Introduction
Pituitary adenomas account for 10–15% of all
intracranial neoplasms and are incidentally identified in <27%
of non-selected autopsies (1). The
clinical presentation of pituitary adenomas depends on the
structural and functional characteristics of the tumor (2). The World Health Organization (WHO)
categorizes pituitary tumors into typical adenomas, atypical
adenomas and pituitary carcinomas; of which, typical adenomas
constitute the major class. However, the WHO classification does
not offer an accurate association between the histopathological
findings and the clinical behavior of the tumor (3). An estimated 35–55% of pituitary adenomas
demonstrate invasion into bones, dura or adjacent structures,
including the cavernous or sphenoid sinuses (4). Clinically defined invasive pituitary
adenomas (IPAs) demonstrate earlier and more frequent recurrences,
and may be resistant to conventional treatments, such as surgery
and radiotherapy (5). Specific
biomarkers that distinguish between aggressive and nonaggressive
pituitary adenomas have not yet been identified, although certain
studies suggest that the Ki-67 proliferation index may be of
diagnostic value (3). The WHO
classification of endocrine tumors indicates that invasion of the
surrounding structures, size at presentation, an elevated mitotic
index, a Ki-67 labeling index of >3% and extensive tumor protein
p53 (p53) expression are indicators of aggressive behavior
(6,7).
However, Ki-67 and p53 labeling index evaluations demonstrate
subjective variability, and the cutoff values are controversial
(8). Clinically, endocrine tumors
present a challenging management problem, with a high frequency of
incomplete resections, tendency for recurrence and notable
morbidity (9).
Previously, several studies attempted to identify
novel molecular markers [fibroblast growth factor receptor 4,
matrix metalloproteinases, Ki-67, p53, cyclooxygenase-2,
galectin-3, angiogenesis molecules and pituitary tumor-transforming
1 (PTTG)] that require additional validation (10–13). In a
previous study, multivariate Cox regression analysis assessed
galectin-3 immunohistochemical expression in ≥30% of neoplastic
cells; galectin-3 messenger RNA expression was indicated to be a
strong predictive factor of recurrence or tumor progression
(P<0.001); and a Ki-67 labeling index of >3% (P=0.019) was
indicated in the 81 cases with available follow-up data (12). PTTG expression may be associated with
tumor invasiveness and microvessel density of pituitary adenomas
(13). Apoptosis and mitoses
represent two adverse and asynchronous events that maintain the
optimal cell numbers; cytogenetic analysis may, therefore, be
useful in defining the biological invasion of pituitary tumors
(14). In addition, predicting the
subsequent risk of disease invasion or drug sensitivity is
challenging. However, mutations in classic oncogenes and
tumor-suppressor genes are rarely associated with these tumors
(3–6,8–15). Nonfunctioning pituitary adenomas
(NFPAs) result in few somatic mutations, which is consistent with
the associated low proliferation rates and benign nature; however,
mechanisms other than somatic mutation are likely to be involved in
the etiology of sporadic NFPAs (16).
The majority mechanisms of endocrine tumorigenesis differ
significantly from those associated with haematological
malignancies and non-endocrine tumors (17). In addition, the genetic events
underpinning the development of invasion or refractory pituitary
adenomas are not yet understood (18).
In order to identify the genetic events that may be
contributing to the invasion of pituitary adenomas, whole-exome
sequencing, which has been successfully used to find variants in
multiple tumor types, was applied (16,19–21).
Through stringent variant calling and filtering parameters, 15
identified variants were mainly associated with cell cycle phase,
cellular component organization and biogenesis at cellular level by
whole-exome sequencing in combination with homozygosity mapping
between IPAs and non-invasive pituitary adenomas (nIPA). The
present study supports the role of somatic variants of the PR
domain (PRDM) gene family, which is known to control cell
proliferation in cancer and in normal development, in IPAs.
Materials and methods
Patients and specimens
Specimens from six IPAs and six nIPAs were obtained
from patients that underwent endoscopic transsphenoidal surgery
between December 2009 and January 2010 at Beijing Tiantan Hospital,
Beijing, China. Informed consent was obtained from all individuals
and ethical approval was obtained form the Institutional Review
Board of Beijing Tiantan Hospital Affiliated to Capital Medical
University. Pituitary adenomas, obtained from 12 patients (5 men
and 7 women; mean age, 40.7 years; range, 16.0–63.0 years) that did
not have a family history of endocrine neoplasia, were
characterized based on presurgical clinical and biochemical
findings, including a pituitary hormone test. This tested for 12
types of pituitary hormone: Growth hormone, adrenocorticotropic
hormone, follicle-stimulating hormone, luteinizing hormone,
estradiol, progesterone, human growth hormone, cortisol, total
triiodothyronine, total thyroxine, thyroid-stimulating hormone and
prolactin (PRL) levels (4 patients in normal range, 8 patients with
increased PRL levels; normal range, 2.5–17 ng/ml). Pituitary
adenomas were also characterized based on morphological and
immunohistochemical analysis of removed tissue samples (Table I). Cases with multiple hormonal
changes according to the clinical and pathological data were
excluded. The following IPA diagnostic criteria were adopted: i)
Knosp classification grade III–IV tumors and Hardy classification
invasive adenomas; ii) tumor cells confirmed via pathology as
invading the sphenoid bone or adjacent dura mater; iii) tumor cells
invading the sphenoid sinus cavity or peripheral vascular and
nerve; iv) Ki-67 labeling index of >3% (22). The tumors did not have atypical
features, and constituted the ‘discovery’ set of tumors for exome
capture and DNA sequence analysis. An additional 28 pituitary
adenomas, histologically confirmed, were obtained from 13 women and
15 men (mean age, 61 years; range, 17–71 years), and these
constituted the ‘validation’ set. For histological analysis, the
tumor specimens were divided into two sections. One was stored in
liquid nitrogen and the other was fixed in 4% paraformaldehyde for
24 h (Sinopharm Chemical Reagent Beijing Co., Ltd., Beijing, China)
within 0.5 h of surgery. After washing for 6 h in flowing water,
the specimens underwent gradient dehydration in alcohol, were
embedded in paraffin wax (Leica Biosystems Richmond Inc., Richmond,
IL, USA) and sectioned at a thickness of 5 µm. Sections were
incubated with primary mouse anti-human monoclonal PRDM2 antibody
(catalog no., ab3791; dilution, 1:200; Abcam, Cambridge, MA, USA)
at 4°C overnight. Next, sections were washed three times with
phosphate-buffered saline (PBS; ZSGB-BIO), then incubated with
DyLight-conjugated AffiniPure secondary antibody (goat anti-mouse
IgG H+L; catalog no., ZF-0313; ZSGB-BIO, Beijing, China) with
fluorescence was added at room temperature for 1 h followed by 3
washes with PBS (5 min each). Streptavidin-Biotin Complex
(ZSGB-BIO) was added for 20 min and then the sections were washed
with PBS. Next, sections were mounted with ProLong Gold Antifade
reagent (ZSGB-BIO) with DAPI (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA USA). Staining was visualized using a
LEICA-TCS-SP5II microscope (Leica, Wetzlar, Germany). The
percentage of DAPI-stained cells exhibiting PRDM2 immunoreactivity
was analyzed in 5 randomly selected high power fields.
 | Table I.Clinical data of patients. |
Table I.
Clinical data of patients.
| Specimen | Tumor sub-type | PRL, ng/ml | Age, years | Gender | Tumor volume,
cm3 | Histology | Ki-67 index, % | Invasive | No. of
variants |
|---|
| 1 | NFPA |
5.8 | 63 | F | 16.4 | (−) | >3 | Yes | 23 |
| 2 | NFPA |
13.2 | 53 | F |
4.8 | (−) | 1–2 | No | 12 |
| 3 | NFPA |
9.4 | 42 | M | 13.2 | (−) | >3 | Yes | 28 |
| 4 | NFPA |
11.7 | 53 | F |
7.4 | (−) | 1–2 | No | 14 |
| 5 | PRL |
182 | 25 | F | 10.7 | PRL (+) | >3 | Yes | 27 |
| 6 | PRL | 1,625 | 16 | F | 12.2 | PRL (+) | >3 | Yes | 37 |
| 7 | PRL | 3,117 | 54 | M | 14.3 | PRL (+) | >3 | Yes | 26 |
| 8 | PRL |
268 | 29 | M |
9.2 | PRL (+) | >3 | Yes | 32 |
| 9 | PRL |
123 | 32 | F |
2.4 | PRL (+) | 1–2 | No | 21 |
| 10 | PRL |
233 | 43 | F |
3.1 | PRL (+) | 1–2 | No | 20 |
| 11 | PRL | 2,899 | 34 | M |
1.9 | PRL (+) | 1–2 | No | 25 |
| 12 | PRL | 2,830 | 44 | M |
4.1 | PRL (+) | 1–2 | No | 19 |
Specimen preparation, exome capture,
DNA sequencing and bioinformatics analysis
Total DNA was extracted from pituitary adenomas
using the QIAamp DNA Mini Kit (Qiagen GmbH, Hilden, Germany). An
aliquot containing 5 µg of genomic DNA was purified and quantified
from each specimen. Exome enrichment was performed by using an ABI
SOLiD optimized SureSelect Human All Exon kit (Agilent
Technologies, Inc., Santa Clara, CA, USA), which included the
exonic sequences of ~18,000 genes, covering a total of 42 Mb of
genomic sequences. The enriched exome libraries were then amplified
by emulsion polymerase chain reaction (ePCR; Ion PI™ Hi-Q™ OT2 200
kit; cat no. a26434), according to the manufacturer's instructions
(Thermo Fisher Scientific, Inc.), and based on a library
concentration of 0.5 pM. The PCR products were then sequenced on a
SOLiD5500 sequencer (Thermo Fisher Scientitic, Inc.), and one
quadrant of a SOLiD sequencing slide was required for each
sample.
Color-space reads were mapped to the hg19 reference
human genome using SOLiDBioScope software (5500 W Series Genetic
Analyzer V2.0; Thermo Fisher Scientitic, Inc.), which is suitable
for a repetitive mapping approach. Single-nucleotide polymorphisms
(SNPs) were then called using the diBayes algorithm with
conservative default call stringency. Known SNPs available from the
Single Nucleotide Polymorphism Database (dbSNP) version 130, which
is maintained by the National Center for Biotechnology Information,
were excluded.
Mutation validation
Primer3 software (version 0.4.0; http://frodo.wi.mit.edu/primer3/) was used to
generate primers for the PCR amplification of variants identified
via exome sequencing or exons covered in additional screening using
a SOLiD5500xl sequencer (Thermo Fisher Scientific, Inc.; Table II). The DNA ladder (DL1000; Takara
Bio, Inc., Otsu, Japan) and ethidium bromide were purchased from
Takara Bio, Inc. Amplification products of an appropriate size were
identified using agarose gel electrophoresis (100 V, 30 min).
Amplicons from 3 normal pituitary and 28 pituitary tumor DNA
molecules coupled with leukocyte were sequenced using forward and
reverse primers. Variants were confirmed by at least two
independent sequences from various primers.
 | Table II.Methylation validation primers. |
Table II.
Methylation validation primers.
| Gene | Forward primer | Reverse primer |
|---|
| PRDM8 |
5′-ATTCCCTTTCAAACGACCAGA-3′ |
5′-AAGAGTTGGATACGTCGTAAA-3′ |
| PRDM2 |
5′-GGCCAAGAAGCGGAGAACT-3′ |
5′-AAGTCACAGCGACTCACCAGC-3′ |
| MGAM |
5′-GGCGGAGTCCTTGCTCTTAT-3′ |
5′-GTATGACAGTGCAGCTTCAGGA-3′ |
| SPANXN2 |
5′-GAGGAGGACGAAGGCCTAGA-3′ |
5′-CTCACTACCAATGGCGATGA-3′ |
| TRIOBP |
5′-CCAGGCTTCCTCCATGACAC-3′ |
5′-TGTGTCCAGCAGGACGATC-3′ |
| ZNF717 |
5′-CCTTTCGCTGTAAGTCATTCCT-3′ |
5′-TCAGAGAACTCATGCTGGCA-3′ |
| PRB3 |
5′-CCCCCACAAGGAGGAAACCA-3′ |
5′-CCACAAGGAGGAAACCAGT-3′ |
| DPCR1 |
5′-TTCTGATTGGACTCCCTCTC-3′ |
5′-TAGTGCGATCTCCTGACCTC-3′ |
| DSPP |
5′-ATCTCTTGTAATTTAGCTACC-3′ |
5′-AATATATTGGTACATCACCA-3′ |
| MX2 |
5′-AGAAGCTTGGACGTGCCAAG-3′ |
5′-AGGGGTCCAGGTCACAGCC-3′ |
| EGFL7 |
5′-TCCTGGGTTGGGTCAGCCATGC-3′ |
5′-AATTGAATGATGTGCAGTTG-3′ |
| LRRC50 |
5′-CGAGACCATCCTAGCCAACAC-3′ |
5′-TGTTCCTTCTGATGTTCGGAT-3′ |
| LRP1B |
5′-AGCCAATTCGAATCCTTGCTA-3′ |
5′-TTGCATGACTAATATACCTGTT-3′ |
| MAST4 |
5′-CTTGAACTCTGCCTCAAGCATTC-3′ |
5′-ACAAGAACTGGTTTGGTAC-3′ |
| RP1L1 |
5′-GCTTGCCCTTGATATCCTTTTAT-3′ |
5′-TTCATCTGCAAACTTAACTCCG-3′ |
| GAPDH |
5′-CAGCTGAGGGACCCATGAA-3′ |
5′-AAGTGGTCATTGAGGGCGAT-3′ |
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from frozen normal pituitary
and pituitary adenomas (~50 mg) using the TRIzol Reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). RT-qPCR was performed
as described previously (23), using
the Applied Biosystems 7500 Fast System (Thermo Fisher Scientific,
Inc.) and the primers indicated in Table III. The fold-change in differential
expression for each gene was calculated using the comparative Cq
method (also known as the 2−ΔΔCq method), as previously
described (24).
 | Table III.Reverse transcription-quantitative
polymerase chain reaction primers. |
Table III.
Reverse transcription-quantitative
polymerase chain reaction primers.
| Gene | Forward primer | Reverse primer |
|---|
| DPCR1 |
5′-AGTGCTGCCTCCTCTTCCTTCTA-3′ |
5′-GGGAGCTCTGGAGGTCTTTGTC-3′ |
| DSPP |
5′-GCATTTGGGCAGTAGCATGG-3′ |
5′-CTGACACATTTGATCTTGCTAGGAG-3′ |
| MGAM |
5′-GGCGGAGTCCTTGCTCTTAT-3′ |
5′-GTATGACAGTGCAGCTTCAGGA-3′ |
| EGFL7 |
5′-ATGTGGATGAATGCAGTGCT-3′ |
5′-TGTCCACTCCTGTCGGGTT-3′ |
| MX2 |
5′-GCCAGGTGGAGAAAGAGATACACAA-3′ |
5′-AGGTCAATGATGGTCAGGTCTGG-3′ |
| LRRC50 |
5′-CGAGACCATCCTAGCCAACAC-3′ |
5′-TGTTCCTTCTGATGTTCGGAT-3′ |
| PRDM2 |
5′-AGCAGCTGCGATTGAGGA-3′ |
5′-CAGAGGTGAAATCTGGCTCACTT-3′ |
| PRDM8 |
5′-ATTCCCTTTCAAACGACCAGA-3′ |
5′-AAGAGTTGGATACGTCGTAAA-3′ |
| LRP1B |
5′-AGCCAATTCGAATCCTTGCTA-3′ |
5′-TTGCATGACTAATATACCTGTT-3′ |
| RP1L1 |
5′-AGAAGCGAGGCTGAAACTTTATCTG-3′ |
5′-TCACACTCGGCTTGGTCTTTG-3′ |
| PRB3 |
5′-CCTCCAGCAAGATGCTACTGATT-3′ |
5′-GGGAGATTCTTCCTGGCTGA-3′ |
| ZNF717 |
5′-CCTTTCGCTGTAAGTCATTCCT-3′ |
5′-TCAGAGAACTCATGCTGGCA-3′ |
| MAST4 |
5′-CTTGAACTCTGCCTCAAGCATTC-3′ |
5′-ACAAGAACTGGTTTGGTAC-3′ |
| SPANXN2 |
5′-GAGGAGGACGAAGGCCTAGA-3′ |
5′-CTCACTACCAATGGCGATGA-3′ |
| TRIOBP |
5′-CCAGGCTTCCTCCATGACAC-3′ |
5′-TGTGTCCAGCAGGACGATC-3′ |
| GAPDH |
5′-TGAAGGGCATTCTGGGATAC-3′ |
5′-TGTGGACACCACCTGTAGGA-3′ |
Immunohistochemical analysis
Pituitary adenomas and pituitary gland specimens
were sectioned to a thickness of 5 µm in paraffin wax (Leica
Biosystems Richmond Inc.). The sections were subjected to gradient
dewaxing, removed of water, treated with fresh 3% hydrogen peroxide
(ZSGB-BIO) at room temperature for 10 min and washed with
phosphate-buffered saline (pH 7.2; ZSGB-BIO) 3 times for 5 min
each. For microwave repair, the specimens were placed in 0.01%
citric acid (pH 6.0; ZSGB-BIO), kept warm in a microwave oven (600
W; LG Electronics Appliances Co., Ltd., Tianjin, China) for 10 min,
allowed to cool to room temperature and washed once with PBS for 10
min. Antibody repair solution was added at room temperature for 10
min, and then washed 3 times with PBS for 5 min each time. PRDM2
antibody (monoclonal; Abcam Inc., Eugene, OR, USA) was added at a
1:200 dilution and incubated at 4°C overnight. The
DyLight™-conjugated AffiniPure secondary antibody with fluorescence
(ZSGB-BIO) was added at room temperature for 1 h, followed by 3
washes with PBS for 5 min each time. Streptavidin-biotin complex
was added for 20 min and then washed with PBS for 5 min. Sections
were mounted with Prolong Gold Antifade reagent with
4′,6-diamidino-2-phenylindole (DAPI; Invitrogen; Thermo Fisher
Scientific, Inc.). Sections were analyzed with a LEICA-TCS-SP5II
(Leica Microsystems GmbH, Wetzlar, Germany) to estimate the
percentage of DAPI-stained cells displaying PRDM2
immunoreactivity.
Statistical analysis
All statistical analyses were performed using SPSS
version 20.0 (IBM SPSS, Armonk, NY, USA). For comparisons, one-way
analyses of variance, χ2 tests, Wilcoxon rank-sum tests
and two-tailed Student's t-tests were performed as appropriate.
Binary logistic regression was performed to identify the
independent factors associated with pituitary adenoma recurrence.
P<0.05 was used to indicate a statistically significant
difference.
Results
Identification of variant genes by
whole-exome sequencing
For the identification of tumor-specific somatic
variants, whole-exome capture using DNA from the discovery set of
six IPAs and six nIPAs yielded excellent target region coverage,
with ~72% of the exome covered to a depth of at least 30-fold
between the somatic variant calling algorithm and confirmatory
sequencing. Several prioritization steps were taken to decrease the
number of genetic variants and to find the potentially pathogenic
variants, as follows: i) Variants should have a deleterious effect
on protein function (as predicted by protein prediction software,
such as PolyPhen-2, MutationTaster and SIFT); ii) variants should
be present at sufficient allele frequency to represent likely
heterozygous or homozygous changes (i.e., present from early in the
tumorigenic process), although deviation from the expected
heterozygous or homozygous allele frequencies may represent either
contamination with normal tissue or the preference of the sequence
and alignment process for the wild-type allele, as previously
reported (25,26); and iii) variants should be involved in
biological processes relevant to tumorigenesis (27). Approximately 90% of single-nucleotide
variants (SNVs) resulted in missense amino-acid changes, whereas
the remaining (~10%) were synonymous changes. Over 70% of the SNVs
occurred as C:G-T:A transitions, and <30% were transversions.
Using stringent variant calling and filtering parameters (16), 233 variants were identified in the
specimens.
In addition, five variants (C8orf79, chr8:12879694;
FSHD region gene 1 family member B, pseudogene, chr20:29632674;
mucin 2, oligomeric mucus/gel-forming, chr11:1092715; mucin 6,
oligomeric mucus/gel-forming, chr11:1018092; and solute carrier
family 5 member 3, chr21:35467473) were present in all specimens,
and 47 were detected in either the IPA or nIPA. Of these, 15 were
somatic variants confirmed by dideoxynucleotide sequencing. Of the
15 confirmed variants, 13 occurred as SNVs, including three
synonymous SNVs, and two comprised insertions (Table IV). The genes with variants were
generally associated with angiogenesis, metabolism, cell cycle
phase, cellular component organization, cytoskeleton and biological
immunity at a cellular level. The genes include: EGF like domain,
multiple 7 (EGFL7), associated with angiogenesis; low density
lipoprotein receptor-related protein 1B (LRP1B) and
maltase-glucoamylase (α-glucosidase) associated with cell
metabolism; dentin sialophospho protein (DSPP), PR domain
containing 2, with ZNF domain, RIZ1 (PRDM2), PR domain containing 8
(PRDM8) and zinc finger protein 717 (ZNF717) associated with cell
proliferation; dynein, axonemal, assembly factor 1 (LRRC50),
microtubule associated serine/threonine kinase and TRIO and F-actin
binding protein (TRIOBP) associated with cytoskeleton; myxovirus
(influenza virus) resistance (MX2) associated with cell cycle
phase; diffuse panbronchiolitis critical region 1 (DPCR1),
proline-rich protein BstNI subfamily 3 (PRB3) and SPANX family
member N2 (SPANXN2) associated with immune response; and KIAA0226
associated with vesicle trafficking.
 | Table IV.Variants identified in sporadic
pituitary adenomas. |
Table IV.
Variants identified in sporadic
pituitary adenomas.
| Gene | Gene name | Exon-ID | Coverage | Variant | Start | End | Reference | Gene type | Protein change | Catalog (case
≥4) | Mutation type |
|---|
| DPCR1 | Diffuse
panbronchiolitis critical region 1 | DPCR1-2 | 104 | SNV |
30919188 |
30919188 | A | C | N983H |
IPA | Missense |
| DSPP | Dentin sialophospho
protein | DSPP-5 | 109 | SNV |
88536900 |
88536900 | A | G | N1029S |
IPA | Missense |
| EGFL7 | EGF-like-domain,
multiple 7 | EGFL7-10 | 38 | SNV | 139565452 | 139565452 | G | A | D208N | nIPA | Missense |
| KIAA0226 | KIAA0226 | KIAA0226-16 | 52 | SNV | 197409451 | 197409451 | C | T | – |
IPA | Synonymous |
| LRRC50 | Dynein, axonemal,
assembly factor 1 | LRRC50-8 | 33 | SN |
84203660 |
84203660 | G | A | – |
IPA | Synonymous |
| LRP1B | Low density
lipoprotein receptor-related protein 1B | LRP1B-7 | 46 | SNV | 141946094 | 141946094 | G | A | V303I | nIPA | Missense |
| MAST4 | Microtubule
associated serine/threonine kinase family member 4 | MAST4-32 | 57 | Insert |
65892764 |
65892764 | – | GCT | – | nIPA | Frame shift |
| MGAM |
Maltase-glucoamylase (α-glucosidase) | MGAM-3 | 37 | SNV | 141708332 | 141708332 | C | A | P52T | nIPA | Missense |
| MX2 | Myxovirus
(influenza virus) resistance | MX2-10 | 33 | SNV |
42771182 |
42771182 | G | A | – |
IPA | Synonymous |
| PRB3 | Proline-rich
protein BstNI subfamily 3 | PRB3-3 | 37 | SNV |
11420621 |
11420621 | G | A | P188W | nIPA | Missense |
| PRDM2 | PR domain
containing 2, with ZNF domian, RIZ1 | PRDM2-10 | 53 | Insert |
14106398 |
14106398 | – | TCC | – |
IPA | Frame shift |
| PRDM8 | PR domain
containing 8 | PRDM8-10 | 32 | SNV |
81122528 |
81122528 | A | G | N102D | nIPA | Missense |
| SPANXN2 | SPANX family,
member N2 | SPANXN2-2 | 50 | SNV | 142796177 | 142796177 | T | C | L167Q | nIPA | Missense |
| TRIOBP | TRIO and F-actin
binding protein | TRIOBP-7 | 36 | SNV |
38119487 |
38119487 | A | C | E308D | nIPA | Missense |
| ZNF717 | Zinc finger protein
717 | ZNF717-5 | 70 | SNV |
75786450 |
75786450 | G | A | T775H | nIPA | Missense |
Furthermore, three variants were indicated in PRDM2
in five separate IPA specimens, including two synonymous and one
frame shift. One mutation was indicated in PRDM8 (missense, N102D)
and PR domain containing 10 (missense, S1018R) in a tumor
separately (data not shown), and four variants (R246Q, G272C, S501F
and A1247G) were indicated in PR domain containing 16 in three
separate IPA specimens (data not shown).
Analysis of the expression of variant
genes by RT-qPCR
RT-qPCR was used to test whether the invasion of
pituitary adenomas was associated with differences in the
expression levels of 15 genes. Expression of DPCR1, KIAA0226, MX2,
PRB3, PRDM2, PRDM8, SPANXN2, TRIOBP and ZNF717 in IPA specimens was
50% decreased compared with in nIPA specimens (Fig. 1). In particular, DPCR1, PRDM2, PRDM8,
SPANXN2 and ZNF717 mRNA levels in IPA specimens were approximately
four-fold decreased compared with in nIPA specimens (P=0.003,
0.007, 0.009 and 0.004, respectively). By contrast, the mRNA levels
of DSPP, EGFL7, LRP1B and LRRC50 in IPA specimens were increased
compared with in nIPA specimens (P=0.041, 0.037, 0.022 and 0.013,
respectively; Fig. 2).
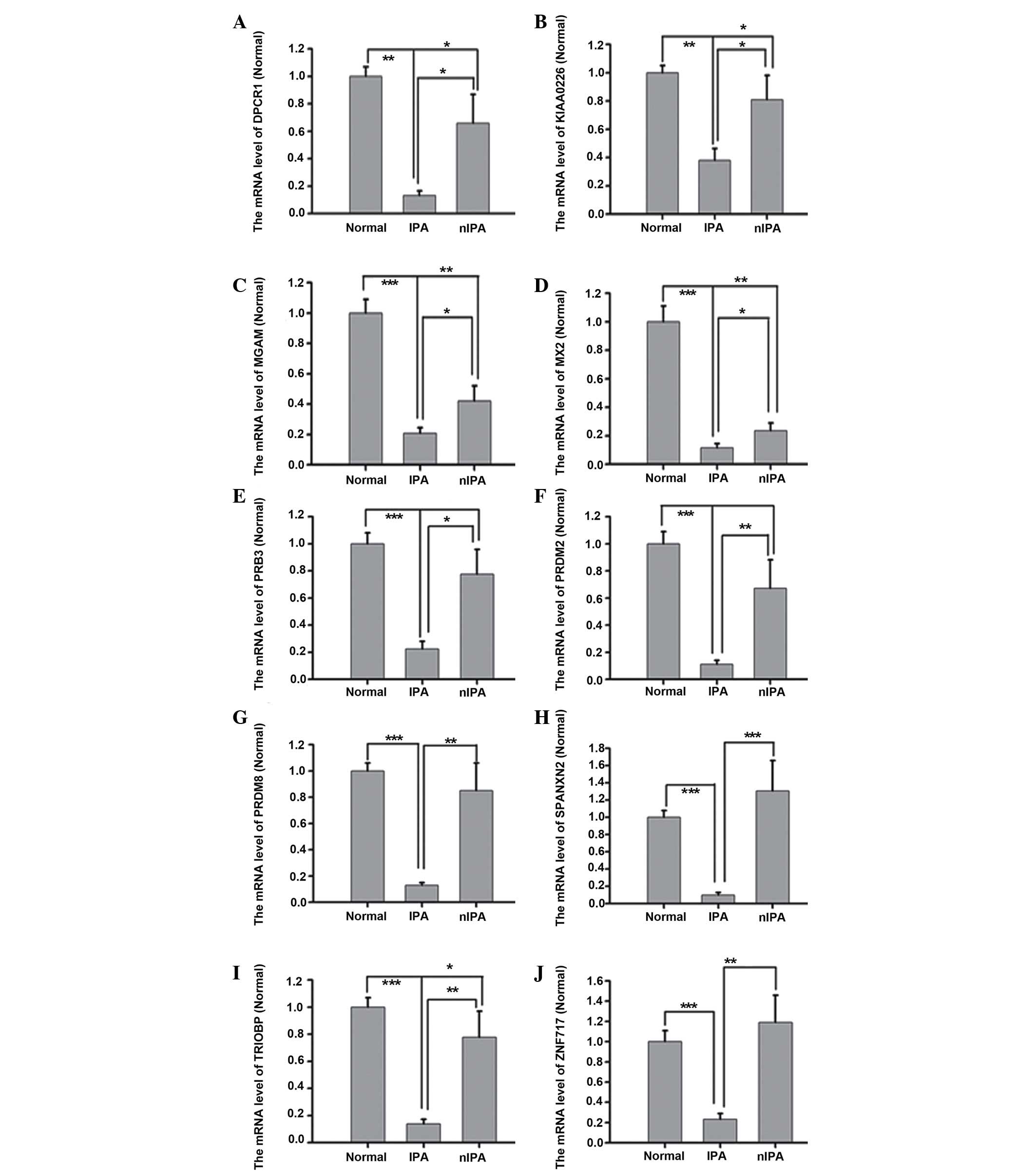 | Figure 1.mRNA levels of (A) DPCR1, (B)
KIAA0226, (C) MGAM, (D) MX2, (E) PRB3, (F) PRDM2, (G) PRDM8, (H)
SPANXN2, (I) TRIOBP and (J) ZNF717 with variants, which are lower
in IPA specimens compared with nIPA specimens. mRNA levels of
DPCR1, KIAA0226, MX2, PRB3, PRDM2, PRDM8, SPANXN2, TRIOBP and
ZNF717 in IPA specimens were 50% decreased compared with nIPA
specimens. In particular, DPCR1, PRDM2, PRDM8, SPANXN2 and ZNF717
mRNA levels in IPA specimens were approximately four-fold lower
compared with nIPA specimens (P<0.01). n=3–8. Groups were
Normal, IPA and nIPA. nIPA, non-invasive pituitary adenoma; IPA,
invasive pituitary adenoma; Normal, normal pituitary; mRNA,
messenger RNA; DPCR1, diffuse panbronchiolitis critical region 1;
MX2, myxovirus (influenza virus) resistance; PRB3, proline-rich
protein BstNI subfamily 3; PRDM2, PR domain containing 2, with ZNF
domain, RIZ1; PRDM8, PR domain containing 8; SPANXN2, SPANX family
member N2; TRIOBP, TRIO and F-actin binding protein; ZNF717, zinc
finger protein 717. *p<0.05; **p<0.01; ***p<0.001. |
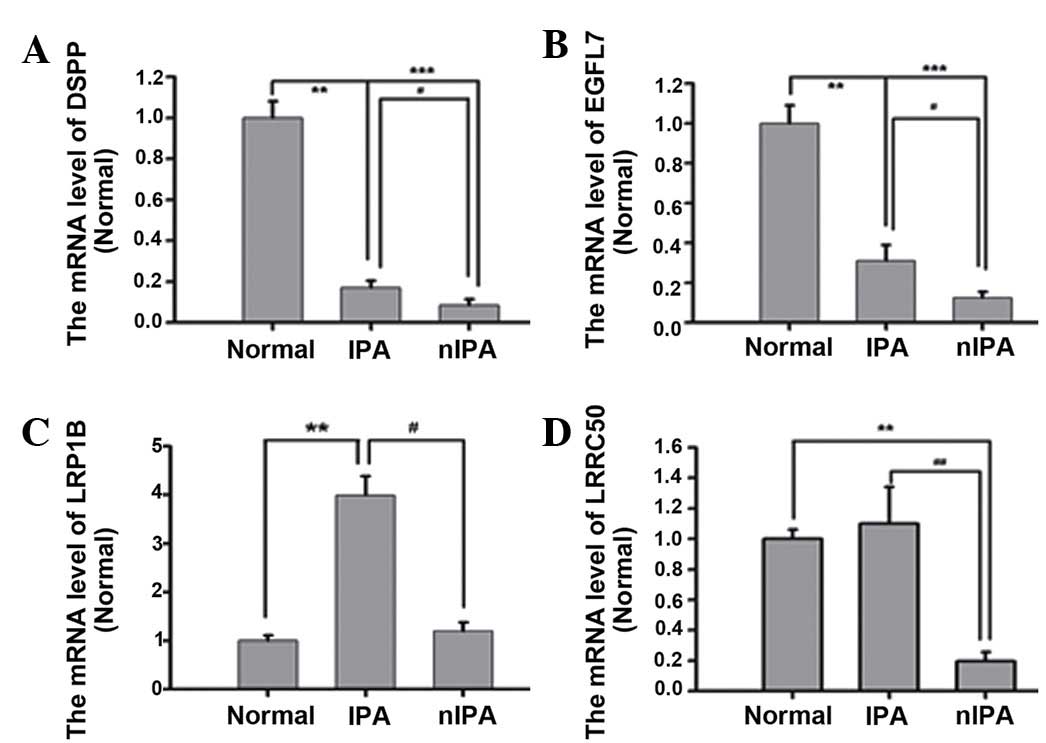 | Figure 2.mRNA level of (A) DSPP, (B) EGFL7,
(C) LRP1B and (D) LRRC50 with variants, which are increased in IPA
specimens compared with nIPA specimens. mRNA levels of DSPP, EGFL7
and LRRC50 were increased in IPA compared with in nIPA specimens
(P<0.01). n=3–8. Groups were Normal, nIPA and IPA. mRNA,
messenger RNA; Normal, normal pituitary; nIPA, non-invasive
pituitary adenoma; IPA, invasive pituitary adenoma; DSPP, dentin
sialophospho protein; EGFL7, EGF like domain, multiple 7; LRRC50,
dynein, axonemal, assembly factor 1. *p<0.05; **p<0.01;
***p<0.001. |
PRDM2 levels are associated with
recurrence in pituitary adenomas
The usual morphological signs of tumor aggression
are poorly associated with the invasive potential of pituitary
tumors, proliferation capacity, tendency of post-surgical
recurrence and global biological behavior (28). PRDM2 contains a PR domain that
demonstrates histone H3 lysine 9 methylation activity (29). Therefore, whether decreased levels of
PRDM family mRNA in pituitary adenomas was associated with certain
clinical parameters was assessed (Table
V). No significant association was indicated between PRDM2 mRNA
levels and age, gender or tumor size. However, decreased PRDM2
protein levels were more frequently observed in recurrent tumors
(Fig. 3). Furthermore, binary
multivariate regression revealed that decreased levels of PRDM2
were independently associated with tumor recurrence (odds ratio,
0.065; 95% confidence interval, 0.050–0.832; P=0.036).
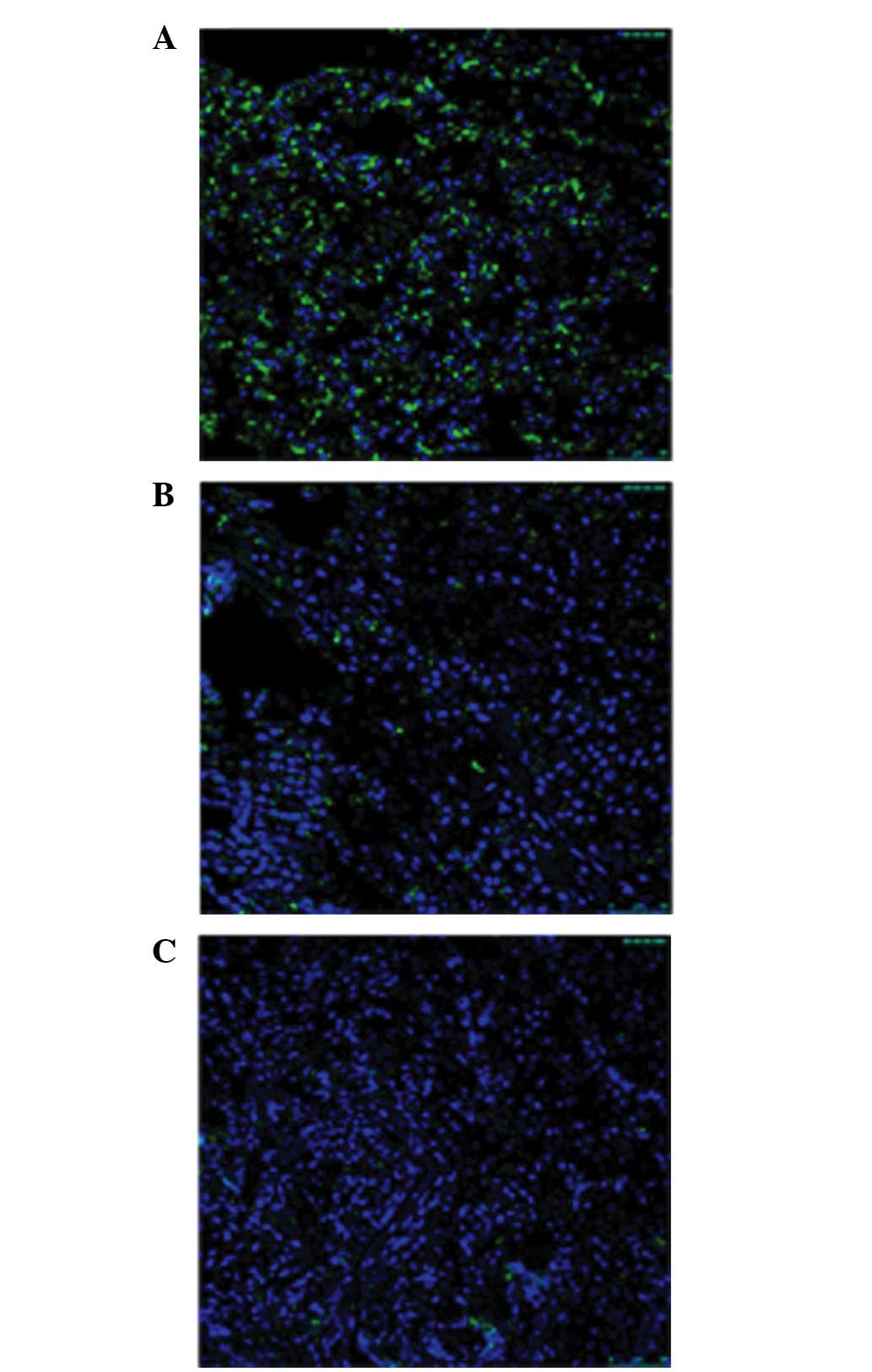 | Figure 3.Confocal images show the number of
PRDM2-positive puncta in recurrent pituitary adenomas. (A) Normal
pituitary; (B) non-invasive pituitary adenoma; (C) inviasive
pituitary adenoma. Green, PRDM2; dilution, 1:200. Blue,
4′,6-diamidino-2-phenylindole; dilution, 1:3,000. Scale bar, 50 µm.
PRDM2, PR domain containing 2, with ZNF domain, RIZ1. |
 | Table V.Association between PRDM2 mRNA level
in pituitary adenomas and various clinical parameters. |
Table V.
Association between PRDM2 mRNA level
in pituitary adenomas and various clinical parameters.
|
| mRNA level |
|
|
|---|
|
|
|
|
|
|---|
| Parameter | Higha | Low | χ2 | P-value |
|---|
| All cases | 12 | 12 |
|
|
| Age, years |
|
| 0.689 | 0.406 |
|
≥50 | 4 | 6 |
|
|
|
<50 | 8 | 6 |
|
|
| Gender |
|
| 0.168 | 0.682 |
|
Male | 6 | 7 |
|
|
|
Female | 6 | 5 |
|
|
| Tumor size, cm |
|
| 0.178 | 0.673 |
| ≥2 | 5 | 4 |
|
|
|
<2 | 7 | 7 |
|
|
|
Recurrenceb |
|
| 6.511 | 0.011 |
|
Yes | 4 | 10 |
|
|
| No | 8 | 2 |
|
|
Discussion
Tumor invasion may be based on clinical,
radiological and pathological features (2–6,8–21,23,27–30).
However, no standard or comparable score on IPA is generally
accepted, except for radiological classification. IPAs are
associated with a poor prognosis, as therapeutic options are
limited. In addition, IPAs tend to recur quickly following initial
treatment, are generally unresponsive to therapy and are a
challenge to manage (5). In the
present study, 15 somatic variants that are mainly associated with
metabolism, cell cycle phase, cellular component organization,
cytoskeleton and biological immunity at a cellular level, but not
with genes previously implicated in pituitary adenomas, were
identified by whole-exome sequencing.
A growing body of evidence suggests a coevolutionary
model of cancer, wherein the cross-talk between tumor cells and the
host determine the malignant potential of individual tumors
(31). Endogenous T cells respond to
and infiltrate tumors, significantly delaying malignant progression
in mouse models (32). Low expression
levels of interleukin-6 and signal transducer and activator of
transcription 3 were indicated to be significant in the dysimmunity
of pituitary adenoma (33). DPCR1,
located between major histocompatibility complex (MHC), class I
(HLA)-B and HLA-A on chromosome 6p21.33, is classified as one of
the HLA molecules. The DPCR1 gene may contain markers for diagnosis
of diffuse pan-bronchiolitis, a bronchiolar disease that affects
human airways (34). To fully escape
the immune system, cancer cells typically mutate to decrease the
expression of antigens, lose expression of HLA proteins or employ
an aberrant antigen processing pathway (35). However, to the best of our knowledge,
the association between DPCR1 variations and the risk of IPA has
not yet been investigated. The mRNA level of DPCR1 is approximately
four-fold lower in IPA compared with in nIPA specimens; therefore,
the invasion of pituitary adenomas is hypothesized to be associated
with the induction of immune-escape via the downregulation of
DPCR1.
The expansion of solid tumors depends on the
continuous growth of novel blood vessels from pre-existing
capillaries. The role of angiogenesis and tumor blood vessels in
the pathogenesis of pituitary tumors remains a mystery. A previous
study indicated the involvement of prolactin during vasculature
remodelling by acting on the endothelial and perivascular cells in
pituitary adenomas (36). The
vascular-specific secreted factor EGFL7 is a component of the
interstitial extracellular matrix (ECM) and regulates the proper
spatial organization of endothelial cells within each filopodia,
affecting the collective movement of the cells (37). A previous study indicated that the
expression of EGFL7 in neural stem cells (NSCs) in vitro
decreased NOTCH-specific signaling and resulted in the decreased
proliferation and self-renewal of NSCs (38). EGFL7 acts as a soluble NOTCH
inhibitor, which is in contrast to the typical NOTCH inhibitory
molecules that are expressed by adjacent cells as transmembrane
proteins (39,40). A previous study indicated that the
differentially expressed genes involved in this pathway were
delta-like 1 homolog, C-terminal binding protein 2, hes family bHLH
transcription factor (HES)1, HES5 and E1A binding protein p300 in
plurihormonal pituitary adenomas (41). Another study used RT-qPCR assays and
western blot analyses to observe upregulated NOTCH3 and jagged 1
(JAG1) in human NFPAs; furthermore, NOTCH3 was positively
associated with JAG1 at the mRNA and protein levels (42). The elevated expression of EGFL7 in
IPAs may be associated with invasive behavior via activation of the
NOTCH pathway.
The attachment, movement and invasion of cancer
cells are facilitated by the actin cytoskeleton and tubulin, as the
structural element of microtubules (8–21,23,27–43). ECM
proteins send information to the cells, which respond with a
specific cytoskeletal organization that modulates specific cellular
patterns of behavior in GH3 tumor pituitary cell (9–21,23,27–44).
Pituitary tumor cells acquire different patterns: mesenchymal, and
leucocyte/amoeboid, the last observed in the invasive adenomas and
amoeboid migration pattern has been associated with high invasion
capacity (10–21,23,27–45).
LRRC50 was identified as a putative ciliary protein in two
independent bioinformatic studies (46,47).
LRRC50 hu255H mutants develop pronephric cysts with an increased
proliferative index, severely reduced brush border, and
disorganized pronephric cilia manifesting impaired localized fluid
flow consistent with ciliary dysfunction (48). LRRC50 to be a novel tumor suppressor
implicated in human seminoma pathogenesis. A pathogenic Gln307Glu
change is significantly enriched in individuals with seminoma
tumors (48). The Arg488Glu mutation
of LRRC50 in IPA is, therefore, hypothesized to be a
loss-of-function mutation due to the increased mRNA levels in IPAs
compared with in nIPAs.
Increasing evidence has demonstrated the primary
roles of tumor suppressors, oncogenes and cell cycle abnormalities
in pituitary tumorigenesis. The PRDM group of proteins is an
evolutionarily conserved protein family with 17 predicted members
in humans; of which, few protein members have been characterized
(49). Numerous studies suggest that
PRDM family proteins interact with a number of chromatin modifying
proteins, and act primarily as negative regulators of transcription
(50–54). PRDM2/RIZ is a binding partner for the
retinoblastoma tumor suppressor protein and is the frequent target
for inactivation in a variety of human tumors, including breast,
liver, and colon cancers (55). Its
tumor suppressor function is directly confirmed by the tumorigenic
phenotype of mice deficient for RIZ1, the PR-containing isoform of
PRDM2. In the present study, PRDM2 and PRDM8 mRNA levels were
approximately five-fold lower in IPA specimens compared with nIPA
specimens (P=0.007 and 0.009, respectively). In addition, binary
multivariate regression revealed that decreased levels of PRDM2
were independently associated with tumor recurrence.
Exome sequencing studies allow the comprehensive
testing of coding variation in an unbiased manner. The results of
the current study demonstrate that whole-exome sequencing will be
particularly valuable for the identification of genes under
conditions in which mapping has been confounded by locus
heterogeneity and uncertainty of the boundaries of diagnostic
classification. Whole-exome sequencing may be useful in the future
for a wide range of applications to medicine.
Acknowledgements
The present study was supported by the Research
Special Fund for Public Welfare Industry of Health of China (grant
no. 201402008), the National Natural Science Foundation of China
(grant no. 81272522) and Beijing Natural Science Foundation of
China (grant no. 7162035).
References
|
1
|
Ezzat S, Asa SL, Couldwell WT, Barr CE,
Dodge WE, Vance ML and McCutcheon IE: The prevalence of pituitary
adenomas: A systematic review. Cancer. 101:613–619. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bronstein MD and Melmed S: Pituitary
tumorigenesis. Arq Bras Endocrinol Metabol. 49:615–625. 2005.(In
Portuguese). View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Al-Shraim M and Asa SL: The 2004 World
Health Organization classification of pituitary tumors: What is
new? Acta Neuropathol. 111:1–7. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Scheithauer BW, Kovacs KT, Laws ER Jr and
Randall RV: Pathology of invasive pituitary tumors with special
reference to functional classification. Journal Neurosur.
65:733–744. 1986. View Article : Google Scholar
|
|
5
|
Colao A, Grasso LF, Pivonello R and
Lombardi G: Therapy of aggressive pituitary tumors. Expert Opin
Pharmacother. 12:1561–1570. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Di Ieva A, Rotondo F, Syro LV, Cusimano MD
and Kovacs K: Aggressive pituitary adenomas-diagnosis and emerging
treatments. Nat Rev Endocrinol. 10:423–435. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lloyd RV, Kovacs K, Young WF Jr, Farrell
WE, Asa SL, Trouillas J, Kontogeorgos G, Sano T, Scheithauer BW and
Horvath E: Tumors of the pituitaryDeLellis RA, Lloyd RV, Heitz PU
and Eng C: World Health Organization Classification of Tumors:
Pathology and Genetics: Tumors of Endocrine Organs. IARC Press;
Lyon: pp. 10–47. 2004
|
|
8
|
Gejman R, Swearingen B and Hedley-Whyte
ET: Role of Ki-67 proliferation index and p53expression in
predicting progression of pituitary adenomas. Hum Pathol.
39:758–766. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zada G, Woodmansee WW, Ramkissoon S,
Amadio J, Nose V and Laws ER Jr: Atypical pituitary adenomas:
Incidence, clinical characteristics, and implications. J Neurosurg.
114:336–344. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mete O, Ezzat S and Asa SL: Biomarkers of
aggressive pituitary adenomas. J Mol Endocrinol. 49:R69–R78. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Melmed S: 2004 World Health Organization
classification of pituitary tumors: What is new? Acta Neuropathol.
111:78–79. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Righi A, Morandi L, Leonardi E, Farnedi A,
Marucci G, Sisto A, Frank G, Faustini-Fustini M, Zoli M, Mazzatenta
D, et al: Galectin-3 expression in pituitary adenomas as a marker
of aggressive behavior. Hum Pathol. 44:2400–2409. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Galland F, Lacroix L, Saulnier P, Dessen
P, Meduri G, Bernier M, Gaillard S, Guibourdenche J, Fournier T,
Evain-Brion D, et al: Differential gene expression profiles of
invasive and non-invasive non-functioning pituitary adenomas based
on microarray analysis. Endocr Relat Cancer. 17:361–371. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schvartzman JM, Sotillo R and Benezra R:
Mitotic chromosomal instability and cancer: Mouse modelling of the
human disease. Nat Rev Cancer. 10:102–115. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pilarski R and Nagy R: Genetic testing by
cancer site: Endocrine system. Cancer J. 18:364–371. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Newey PJ, Nesbit MA, Rimmer AJ, Head RA,
Gorvin CM, Attar M, Gregory L, Wass JA, Buck D, Karavitaki N, et
al: Whole-exome sequencing studies of nonfunctioning pituitary
adenomas. J Clin Endocrinol Metab. 98:E796–E800. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Asa SL and Ezzat S: The pathogenesis of
pituitary tumours. Nat Rev Cancer. 2:836–849. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhan X, Desiderio DM, Wang X, Zhan X, Guo
T, Li M, Peng F, Chen X, Yang H, Zhang P, et al: Identification of
the proteomic variations of invasive relative to non-invasive
nonfunctional pituitary adenomas. Electrophoresis. 35:2184–2194.
2014.PubMed/NCBI
|
|
19
|
Yoshida K, Sanada M, Shiraishi Y, Nowak D,
Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et
al: Frequent pathway mutations of splicing machinery in
myelodysplasia. Nature. 478:64–69. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Agrawal N, Frederick MJ, Pickering CR,
Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et
al: Exome sequencing of head and neck squamous cell carcinoma
reveals inactivating mutations in NOTCH1. Science. 333:1154–1157.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ni X, Zhuo M, Su Z, Duan J, Gao Y, Wang Z,
Zong C, Bai H, Chapman AR, Zhao J, et al: Reproducible copy number
variation patterns among single circulating tumor cells of lung
cancer patients. Proc Natl Acad Sci USA. 110:21083–21088. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Enseñat J, Ortega A, Topcewski T, Vilalta
J, Obiols G, Mesa J and Sahuquillo J: Predictive value of the Knosp
classification in grading the surgical resection of invasive
pituitary macroadenomas. A prospective study of 23 cases.
Neurocirugia (Astur). 17:519–26. 2006.(In Spanish). View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang F, Gao H, Li C, Bai J, Lu R, Cao L,
Wu Y, Hong L, Wu Y, Lan X and Zhang Y: Low levels of PRB3 mRNA are
associated with dopamine-agonist resistance and tumor recurrence in
prolactinomas. J Neurooncol. 116:83–88. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiao Y, Shi C, Edil BH, de Wilde RF,
Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA,
et al: DAXX/TRX, MEN1, and mTOR pathway genes are frequently
altered in pancreatic neuroendocrine tumors. Science.
331:1199–1203. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Newey PJ, Nesbit MA, Rimmer AJ, Attar M,
Head RT, Christie PT, Gorvin CM, Stechman M, Gregory L, Mihai R, et
al: Whole-exome sequencing studies of nonhereditary (sporadic)
parathyroid adenomas. J Clin Endocrinol Metab. 97:E1995–E2005.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barbieri CE, Baca SC, Lawrence MS,
Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van
Allen E, Stransky N, et al: Exome sequencing identifies recurrent
SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet.
44:685–689. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Popescu MN, Ionescu E, Iovănescu LC, Cotoi
BV, Popescu AI, Gănescu AE, Glodeanu A, Geormăneanu C, Moraru A and
Pătraşcu A: Clinical aggression of prolactinomas: Correlations with
invasion and recurrence. Rom J Morphol Embryol. 54:1075–1080.
2013.PubMed/NCBI
|
|
29
|
Varier RA and Timmers HT: Histone lysine
methylation and demethylation pathways in cancer. Biochimica
Biophysica Acta. 1815:75–89. 2011.
|
|
30
|
Tanase C, Ogrezeanu I and Badiu C:
Pituitary tumor classification: Functionality, invasiveness and
aggressivenessMolecular Pathology of Pituitary Adenomas. Elsevier;
Amsterdam, Netherlands: pp. 1–18. 2011
|
|
31
|
Dhodapkar MV: Personalized
immune-interception of cancer and the battle of two adaptive
systems-when is the time right? Cancer Prev Res (Phila). 6:173–176.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
DuPage M, Cheung AF, Mazumdar C, Winslow
MM, Bronson R, Schmidt LM, Crowley D, Chen J and Jacks T:
Endogenous T cell responses to antigens expressed in lung
adenocarcinomas delay malignant tumor progression. Cancer Cell.
19:72–85. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang W, Xu Z, Fu L, Liu W and Li X:
Pathogenesis analysis of pituitary adenoma based on gene expression
profiling. Oncol Lett. 8:2423–2430. 2014.PubMed/NCBI
|
|
34
|
Shen FF, Yue WB, Zhou FY, Pan Y, Zhao XK,
Jin Y, Song X, Li B, Han XN, Tang S, et al: Variations in the MHC
region confer risk to esophageal squamous cell carcinoma on the
subjects from high-incidence area in northern China. PloS One.
9:e904382014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee JS, Bae JS, Kim JH, Kim JY, Park TJ,
Pasaje CF, Park BL, Cheong HS, Uh ST, Park JS, et al: Effect of
diffuse panbronchiolitis critical region 1 polymorphisms on the
risk of aspirin-exacerbated respiratory disease in Korean
asthmatics. Respir Care. 57:758–763. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Osamura RY, Kajiya H, Takei M, Egashira N,
Tobita M, Takekoshi S and Teramoto A: Pathology of the human
pituitary adenomas. Histochem Cell Biol. 130:495–507. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schmidt M, Paes K, De Mazière A, Smyczek
T, Yang S, Gray A, French D, Kasman I, Klumperman J, Rice DS and Ye
W: EGFL7 regulates the collective migration of endothelial cells by
restricting their spatial distribution. Development. 134:2913–2923.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schmidt MH, Bicker F, Nikolic I, Meister
J, Babuke T, Picuric S, Müller-Esterl W, Plate KH and Dikic I:
Epidermal growth factor-like domain 7 (EGFL7) modulates Notch
signalling and affects neural stem cell renewal. Nat Cell Biol.
11:873–880. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bambino K, Lacko LA, Hajjar KA and
Stuhlmann H: Epidermal growth factor-like domain 7 is a marker of
the endothelial lineage and active angiogenesis. Genesis.
52:657–670. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Louvi A and Artavanis-Tsakonas S: Notch
signalling in vertebrate neural development. Nat Rev Neurosci.
7:93–102. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jiang Z, Gui S and Zhang Y: Analysis of
differential gene expression in plurihormonal pituitary adenomas
using bead-based fiber-optic arrays. J Neurooncol. 108:341–348.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lu R, Gao H, Wang H, Cao L, Bai J and
Zhang Y: Overexpression of the Notch3 receptor and its ligand
Jagged1 in human clinically non-functioning pituitary adenomas.
Oncol Lett. 5:845–851. 2013.PubMed/NCBI
|
|
43
|
Howard J and Hyman AA: Dynamics and
mechanics of the microtubule plus end. Nature. 422:753–758. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Azorín E, Romero-Pérez B, Solano-Agama C,
de la Vega MT, Toriz CG, Reyes-Márquez B, González-Pozos S,
Rosales-García VH, Del Pliego MG, Sabanero M and Mendoza-Garrido
ME: GH3 tumor pituitary cell cytoskeleton and plasma membrane
arrangement are determined by extracellular matrix proteins:
Implications on motility, proliferation and hormone secretion. Int
J Physiol Pathophysiol Pharmacol. 6:66–83. 2014.PubMed/NCBI
|
|
45
|
delPliego MG, Aguirre-Benítez E,
Paisano-Cerón K, Valdovinos-Ramírez I, Rangel-Morales C,
Rodríguez-Mata V, Solano-Agama C, Martín-Tapia D, de la Vega MT,
Saldoval-Balanzario M, et al: Expression of Eag1 K+ channel and
ErbBs in human pituitary adenomas: Cytoskeleton arrangement
patterns in cultured cells. Int J Clin Exp Pathol. 6:458–468.
2013.PubMed/NCBI
|
|
46
|
Stolc V, Samanta MP, Tongprasit W and
Marshall WF: Genome-wide transcriptional analysis of flagellar
regeneration in Chlamydomonas reinhardtii identifies orthologs of
ciliary disease genes. Proc Natl Acad Sci USA. 102:3703–3707. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Avidor-Reiss T, Maer AM, Koundakjian E,
Polyanovsky A, Keil T, Subramaniam S and Zuker CS: Decoding cilia
function: Defining specialized genes required for compartmentalized
cilia biogenesis. Cell. 117:527–539. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Van Rooijen E, Giles RH, Voest EE, van
Rooijen C, Schulte-Merker S and van Eeden FJ: LRRC50, a conserved
ciliary protein implicated in polycystic kidney disease. J Am Soc
Nephrol. 19:1128–1138. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Basten SG, Davis EE, Gillis AJ, van
Rooijen E, Stoop H, Babala N, Logister I, Heath ZG, Jonges TN,
Katsanis N, et al: Mutations in LRRC50 predispose zebrafish and
humans to seminomas. PLoS Genet. 9:e10033842013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hohenauer T and Moore AW: The Prdm family:
Expanding roles in stem cells and development. Development.
139:2267–2282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Davis CA, Haberland M, Arnold MA,
Sutherland LB, McDonald OG, Richardson JA, Childs G, Harris S,
Owens GK and Olson EN: PRISM/PRDM6, a transcriptional repressor
that promotes the proliferative gene program in smooth muscle
cells. Mol Cell Biol. 26:2626–2636. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kajimura S, Seale P, Tomaru T,
Erdjument-Bromage H, Cooper MP, Ruas JL, Chin S, Tempst P, Lazar MA
and Spiegelman BM: Regulation of the brown and white fat gene
programs through a PRDM16/CtBP transcriptional complex. Genes Dev.
22:1397–1409. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Izutsu K, Kurokawa M, Imai Y, Maki K,
Mitani K and Hirai H: The corepressor CtBP interacts with Evi-1 to
repress transforming growth factor beta signaling. Blood.
97:2815–2822. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yu J, Angelin-Duclos C, Greenwood J, Liao
J and Calame K: Transcriptional repression by blimp-1 (PRDI-BF1)
involves recruitment of histone deacetylase. Mol Cell Biol.
20:2592–2603. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tam W, Gomez M, Chadburn A, Lee JW, Chan
WC and Knowles DM: Mutational analysis of PRDM1 indicates a
tumor-suppressor role in diffuse large B-cell lymphomas. Blood.
107:4090–4100. 2006. View Article : Google Scholar : PubMed/NCBI
|

















