H,
et al: Circulating exosomal microRNAs as biomarkers of colon
cancer. Plos One. 9:e929212014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bretthauer M: Evidence for colorectal
cancer screening. Best Pract Res Clin Gastroenterol. 24:417–425.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Atkin WS, Edwards R, Kralj-Hans I,
Wooldrage K, Hart AR, Northover J, Parkin DM, Wardle J, Duffy SW
and Cuzick J: UK Flexible Sigmoidoscopy Trial Investigators:
Once-only flexible sigmoidoscopy screening in prevention of
colorectal cancer: A multicentre randomised controlled trial.
Lancet. 375:1624–1633. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen CC, Yang SH, Lin JK, Lin TC, Chen WS,
Jiang JK, Wang HS and Chang SC: Is it reasonable to add
preoperative serum level of CEA and CA19-9 to staging for
colorectal cancer? J Surg Res. 124:169–174. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mencía A, Modamio-Høybjør S, Redshaw N,
Morín M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I,
Steel KP, Dalmay T, et al: Mutations in the seed region of human
miR-96 are responsible for nonsyndromic progressive hearing loss.
Nat Genet. 41:609–613. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li H, Xie S, Liu M, Chen Z, Liu X, Wang L,
Li D and Zhou Y: The clinical significance of downregulation of
mir-124-3p, mir-146a-5p, mir-155-5p and mir-335-5p in gastric
cancer tumorigenesis. Int J Oncol. 45:197–208. 2014.PubMed/NCBI
|
|
13
|
Cao J, Cai J, Huang D, Han Q, Yang Q, Li
T, Ding H and Wang Z: miR-335 represents an invasion suppressor
gene in ovarian cancer by targeting Bcl-w. Oncol Rep. 30:701–706.
2013.PubMed/NCBI
|
|
14
|
Sun Z, Zhang Z, Liu Z, Qiu B, Liu K and
Dong G: MicroRNA-335 inhibits invasion and metastasis of colorectal
cancer by targeting ZEB2. Med Oncol. 31:9822014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Farh KK, Grimson A, Jan C, Lewis BP,
Johnston WK, Lim LP, Burge CB and Bartel DP: The widespread impact
of mammalian MicroRNAs on mRNA repression and evolution. Science.
310:1817–1821. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baskerville S and Bartel DP: Microarray
profiling of microRNAs reveals frequent coexpression with
neighboring miRNAs and host genes. RNA. 11:241–247. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Montecalvo A, Larregina AT, Shufesky WJ,
Stolz DB, Sullivan ML, Karlsson JM, Baty CJ, Gibson GA, Erdos G,
Wang Z, et al: Mechanism of transfer of functional microRNAs
between mouse dendritic cells via exosomes. Blood. 119:756–766.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ogawa Y, Kanai-Azuma M, Akimoto Y,
Kawakami H and Yanoshita R: Exosome-like vesicles with dipeptidyl
peptidase IV in human saliva. Biol Pharm Bull. 31:1059–1062. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu G, Drescher KM and Chen XM: Exosomal
miRNAs: Biological properties and therapeutic potential. Front
Genet. 3:562012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Théry C, Ostrowski M and Segura E:
Membrane vesicles as conveyors of immune responses. Nat Rev
Immunol. 9:581–593. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pegtel DM, Cosmopoulos K, Thorley-Lawson
DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD,
Würdinger T and Middeldorp JM: Functional delivery of viral miRNAs
via exosomes. Proc Natl Acad Sci USA. 107:6328–6333. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Skog J, Würdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky
AM and Breakefield XO: Glioblastoma microvesicles transport RNA and
proteins that promote tumour growth and provide diagnostic
biomarkers. Nat Cell Biol. 10:1470–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Igaz I and Igaz P: Tumor surveillance by
circulating microRNAs: A hypothesis. Cell Mol Life Sci.
71:4081–4087. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and Computational Biology Solutions
using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irizarry
R and Huber W: Springer; New York, PA: pp. 397–420. 2005,
View Article : Google Scholar
|
|
27
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37:(Database Issue). D105–D110.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huangda W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hulsegge I, Kommadath A and Smits MA:
Globaltest and GOEAST: Two different approaches for gene ontology
analysis. BMC Proc. 3(Suppl 4): S102009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kanehisa M: The KEGG database. Novartis
Found Symp. 247:91–101; discussion 101–103, 119–128, 244–252. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huangda W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9. 1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:(Database Issue). D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Patil A and Nakamura H: Filtering
high-throughput protein-protein interaction data using a
combination of genomic features. BMC Bioinformatics. 6:1002005.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Takahashi M, Sung B, Shen Y, Hur K, Link
A, Boland CR, Aggarwal BB and Goel A: Boswellic acid exerts
antitumor effects in colorectal cancer cells by modulating
expression of the let-7 and miR-200 microRNA family.
Carcinogenesis. 33:2441–2449. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen F, Chen C, Yang S, Gong W, Wang Y,
Cianflone K, Tang J and Wang DW: Let-7b inhibits human cancer
phenotype by targeting cytochrome P450 epoxygenase 2J2. Plos One.
7:e391972012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yan X, Liang H, Deng T, Zhu K, Zhang S,
Wang N, Jiang X, Wang X, Liu R, Zen K, et al: The identification of
novel targets of miR-16 and characterization of their biological
functions in cancer cells. Mol Cancer. 12:922013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ma Q, Wang X, Li Z, Li B, Ma F, Peng L,
Zhang Y, Xu A and Jiang B: microRNA-16 represses colorectal cancer
cell growth in vitro by regulating the p53/survivin signaling
pathway. Oncol Rep. 29:1652–1658. 2013.PubMed/NCBI
|
|
39
|
Yang Y, Wang F, Shi C, Zou Y, Qin H and Ma
Y: Cyclin D1 G870A polymorphism contributes to colorectal cancer
susceptibility: Evidence from a systematic review of 22
case-control studies. Plos One. 7:e368132012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Probst-Hensch NM, Sun CL, Van Den Berg D,
Ceschi M, Koh WP and Yu MC: The effect of the cyclin D1 (CCND1)
A870G polymorphism on colorectal cancer risk is modified by
glutathione-S-transferase polymorphisms and isothiocyanate intake
in the Singapore Chinese Health Study. Carcinogenesis.
27:2475–2482. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J,
Xing R, Sun Z and Zheng X: miR-16 family induces cell cycle arrest
by regulating multiple cell cycle genes. Nucleic Acids Res.
36:5391–5404. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Schultz J, Lorenz P, Gross G, Ibrahim S
and Kunz M: MicroRNA let-7b targets important cell cycle molecules
in malignant melanoma cells and interferes with
anchorage-independent growth. Cell Res. 18:549–557. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu J, Ji X, Zhu L, Jiang Q, Wen Z, Xu S,
Shao W, Cai J, Du Q, Zhu Y and Mao J: Up-regulation of
microRNA-1290 impairs cytokinesis and affects the reprogramming of
colon cancer cells. Cancer Lett. 329:155–163. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Behrens J: The role of the Wnt signalling
pathway in colorectal tumorigenesis. Biochem Soc Trans. 33:672–675.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Karim RZ, Tse GM, Putti TC, Scolyer RA and
Lee CS: The significance of the Wnt pathway in the pathology of
human cancers. Pathology. 36:120–128. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zeller KI, Jegga AG, Aronow BJ, O'Donnell
KA and Dang CV: An integrated database of genes responsive to the
Myc oncogenic transcription factor: Identification of direct
genomic targets. Genome Biol. 4:R692003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pelengaris S, Khan M and Evan G: c-MYC:
More than just a matter of life and death. Nat Rev Cancer.
2:764–776. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
48
|
González V and Hurley LH: The c-MYC NHE
III(1): Function and regulation. Annu Rev Pharmacol Toxicol.
50:111–129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hermeking H: The MYC oncogene as a cancer
drug target. Curr Cancer Drug Targets. 3:163–175. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li XM, Wang AM, Zhang J and Yi H:
Down-regulation of miR-126 expression in colorectal cancer and its
clinical significance. Med Oncol. 28:1054–1057. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang Y, Wang X, Xu B, Wang B, Wang Z,
Liang Y, Zhou J, Hu J and Jiang B: Epigenetic silencing of miR-126
contributes to tumor invasion and angiogenesis in colorectal
cancer. Oncol Rep. 30:1976–1984. 2013.PubMed/NCBI
|
|
52
|
Liu P, Cheng H, Roberts TM and Zhao JJ:
Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev
Drug Discov. 8:627–644. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhu N, Zhang D, Xie H, Zhou Z, Chen H, Hu
T, Bai Y, Shen Y, Yuan W, Jing Q and Qin Y: Endothelial-specific
intron-derived miR-126 is down-regulated in human breast cancer and
targets both VEGFA and PIK3R2. Mol Cell Biochem. 351:157–164. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kang HY, Lin HK, Hu YC, Yeh S, Huang KE
and Chang C: From transforming growth factor-beta signaling to
androgen action: Identification of Smad3 as an androgen receptor
coregulator in prostate cancer cells. Proc Natl Acad Sci USA.
98:3018–3023. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Bierie B and Moses HL: TGF-beta and
cancer. Cytokine Growth Factor Rev. 17:29–40. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Han SU, Kim HT, Seong DH, Kim YS, Park YS,
Bang YJ, Yang HK and Kim SJ: Loss of the Smad3 expression increases
susceptibility to tumorigenicity in human gastric cancer. Oncogene.
23:1333–1341. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Liu G, Fang Y, Zhang H, Li Y, Li X, Yu J
and Wang X: Computational identification and microarray-based
validation of microRNAs in Oryctolagus cuniculus. Mol Biol Rep.
37:3575–3581. 2010. View Article : Google Scholar : PubMed/NCBI
|