Introduction
Non-small cell lung cancer (NSCLC) is the most
common type of lung cancer and the leading cause of
cancer-associated mortality worldwide, which mostly results from
local invasion or distant metastases, rather than being due to
primary tumors (1). The 5-year
survival rate of advanced lung cancer has been reported to be
<15% (2). It has been recognized
that certain tumors present a predilection for metastasis to
specific organs (3,4), being bone, brain, liver and lung the
most frequent target organs of metastasis (4). The mechanism of tumor development in
lung cancer is likely to be associated with genetic diseases that
occur when certain genes are mutated or exhibit abnormalities such
as deletions (3). Novel strategies
for the treatment of lung cancer require the identification of
novel molecules that are able to inhibit the invasiveness and
metastasis of lung cancer cells (5).
Small interfering (si)RNA molecules consist of 21–23
nucleotides in length and display a characteristic and highly
specific structure containing 2–3 nucleotides in the 3′ overhangs
and 5′ phosphate and 3′ hydroxyl groups to prevent erroneous gene
silencing, in addition to a sense (passenger) strand and an
antisense (guide) strand (6). The
remarkable advances in biology and cancer therapy that had occurred
during the past recent decades have led to the development of siRNA
technology, which is an effective method to sequence-specific
knockdown or reduce gene function through homology-dependent
degradation of the corresponding target messenger (m)RNA, via a
phenomenon known as RNA interference (RNAi), which is one of the
most rapidly growing topics of research for the treatment of
various diseases, including cancer (6,7). Despite
the caveats and ongoing challenges such as off-target effects and
false discovery rates, RNAi remains a powerful approach for reverse
genetics in large-scale functional analysis conducted in cultured
cells and in various in vivo systems (6). Furthermore, the development of siRNA
therapeutics is rapidly gaining momentum at present (6).
Lung cancer is the leading cause of
cancer-associated mortality and morbidity worldwide (8), and NSCLC is responsible for almost 80%
of all lung cancer-associated mortalities, mostly resulting from
tumor invasion and metastasis (8,9).
Fibroblast growth factor receptor 3 (FGFR3) is a member of the
transmembrane receptor tyrosine kinase (RTK) family, which
interacts with FGF ligands and participates in the regulation of
cell proliferation, differentiation and tumorigenesis (10,11).
Previous studies demonstrated that the expression levels of
FGFR3 were significantly upregulated in metastasis of NSCLC,
compared with normal lung tissues, according to the results of
quantitative analysis of FGFR3 expression in humans (3,10,12). In addition, FGFR3 has been
demonstrated to be involved in the RAS/RAF/mitogen-activated
protein kinase (MAPK) kinase (MEK)/MAPK signaling pathway through
the activation of p90 ribosomal S6 kinase (13).
Migration and invasion are critical steps in the
initial progression of cancer that facilitate metastasis (14,15). In
particular, the critical steps required for the initiation and
progression of tumor invasion are epithelial-mesenchymal transition
(EMT) and extracellular matrix (ECM) degradation (14). E-cadherin is an important marker of
EMT (15), while matrix
metalloproteinase (MMP)9 is a member of the MMP family of enzymes,
which are involved in the degradation of the ECM (16,17).
Increased levels of MMP9 have been correlated with tumor
aggressiveness in numerous types of cancer (18).
RNAi is a popular method for exploring gene
function, which suppresses the expression of a specific gene of
interest in transfected mammalian cell cultures (19). In the present study, siRNA-FGFR3
significantly inhibited the expression of FGFR3 by directly
targeting FGFR3 in A549 cells. The present authors hypothesized
that siRNA-FGFR3 was able to silence FGFR3 and inhibit the
migration of A549 cells by decreasing the expression levels of
E-caherin and MMP9 in these cells. To test the above hypothesis,
cell invasion ability was assessed by Transwell invasion assay,
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and western blot analysis in A549 cells transfected with
siRNA-FGFR3.
Materials and methods
Cell line and culture
A549 lung cancer cells were obtained from the
Shanghai Cell Bank Chinese Academy of Sciences (Shanghai, China).
Cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Hyclone; GE Healthcare Life Sciences, Logan, UT,
USA), 40,000 mU/ml penicillin (Mediatech, Inc.; Corning Life
Sciences, Manassas, VA, USA) and 40 µg/ml streptomycin (Mediatech,
Inc.; Corning Life Sciences), at 37°C in a humidified incubator
with 5% CO2. Upon reaching 90% confluence, the cells
were dissociated using 0.25% trypsin (Hyclone, Shanghai, China),
and subcultured.
siRNA transfection
Cells were transfected with siRNAs targeting FGFR3
or with control siRNA using Lipofectamine® 2000
(Lipo2000) (Invitrogen; Thermo Fisher Scientific, Inc.). The
transfection was performed at 37°C in a humidified incubator with
5% CO2. The siRNAs were designed and synthesized by
Shanghai GenePharma Co., Ltd. (Shanghai, China), and their
sequences were queried with Basic Local Alignment Search Tool
[National Center for Biotechnology Information (NCBI), Bethesda,
MD, USA], and identified as FGFR3-specific sequence, once homology
with other genes was excluded. In total, 3 groups of cells were
transfected with siRNAs whose sequence was specific for FGFR3
(namely, siRNA-855, siRNA-1447 and siRNA-2076). Another group of
cells was transfected with a fluorescein amidite (FAM)-labeled
nonspecific siRNA, that served as negative control (NC)siRNA. The
sequences of the siRNAs used in the present study were as follows:
siRNA-855 sense, 5′-GCAUUGGAGGCAUCAAGCUTT-3′ and anti-sense,
5′-AGCUUGAUGCCUCCAAUGCTT-3′; siRNA-1447 sense,
5′-GCUGAAAGACGAUGCCACUTT-3′ and anti-sense,
5′-AGUGGCAUCGUCUUUCAGCTT-3′; siRNA-2076 sense,
5′-GCACACACGACCUGUACAUTT-3′ and anti-sense,
5′-AUGUACAGGUCGUGUGUGCTT-3′; and NC-siRNA sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and anti-sense,
5′-ACGUGACACGUUCGGAGAATT-3′. When A549 cells seeded into 6-well
plates reached 80% confluence, transfection was conducted by mixing
5 µl siRNA with 5 µl Lipo2000 in a final volume of 2,000 µl
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing
10% serum without antibiotics, according to the manufacturer's
protocol. Cell morphology and transfection efficiency were
evaluated at 6 h following transfection. Cells were divided into 5
groups termed A-E, and transfected with siRNA-855, siRNA-1447,
siRNA-2076, NC-siRNA and blank control, respectively. Transfections
were performed in triplicate and the experiment was repeated ≥3
times.
Transwell invasion assay
Cell invasion was assessed by Transwell invasion
assay using 8-µm Matrigel-coated invasion chambers (Costar; Corning
Life Sciences), according to the manufacturer's protocol. Briefly,
A549 cells (0.5×105 cells/well) were suspended in 200 µl
serum-free medium and seeded on the upper chamber of a 24-well
Transwell migration chamber (Costar; Corning Life Sciences). The
lower compartment was filled with 750 µl complete medium. Following
16-h culture at 37°C in a humidified incubator with 5%
CO2, those cells that were retained on top of the
membrane were removed with a cotton swab, while those cells that
had migrated to the bottom side (invaded cells) were fixed with 10%
formalin (Costar; Corning Life Sciences), stained with 0.1% crystal
violet (Costar; Corning Life Sciences) and observed under a light
microscope (Olympus BX41; Olympus Corporation, Tokyo, Japan) at
200x magnification. The number of penetrating cells present in 9
random fields was counted, and the experiment was repeated ≥3
times.
RT-qPCR
Total RNA was isolated from A549 cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) at 36 h
post-transfection, according to the manufacturer's protocol. The
concentration and purity of the extracted RNA was quantified by its
absorbance at 260 nm using a BioPhotometer (D30; Eppendorf,
Hamburg, Germany). RT-qPCR was conducted with SYBR Green II
(catalogue number RR037A; Takara Bio, Inc., Otsu, Japan), according
to the manufacturer's protocol. The experiment was repeated ≥3
times. The PCR primers were designed and synthesized by Sangon
Biotech Co., Ltd. (Shanghai China), and their sequences were as
follows: FGFR3 sense, 5′-TCAGGGTGGTCTCTTCTTGG-3′ and anti-sense,
5′-CGTCGCTGGGTTAACAAAT-3′; MMP9 sense, 5′-TTCCAAACCTTTGAGGGCGA-3′
and anti-sense, 5′-GCAAAGGCGTCGTCAATCAC-3′; E-cadherin sense,
5′-GGGTTATTCCTCCCATCAGC-3′ and anti-sense,
5′-GTCACCTTCAGCCATCCTGT-3′; and glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) sense, 5′-GATCATCAGCAATGCCTCCTG-3′ and
anti-sense, 5′-GAGTCCTTCCACGATACCAAAG-3′. The sequence of the
specific primers for FGFR3, MMP9, E-cadherin and GAPDH were based
on the corresponding GenBank sequence (NCBI) (FGFR3 GeneID,
NM_000142.4 GI:254028235; MMP9 GeneID, NM_004994.2 GI:74272286;
E-cadherin GeneID, NM_004360.3 GI:169790842; and GAPDH GeneID,
NM_001256799.1 GI:378404907). PCR was performed under the following
conditions: Initial denaturation at 95°C for 5 min, followed by 35
cycles of 95°C for 30 sec, 55°C for 1 min and 72°C for 1 min, with
a final extension step at 72°C for 5 min. GAPDH served as an
internal control. To ensure the specificity of each set of primers,
the melting point temperatures of the amplicons generated by PCR
were evaluated using High Resolution Melt Software, a
first-derivative primer melting curve analysis software
(LightCycler 480; Roche Diagnostics, Indianapolis, IN, USA). The
mRNA expression levels of FGFR3, MMP9 and E-cadherin were
normalized to those of GAPDH, and relative quantification was
performed using the comparative cycle threshold method (2-ΔΔCq)
(20). All PCR experiments were
repeated ≥3 times.
Western blot analysis
To analyze the protein expression levels of FGFR3,
MMP9 and E-cadherin in A549 cells, transfected and untransfected
cells were rinsed twice with precooled phosphate-buffered saline
(Costar; Corning Life Sciences), prior to being homogenized in
radioimmunoprecipitation assay (RIPA) buffer (Costar; Corning Life
Sciences) supplemented with a protease and phosphatase inhibitor
cocktail (Roche Diagnostics GmbH, Mannheim, Germany) at 36 h
post-transfection. Cell lysates were centrifuged (GTR 21-1;
Shanghai Medical Instruments Co., Ltd., Shanghai, China) at 14,000
× g for 10 min at 4°C, and the supernatants were then mixed with 5X
sodium dodecyl sulfate (SDS; Costar; Corning Life Sciences) loading
sample buffer, boiled for 5 min and separated using 10%
SDS-polyacrylamide gel electrophoresis at 60 V for 5 h. Total
cellular protein levels were quantified by Bradford assay (21) using standard protein solution and
Coomassie Bri-liant Blue (R-250) (Generay Biotech Co., Ltd.,
Shanghai, China). Following electrophoresis, the proteins were
transferred to PVDF membranes (Shanghai Kehua Bio-Engineering Co.,
Ltd., Shanghai, China) by electrophoretic transfer. Membranes were
blocked in 5% skimmed milk for 2 h, rinsed with Tris-buffered
saline containing Tween 20, and incubated overnight at 4°C with
rabbit anti-human monoclonal FGFR3 (cat. no. ab137084; Abcam,
Cambridge, MA, USA), MMP9 (cat. no. ab76003; Abcam) and E-cadherin
(cat. no. 3195; Cell Signaling Technology, Inc., Danvers, MA, USA)
primary antibodies (1:10,000 dilution in 5% skimmed milk).
Following 3 washes with Tris-buffered saline containing Tween 20,
membranes were incubated with a secondary antibody [horseradish
peroxidase-conjugated goat anti-rabbit immunoglobulin (Ig)G] (cat.
no. CW0103M; Beijing Kang Century Biotechnology Co., Ltd., Beijing,
China) for 12 h at 4°C. Goat anti-mouse GAPDH (cat. no. CW0101M;
Beijing Kang Century Biotechnology Co., Ltd.) was used as an
internal standard. Signals were detected with a Novex®
enhanced chemiluminescence (ECL) substrate reagent kit (Thermo
Fisher Scientific, Inc.) for 1 h at 37°C. For that purpose,
developer A and B (obtained from the ECL Western Blotting kit;
Costar; Corning Life Sciences) were mixed at a 1:1 ratio, prior to
be added to the blots, which were subsequently exposed and imaged.
The relative intensity of the bands was analyzed using ImageJ
software (National Institutes of Health, Bethesda, MA, USA). All
experiments were performed in triplicate.
Statistical analysis
Results were analyzed with SPSS statistical software
version 18.0 (SPSS, Inc., Chicago, IL, USA). Data are represented
as the mean ± standard deviation of 5 replicates. Differences
between multiple sets of data were compared by analysis of
variance, while differences between 2 groups were compared by
homogeneity of variance test. The significance level was considered
to be α=0.05, and P<0.05 was considered to indicate a
statistically significant difference.
Results
Morphology of A549 cells and
transfection efficiency
Morphological changes in transfected A549 cells were
observed under an inverted microscope at 6 h post-transfection.
Prior to transfection, cells were adherent, fusiform and exhibited
adequate growth, moderate size and a clear nucleolus, whereas 6 h
following transfection cells were irregular, poorly adherent and
exhibited shrinkage. Transfection efficiency was determined under
an inverted fluorescence microscope (BX43; Olympus Corporation,
Tokyo, Japan), based on the transfection efficiency exhibited by
the FAM-siRNA-transfected cells (70–80%; Fig. 1A and B).
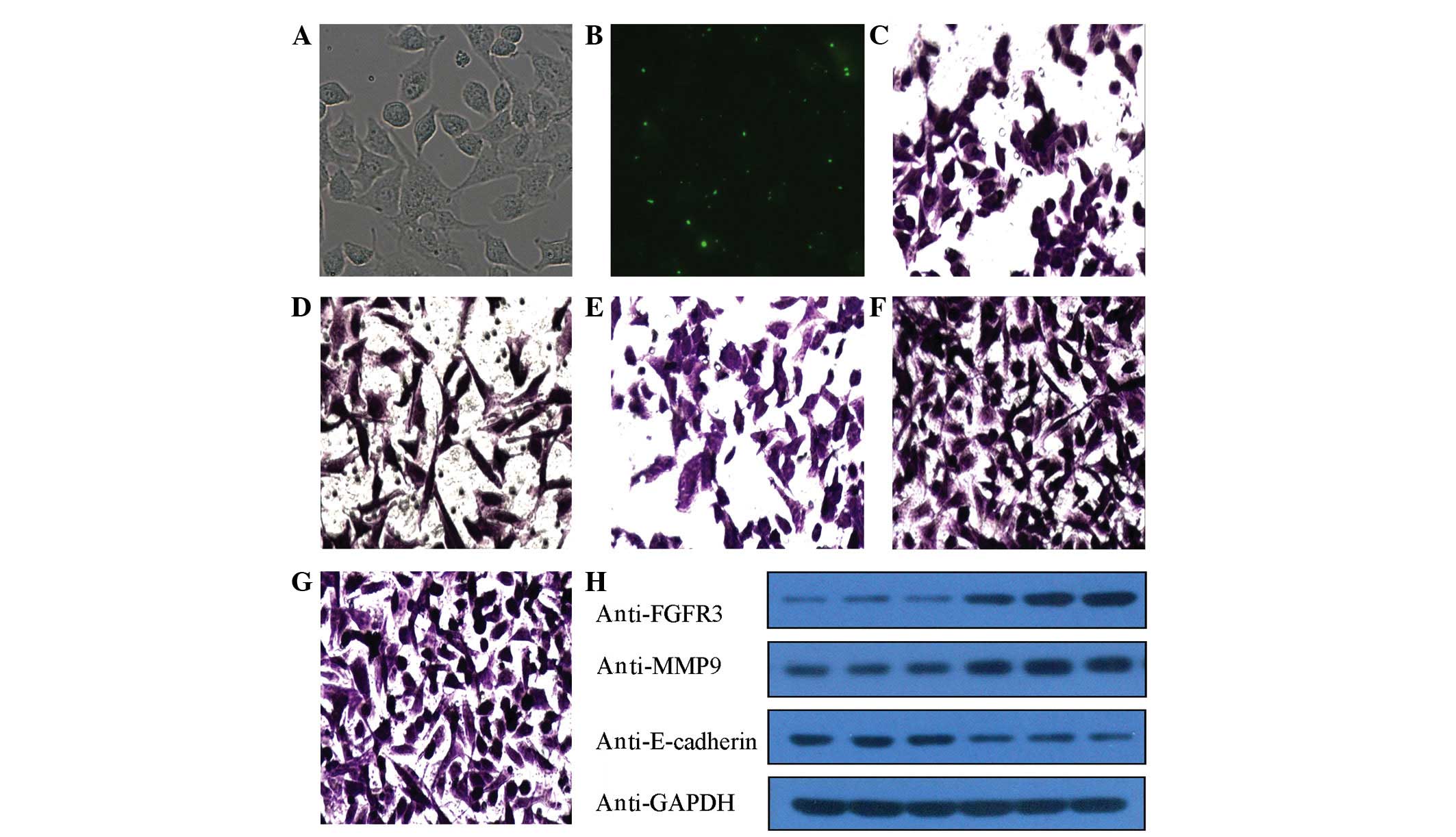 | Figure 1.(A and B) Following transfection with
small interfering RNA labeled with fluorescein amidite for 6 h,
green fluorescence emission produced by blue light excitation was
observed in the transfected A549 cells under an inverted
fluorescence microscope. The transfection efficiency was determined
to be 70–80%. (A) shows image under high power microscope
(magnification, ×200) and (B) shows fluorescence microscopy
(magnification, ×200). Following transfection for 6 h, A549 cells
in (C) group A, (D) group B, (E) group C, (F) group D and (G) group
E were subjected to Transwell assay. Cells that migrated to the
bottom chamber (invaded cells) were fixed with 10% formalin,
stained with 0.1% crystal violet and observed under 200x
magnification. (H) Compared with D and E groups, the protein
expression levels of fibroblast growth factor receptor 3 and matrix
metalloproteinase 9 were downregulated in groups A-C, whereas the
expression of E-cadherin protein was upregulated in these
groups. |
siRNA-FGFR3 inhibits A549
invasion
To investigate the inhibitory effect of siRNA-855,
siRNA-1447 and siRNA-2076 on the invasion of A549 cells, an
invasion assay was performed with Matrigel upon inhibiting FGFR3
expression in A549 cells in vitro via siRNA-FGFR3
transfection. The results indicated that the inhibition of FGFR3
mediated by siRNA transfection led to a decrease in the invasion of
siRNA-FGFR3-transfected cells (A-C groups), compared with the
NC-siRNA and blank control groups (P<0.01, Table I). These results suggest that
siRNA-855, siRNA-1447 and siRNA-2076 are involved in the migration
of A549 cells. In addition, siRNA-FGFR3 inhibited the invasion of
A549 cells transfected with siRNA-855, siRNA-1447 and siRNA-2076,
compared with the blank control group. Following transfection of
A549 cells with siRNA for 36 h, the results of the subsequent
Transwell invasion assay demonstrated that the average number of
invasive cells per high-power field in the siRNA-855, siRNA-1447
and siRNA-2076 groups was significantly reduced, compared with the
blank control group (Fig. 1;
magnification, ×200). Thus, the number of invasive cells in the
above groups was 0.36, 0.39 and 0.38 times the number of invasive
cells in the blank control group, respectively (Table I). Thus, the rate of inhibition of
invasion for groups A, B and C was determined to be 63.9, 61.4 and
61.8% (P=0.000004, P=0.000006 and P=0.00005, respectively; Table I). No significant differences were
observed for the NC-siRNA group, where the number of invasive cells
was determined to be 0.97 times that of the blank control group
(P=0.745000; Fig. 1 and Table I).
 | Table I.Effect of siRNA transfection on the
invasion ability of A549 cells. |
Table I.
Effect of siRNA transfection on the
invasion ability of A549 cells.
| Group | No. of penetrated
cells/high-power field | No. of
transfected/blank cells | Invasion inhibition
rate (%) | P-value |
|---|
| siRNA-855 | 32.49±8.25 | 0.36 | 63.9 | 0.000004 |
| siRNA-1447 | 34.68±11.48 | 0.39 | 61.4 | 0.000006 |
| siRNA-2076 | 34.29±9.44 | 0.38 | 61.8 | 0.000005 |
| NC-siRNA | 87.82±3.98 | 0.97 |
2.0 | 0.745000 |
| Blank | 89.95±0.73 | – | – | – |
mRNA expression of FGFR3, MMP9 and
E-cadherin was altered in siRNA-transfected A549 cells
The Real-time PCR amplification efficiency was 100%,
the 2-ΔΔCq method was applied to quantify fold-changes
in the expression levels of FGFR3, MMP9 and E-cadherin in each cell
group. As indicated in Tables
II–IV, the number of mRNA
corresponding to the FGFR3 gene was significantly reduced in the A,
B and C groups, compared with the E group (P=0.000006, P=0.000004
and P=0.000004, respectively). Compared with the E group, the mRNA
expression levels of E-cadherin in the siRNA-transfected groups
increased significantly (P=0.000005, P=0.000003 and P=0.000003 for
groups A, B and C, respectively), while the mRNA expression levels
of MMP9 in these groups were significantly reduced (P=0.000007,
P=0.000005 and P=0.000004 for groups A, B and C, respectively).
Compared with the blank control group, the cDNA copy number of
FGFR3, MMP9 and E-cadherin in the NC-siRNA group did not change
significantly (P>0.05; Tables
II–IV). These results indicated
that siRNA transfection with Lipo2000 markedly inhibited the mRNA
expression of FGFR3 in a specific manner, which led to a marked
reduction in the mRNA expression levels of MMP9 and a marked
increase in the mRNA expression levels of E-cadherin in
siRNA-FGFR3-transfected A549 cells.
 | Table II.RNA interference inhibited the
messenger RNA expression of fibroblast growth factor receptor 3 in
A549 cells. |
Table II.
RNA interference inhibited the
messenger RNA expression of fibroblast growth factor receptor 3 in
A549 cells.
| Group | siRNA-855 | siRNA-1447 | siRNA-2076 | NC-siRNA |
|---|
| Fold-change | 0.22±0.89 | 0.26±0.12 | 0.26±0.09 | 0.99±0.03 |
| P-value | 0.000006 | 0.000004 | 0.000004 | 0.895000 |
| siRNA-855 | 32.49±8.25 | 0.36 | 63.9 | 0.000004 |
| siRNA-1447 | 34.68±11.48 | 0.39 | 61.4 | 0.000006 |
| siRNA-2076 | 34.29±9.44 | 0.38 | 61.8 | 0.000005 |
| NC-siRNA | 87.82±3.98 | 0.97 |
2.0 | 0.745000 |
| Blank | 89.95±0.73 | 0.95 |
2.3 | 0.807000 |
 | Table IV.RNA interference affected the
messenger RNA expression of E-cadherin in A549 tells. |
Table IV.
RNA interference affected the
messenger RNA expression of E-cadherin in A549 tells.
| Group | siRNA-855 | siRNA-1447 | siRNA-2076 | NC-siRNA |
|---|
| Fold-change | 2.35±0.58 | 2.13±0.32 | 2.12±0.20 | 1.06±0.15 |
| P-value | 0.000005 | 0.000003 | 0.000003 | 0.803000 |
| siRNA-855 | 32.49±8.25 | 0.36 | 63.9 | 0.000004 |
| siRNA-1447 | 34.68±11.48 | 0.39 | 61.4 | 0.000006 |
| siRNA-2076 | 34.29±9.44 | 0.38 | 61.8 | 0.000005 |
| NC-siRNA | 87.82±3.98 | 0.97 |
2.0 | 0.745000 |
| Blank | 89.95±0.73 | 0.95 |
2.3 | 0.807000 |
Protein expression of FGFR3, MMP9 and
E-cadherin was altered in siRNA-transfected A549 cells
Western blot analysis demonstrated that, compared
with groups D and E, the protein levels of FGFR3 and MMP9 were
downregulated in A-C groups, while the protein levels of E-cadherin
were upregulated in these groups, according to the lower and higher
gray, respectively, of the protein bands in the A-C groups
(Fig. 1H). ImageJ software was used
to analyze the gray values of the different protein bands. Compared
with the E group, the FGFR3/GAPDH and MMP9/GAPDH ratios in the A-C
groups were significantly decreased, while the E-cadeherin/GAPDH
ratio in these groups was significantly increased, compared with
the E group (P<0.01; Tables
V–VII). No significant
differences in FGFR3, MMP-9 and E-cadherin expression were observed
between the A-C groups or between groups D and E (P>0.01;
Tables V–VII). These results suggest that the
protein expression of FGFR3, encoded by FGFR3 mRNA, was inhibited,
possibly due to the specific degradation of FGFR3 mRNA induced by
siRNA-FGFR3, which led to a decrease in the protein expression
levels of MMP9 and an increase in the protein expression levels of
E-cadherin.
 | Table V.Ratio of FGFR3/GAPDH protein
expression in each group. |
Table V.
Ratio of FGFR3/GAPDH protein
expression in each group.
| Group | siRNA-855 | siRNA-1447 | siRNA-2076 | NC-siRNA | Blank |
|---|
| FGFR3/GAPDH | 0.200±0.026 | 0.190±0.020 | 0.187±0.038 | 0.553±0.087 | 0.620±0.0095 |
| P-value | 0.000009 | 0.000007 | 0.000006 | 0.218000 | 0.541000 |
| siRNA-855 | 32.49±8.25 | 0.36 | 63.9 | 0.000004 | – |
| siRNA-1447 | 34.68±11.48 | 0.39 | 61.4 | 0.000006 | – |
| siRNA-2076 | 34.29±9.44 | 0.38 | 61.8 | 0.000005 | – |
| NC-siRNA | 87.82±3.98 | 0.97 |
2.0 | 0.745000 | – |
| Blank | 89.95±0.73 | – | – | – | – |
 | Table VII.Ratio of MMP9/GAPDH protein
expression in each group. |
Table VII.
Ratio of MMP9/GAPDH protein
expression in each group.
| Group | siRNA-855 | siRNA-1447 | siRNA-2076 | NC-siRNA | Blank |
|---|
| MMP9/GAPDH | 0.363±0.055 | 0.357±0.080 | 0.360±0.134 | 0.770±0.120 | 0.750±0.066 |
| P-value | 0.000600 | 0.001000 | 0.001000 | 0.804000 | – |
Discussion
The process of carcinoma metastasis consists of a
series of sequential steps (4).
Initially, tumor cells are detached from the ECM, and the
surrounding tissue is then invaded (4). Losing cell-cell adhesion and gaining
motility are important for carcinoma cells to separate from
neighboring cells and invade other tissues (4). EMT is a multistage process involving
major alterations in cell morphology, cell-cell and cell-matrix
adhesions. ETM is important for the motile and invasive
capabilities of cells, which are essential in malignant tumor
progression and metastasis (14). A
hallmark of EMT is the loss of expression of E-cadherin, which is a
key cell-cell adhesion molecule (22). It has been demonstrated that the
ability of invasion and metastasis of certain invasive carcinoma
cells may be altered by forcing the expression of E-cadherin
(22–24).
ECM dysregulation and remodeling are essential for
the progression of neoplastic processes, and MMP-mediated ECM
degradation has been demonstrated to lead to cancer cell invasion
and metastasis (25). Previous
studies on stromal cells undergoing EMT have revealed that EMT may
be the most critical and abnormal signaling pathway that occurs
during development and metastasis of lung cancer (22). MMP9 has been identified as a critical
component of the multifunctional MMP family of enzymes, which act
as ECM-degrading proteases and may promote metastasis (26,27). The
hypothetical role of MMP9 as a metastasis promoter is supported by
a previous study (28).
The FGFR3 gene is located in the 4p16.3 region of
the chromosome, which consists of 19 exons and 18 introns expanding
across 16.5 kb and encodes a 4.4-kb mRNA molecule (29–31). FGFR3
is a member of the FGFR family of tyrosine kinase receptors, and
consists of an extracellular domain that includes a signal peptide,
followed by 3 Ig-like domains, an acidic box, a transmembrane
domain and an intracellular tyrosine kinase domain (32). FGFR3 is activated by binding of the
FGF ligand to the extracellular Ig-like domains II and III
(3). Subsequently, the
trans-autophosphorylation of tyrosine residues in the cytoplasmic
domain of FGFR3 stimulates the intrinsic catalytic activity of the
receptor, and leads to the activation of downstream signaling
pathways (13,33). FGFR3 has been demonstrated to be
involved in the RAS/RAF/MEK/MAPK signaling pathway through the
activation of p90 ribosomal S6 kinase (13,33).
Previous studies have reported that FGFR3 is
overexpressed in lung cancer and bone metastasis of lung
adenocarcinoma; thus, FGFR3 may be regarded as a potential target
for cancer therapy (3,10,11).
However, the effect of silencing FGFR3 on the inhibition of
invasion of lung carcinoma remains unknown. Therefore, the present
study aimed to inhibit the expression of FGFR3 in lung cancer cells
in order to evaluate its effects on the invasion ability of NSCLC.
For that purpose, the association between FGFR3, E-cadherin and
MMP9 expression in A549 cells was investigated by siRNA targeting
FGFR3. In addition, the mRNA and protein expression levels of
FGFR3, E-cadherin and MMP9 were analyzed in siRNA-FGFR3-transfected
and control groups.
Previous in vivo and in vitro studies
have demonstrated that the upregulation of FGFR3 increased cancer
cell invasion and metastases in breast (12) and lung cancer (10). A hallmark of cancer is its
self-sufficiency in terms of growth signals, which is frequently
driven by RTK-dependent growth factor signaling pathways (34). These pathways are important during the
development and adult life of multicellular organisms (2,3). FGFR3
belongs to the RTK family, and FGF/FGFR3 signaling has been
frequently observed to occur in certain NSCLC cell lines via the
Ras/RAF/MEK/MAPK signaling pathway, which regulates key cellular
processes such as cell proliferation, migration and survival
(25,35). A previous study has reported that FGFR
signaling is active and important in cells undergoing EMT, which is
induced by transforming growth factor-β (14). EMT is involved in the progression and
metastasis of cancer (14). In the
present study, marked upregulation of E-cadherin and downregulation
of MMP9 via siRNA targeting FGFR3 was observed in lung cancer A549
cells. Furthermore, the results of Transwell assay demonstrated
that the invasion ability of siRNA-FGFR3-transfected cells
significantly decreased, compared with control cells. Taken
together, these results indicate that FGFR3 is associated with
tumor invasion and may promote the expression of E-cadherin and
MMP9, which are associated with EMT and ECM remodeling,
respectively, in lung cancer. However, the direct mechanism between
FGFR3 and MMPs remains to be elucidated. Therefore, siRNA targeting
FGFR3 may act as a tumor suppressor through the regulation of
FGFR3. Thus, RNAi technology may serve as a potential target of
NSCLC therapy.
In conclusion, the present study suggests that
downregulation of FGFR3 via siRNA-FGFR3 in NSCLC may lead to a
decrease in tumor cell invasion. In addition, the expression of
FGFR3 was observed to be significantly correlated with the
expression of MMP9 and E-cadherin in the present study. According
to these results, FGFR3 may be an important marker of NSCLC
metastasis and a potential therapeutic target. Further studies are
required to clarify the mechanisms of FGFR3 in EMT and ECM
remodeling in NSCLC.
Acknowledgements
The present study was supported by the Natural
Science Foundation of Shandong Province (Jinan, China; grant no.
W2012FZ078) and Wu Jieping Medical Foundation (Beijing, China;
grant no. 320.6750.13210).
References
|
1
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: The impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, DeSantis C, Virgo K, Stein K,
Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al:
Cancer treatment and survivorship statistics, 2012. CA Cancer J
Clin. 62:220–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dat T, Matsuo T, Yoshimaru T, Kakiuchi S,
Goto H, Hanibuchi M, Kuramoto T, Nishioka Y, Sone S and Katagiri T:
Identification of genes potentially involved in bone metastasis by
genome-wide gene expression profile analysis of non-small cell lung
cancer in mice. Int J Oncol. 40:1455–1469. 2012.PubMed/NCBI
|
|
4
|
Langley RR and Fidler IJ: The seed and
soil hypothesis revisited - the role of tumor-stroma interactions
in metastasis to different organs. Int J Cancer. 128:2527–2535.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Larsen JE and Minna JD: Molecular biology
of lung cancer: Clinical implications. Clin Chest Med. 32:703–740.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Miele E, Spinelli GP, Miele E, Di Fabrizio
E, Ferretti E, Tomao S and Gulino A: Nanoparticle-based delivery of
small interfering RNA: Challenges for cancer therapy. Int J
Nanomedicine. 7:3637–3657. 2012.PubMed/NCBI
|
|
7
|
Joo MK, Yhee JY, Kim SH and Kim K: The
potential and advances in RNAi therapy: Chemical and structural
modifications of siRNA molecules and use of biocompatible
nanocarriers. J Control Release. 193:113–121. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen Y, Gu H, Zhang DS, Li F, Liu T and
Xia W: Highly effective inhibition of lung cancer growth and
metastasis by systemic delivery of siRNA via multimodal mesoporous
silica-based nanocarrier. Biomaterials. 35:10058–10069. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shi L, Zhang B, Sun X, Lu S, Liu Z, Liu Y,
Li H, Wang L, Wang X and Zhao C: MiR-204 inhibits human NSCLC
metastasis through suppression of NUAK1. Br J Cancer.
111:2316–2327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kang J, Lee SY, Lee SY, Kim YJ, Park JY,
Kwon SJ, Na MJ, Lee EJ, Jeon HS and Son JW: MicroRNA-99b acts as a
tumor suppressor in non-small cell lung cancer by directly
targeting fibroblast growth factor receptor 3. Exp Ther Med.
3:149–153. 2012.PubMed/NCBI
|
|
11
|
Lamont FR, Tomlinson DC, Cooper PA,
Shnyder SD, Chester JD and Knowles MA: Small molecule FGF receptor
inhibitors block FGFR-dependent urothelial carcinoma growth in
vitro and in vivo. Br J Cancer. 104:75–82. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Smid M, Wang Y, Klijn JG, Sieuwerts AM,
Zhang Y, Atkins D, Martens JW and Foekens JA: Genes associated with
breast cancer metastatic to bone. J Clin Oncol. 24:2261–2267. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kang S, Dong S, Gu TL, Guo A, Cohen MS,
Lonial S, Khoury HJ, Fabbro D, Gilliland DG, Bergsagel PL, et al:
FGFR3 activates RSK2 to mediate hematopoietic transformation
through tyrosine phosphorylation of RSK2 and activation of the
MEK/ERK pathway. Cancer Cell. 12:201–214. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang X, Lu H, Urvalek AM, Li T, Yu L,
Lamar J, DiPersio CM, Feustel PJ and Zhao J: KLF8 promotes human
breast cancer cell invasion and metastasis by transcriptional
activation of MMP9. Oncogene. 30:1901–1911. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tu CL and You M: Obligatory roles of
filamin A in E-cadherin-mediated cell-cell adhesion in epidermal
keratinocytes. J Dermatol Sci. 73:142–151. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiang XB, Wang JS, Liu DH, Yuan WS and Shi
ZS: Overexpression of matrix metalloproteinase-9 is correlated with
carotid intraplaque hemorrhage in a swine model. J Neurointerv
Surg. 5:473–477. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stenvold H, Donnem T, Andersen S, Al-Saad
S, Al-Shibli K, Busund LT and Bremnes RM: Overexpression of matrix
metalloproteinase-7 and −9 in NSCLC tumor and stromal cells:
Correlation with a favorable clinical outcome. Lung Cancer.
75:235–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moss LA Shuman, Jensen-Taubman S and
Stetler-Stevenson WG: Matrix metalloproteinases: Changing roles in
tumor progression and metastasis. Am J Pathol. 181:1895–1899. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang B, Xie C, Zhong J, Chen H, Zhang H
and Wang X: A549 cell proliferation inhibited by RNAi mediated
silencing of the Nrf2 gene. Biomed Mater Eng. 24:3905–3916.
2014.PubMed/NCBI
|
|
20
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ong KJ, MacCormack TJ, Clark RJ, Ede JD,
Ortega VA, Felix LC, Dang MK, Ma G, Fenniri H, Veinot JG and Goss
GG: Widespread nanoparticle-assay interference: implications for
nanotoxicity testing. PLoS One. 9:e906502014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nijkamp MM, Span PN, Hoogsteen IJ, van der
Kogel AJ, Kaanders JH and Bussink J: Expression of E-cadherin and
vimentin correlates with metastasis formation in head and neck
squamous cell carcinoma patients. Radiother Oncol. 99:344–348.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen ZL, Zhao XH, Wang JW, Li BZ, Wang Z,
Sun J, Tan FW, Ding DP, Xu XH, Zhou F, et al: MicroRNA-92a promotes
lymph node metastasis of human esophageal squamous cell carcinoma
via E-cadherin. J Biol Chem. 286:10725–10734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu Q, Wang YD, Jing SH, Wang XL, Cheng YJ
and Wu FP: Expression of E-cadherin in nasopharyngeal carcinoma and
its relationship with cervical lymph node metastasis. Zhonghua
Zhong Liu Za Zhi. 32:425–428. 2010.(In Chinese). PubMed/NCBI
|
|
25
|
Kessenbrock K, Plaks V and Werb Z: Matrix
metalloproteinases: Regulators of the tumor microenvironment. Cell.
141:52–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Reeves CV, Wang X, Charles-Horvath PC,
Vink JY, Borisenko VY, Young JA and Kitajewski JK: Anthrax toxin
receptor 2 functions in ECM homeostasis of the murine reproductive
tract and promotes MMP activity. PLoS One. 7:e348622012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
de Oliveira Demarchi AC, Zambuzzi WF,
Paiva KB, da Silva-Valenzuela M, Nunes FD, de Cássia Sávio Figueira
R, Sasahara RM, Demasi MA, Winnischofer SM, Sogayar MC and
Granjeiro JM: Development of secondary palate requires strict
regulation of ECM remodeling: Sequential distribution of RECK,
MMP-2, MMP-3, and MMP-9. Cell Tissue Res. 340:61–69. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu J, Ping W, Zu Y and Sun W:
Correlations of lysyl oxidase with MMP2/MMP9 expression and its
prognostic value in non-small cell lung cancer. Int J Clin Exp
Pathol. 7:6040–6047. 2014.PubMed/NCBI
|
|
29
|
Liao RG, Jung J, Tchaicha J, Wilkerson MD,
Sivachenko A, Beauchamp EM, Liu Q, Pugh TJ, Pedamallu CS, Hayes DN,
et al: Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung
squamous cell carcinoma. Cancer Res. 73:5195–5205. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Q, Jiang HO, Quan QL, Li J, He T and
Huang XS: Mutation analysis of FGFR3 gene in a family featuring
hereditary dwarfism. Zhonghua Yi Xue Yi Chuan Xue Za Zhi.
28:705–707. 2011.(In Chinese). PubMed/NCBI
|
|
31
|
Sahlin P, Tarnow P, Martinsson T and
Stenman G: Germline mutation in the FGFR3 gene in a TWIST1-negative
family with Saethre-Chotzen syndrome and breast cancer. Genes
Chromosomes Cancer. 48:285–288. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Haugsten EM, Wiedlocha A, Olsnes S and
Wesche J: Roles of fibroblast growth factor receptors in
carcinogenesis. Mol Cancer Res. 8:1439–1452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Blick C, Ramachandran A, Wigfield S,
McCormick R, Jubb A, Buffa FM, Turley H, Knowles MA, Cranston D,
Catto J and Harris AL: Hypoxia regulates FGFR3 expression via
HIF-1α and miR-100 and contributes to cell survival in non-muscle
invasive bladder cancer. Br J Cancer. 109:50–59. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Schlessinger J: Cell signaling by receptor
tyrosine kinases. Cell. 103:211–225. 2000. View Article : Google Scholar : PubMed/NCBI
|















