Introduction
Liver cancer is one of the most common malignancies,
worldwide, with the highest morbidity rates in China (1). Due to difficult treatment, poor
prognosis, relapse and metastasis following surgery and the high
mortality rate, liver cancer presents a significant health problem
(2). Thus, liver cancer-related genes
and their mechanisms of action have received increasing attention
in recent years. Notably, special AT-rich DNA-binding protein-1
(SATB1) has been investigated due to its association with tumor
growth and metastasis. SATB1 is involved with the growth and
differentiation of T cells, as well as tumor growth and metastasis.
SATB1 regulates gene transcription and expression by recruiting
chromatin remodeling complexes and anchoring specialized DNA
sequences, such as nuclear molecular attachment region-binding
protein (3–12). A previous study demonstrated that the
body, spleen, thymus and lymph nodes of SATB1 gene knockout mice
are significantly smaller those of normal mice, and this difference
increases with age, resulting in death 3 weeks after birth
(7). These results suggest that SATB1
is predominantly expressed in the thymus and that it exhibits an
important function in the growth and differentiation of T cells
(7). A previous study revealed that
SATB1 is involved in the development of tumors and is closely
associated with tumor invasion and metastasis by regulating the
cell cycle, inducing cell differentiation and inhibiting apoptosis
(10). The function of SATB1 has been
confirmed in several tumor types, including breast cancer (13–15),
epithelial ovarian cancer (16),
gastric cancer (17), colorectal
carcinoma (18–20), glioma (21), Hodgkin's lymphoma (22), small cell lung cancer (23), prostate cancer (24,25) and
liver cancer (26). Furthermore, a
previous study has reported that both the mRNA and protein
expression of SATB1 are upregulated when compared with
carcinoma-adjacent and normal liver tissue (26). SATB1 expression levels are also
hypothesized to correlate with tumor size, degree of
differentiation, portal invasion, lymph node metastases and
hepatitis B virus infection (26).
However, the association between SATB1 expression and liver cancer
metastasis and metastasis-associated biological behavior, as well
as the clinicopathological significance of immunohistochemical
SATB1 expression in liver cancer remains unclear. In the present
study, the function of SATB1 expression in liver cancer was
investigated using histology and cytology.
Materials and methods
Tumor specimens and cell culture
A total of 60 primary hepatocellular carcinoma
tissue samples and 60 adjacent normal liver tissue samples were
obtained from patients that underwent surgical resection without
prior chemotherapy or radiation at the Second Affiliated Hospital
of Nanchang University (Nanchang, China) between January 2010 and
December 2013 (Table I) after
obtaining patient consent. All adjacent normal liver tissues were
resected ≥1 cm from the tumor margin. All tissues were fixed in
formaldehyde and histopathological classification, according to the
World Health Organization Classification of Tumors of the Digestive
System (27), was confirmed by
hematoxylin and eosin staining. MHCC-97H (high metastatic
potential) and HepG2 (low metastatic potential) human liver cancer
cell lines were obtained from the Molecular Biology Center of the
Second Affiliated Hospital of Nanchang University and were cultured
in Dulbecco's modified Eagle's medium containing 15% fetal bovine
serum (Biological Industries Israel, Beit-Haemek, Israel) at 37°C
in an atmosphere of 5% CO2 for 48 h. This study was
approved by the ethics committee of the Second Affiliated Hospital
of Nanchang University.
 | Table I.Association between SATB1 expression
and clinicopathological features of 60 hepatocellular carcinoma
patients. |
Table I.
Association between SATB1 expression
and clinicopathological features of 60 hepatocellular carcinoma
patients.
| Parameter | Patients, n | Positive SATB1
expression, n (%) | Negative SATB1
expression, n (%) | P-value |
|---|
| Gender |
|
|
| 1.000 |
| Male | 45 | 30 (66.67) | 15 (33.33) |
|
|
Female | 15 | 10 (66.67) | 5
(33.33) |
|
| Age, years |
|
|
| 0.232 |
| ≥55 | 18 | 14 (77.78) | 4
(22.22) |
|
|
<55 | 42 | 26 (61.90) | 16 (38.10) |
|
| Tumor number |
|
|
| 0.839 |
|
>1 | 17 | 11 (64.71) | 6
(35.29) |
|
| 1 | 43 | 29 (67.44) | 14 (32.56) |
|
| Tumor size, cm |
|
|
| 0.001 |
| ≥5 | 36 | 30 (83.33) | 6
(16.67) |
|
|
<5 | 24 | 10 (41.67) | 14 (58.33) |
|
| Tumor
differentiation |
|
|
| 0.000 |
|
Moderate and poor | 40 | 33 (82.50) | 7
(17.50) |
|
|
Well | 20 | 7 (35.00) | 13 (65.00) |
|
| AFP, ng/ml |
|
|
| 0.449 |
|
≥100 | 22 | 16 (72.73) | 6
(27.27) |
|
|
<100 | 38 | 24 (63.16) | 14 (36.84) |
|
| Hemorrhage and/or
necrosis |
|
|
| 0.010 |
|
Present | 26 | 24 (92.31) | 2
(7.69) |
|
|
Absent | 34 | 16 (47.06) | 18 (52.94) |
|
| Invasion and/or
metastases |
|
|
| 0.022 |
|
Present | 19 | 17 (89.47) | 2
(10.53) |
|
|
Absent | 41 | 23 (56.10) | 18 (43.90) |
|
| TNM stage |
|
|
| 0.008 |
|
I–II | 22 | 7
(31.80) | 15 (68.20) |
|
|
III–IV | 38 | 33 (86.84) | 5
(13.16) |
|
Immunohistochemistry (IHC)
To analyze SATB1 expression, IHC was performed using
10% formalin-fixed 4-µm tissue sections of hepatocellular carcinoma
and adjacent normal liver tissues. The primary antibody used in the
present study was anti-SATB1 (clone, EPR3895; 1:100; Epitomics,
Burlingame, CA, USA). Horseradish peroxidase-conjugated
anti-sheep/rabbit immunoglobulin (Ig)G (cat. no. PV6001; 1:500;
Zhongshan Jinqiao Biotechnology Co., Ltd., Beijing, China) was used
as the secondary antibody. Stromal lymphocytes served as internal
positive controls, with adjacent normal liver tissue and
phosphate-buffered saline buffer replacing the primary antibody as
a negative control (16,18,19). The
estimated percentage of cells exhibiting nuclear SATB1 expression
was denoted as negative, weak, moderate or strong (Fig. 1; <1% positive SATB1 expression,
negative staining; 1–50% positive SATB1 expression, weak staining;
51–75% positive SATB1 expression, moderate staining; >75%
positive SATB1 expression, strong staining) and the combined
nuclear score was calculated by multiplying the fraction of cells
with positive SATB1 expression by the intensity (16). Histological staining for tumor size,
tumor differentiation, invasion and metastases, α-fetoprotein and
TNM stage were performed as previously described (19,26).
Reverse transcription-polymerase chain
reaction (PCR)
Total RNA was extracted from cells using TRIzol
reagent (TransGen Biotech, Inc., Beijing, China) according to the
manufacturer's protocol. A 0.4 µg aliquot of RNA was
reverse-transcribed to cDNA using the PrimeScript RT reagent kit
(Takara Bio, Dalian, China) in a 20 µl volume containing 1 µl cDNA.
A total of 10 µl PCR SuperMix (TransGen Biotech, Inc.) was used in
subsequent reaction using the following primers: Forward,
5′-AGAGGAAGGCTTGGGAGTA-3′ and reverse, 5′-GGGCAGCAGAGCTATGTG-3′ for
SATB1; forward, 5′-TGATGACATCAAGAAGGTGGTGAAG-3 and reverse,
5′-TCCTTGGAGGCCATGTGGGCCA-3′ for GAPDH. PCR was performed using a
MyCycler™ Thermal Cycler System (Bio-Rad Laboratories, Hercules,
CA, USA) under the following conditions: Amplification at 95°C for
5 min, proceeded by 35 cycles at 95°C for 1 min, 60°C for 30 sec
and 72°C for 30 sec, followed by a final extension step at 72°C for
10 min. Negative (no cDNA) and reverse transcription (no reverse
transcription) controls were used. All experiments were performed
in triplicate.
Western blot analysis
Nucleoprotein was extracted from cells using
Nuclear-Cytosol Extraction Kit (Applygen Technologies, Inc.,
Beijing, China). The protein concentration was determined by
spectrophotometer (Eppendorf, Hamburg, Germany) at a wavelength of
280 nm. Protein samples (15 µg) were separated by 6% sodium dodecyl
sulfate polyacrylamide gel electrophoresis and subsequently
transferred to polyvinylidene fluoride membranes, followed by
incubation with the primary SATB1 antibody (1:1,000; catalog no.,
EPR3895) at 4°C overnight. The membranes were washed with
Tris-buffered saline and Tween 20 and incubated with biotinylated
anti-rabbit IgG (1:10,000; catalog no., HS101-01) for 1 h at room
temperature. The intensity of the protein bands was analyzed using
Image-Pro Plus image analysis software (Media Cybernetics,
Rockville, MD, USA).
Transwell migration and wound-healing
assays
In vitro Transwell migration and
wound-healing assays were performed as described previously
(28,29). The cells adhering to the lower surface
of the membrane were stained using hematoxylin and eosin, and the
number of cells was calculated in 5 random high-power fields using
a DM1000 Leica microscope (Leica, Wetzlar, Germany). The areas of
interest were analyzed by ImageJ software (National Institutes of
Health, Bethesda, MD, USA).
Silencing and upregulation of SATB1
expression
Three silencing plasmids (GV102-SATB1-RNAi
(NM_002971): Sequence 1, 5′-AATGCTCTGAAGGACTTAC-3′; sequence 2,
5′-ACTGTCTTACGTGACAGAT-3′; and sequence 3,
5′-TTCCATTTATGATGAGATT-3′), defined as sh1, sh2 and sh3,
respectively and a corresponding control (CON036) were purchased
from GeneChem Co., Ltd., (Shanghai, China). An overexpression
plasmid, GV144-SATB1, defined as HepG2-up and a corresponding
control (CON107) were also purchased from GeneChem Co., Ltd. At 80%
confluence, MHCC-97H hepatic carcinoma cells were transfected with
GV102-SATB1-RNAi or CON036 using TurboFect transfection reagent
(Thermo, USA) and HepG2 hepatic carcinoma cells were transfected
with GV144-SATB1 or CON107. Transgene expression was analyzed after
24–48 h.
Statistical analysis
All assays were repeated four times and all in
vitro experiments were performed in triplicate. The data are
presented as the mean ± standard error of the mean. The
χ2 test was performed to analyze associations between
SATB1 expression (nuclear score) and hepatocellular
carcinoma-associated biological parameters. The Student's
t-test was used to compare differences between groups. All
statistical analyses were performed using SPSS 13.0 (SPSS, Inc.,
Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
SATB1 expression is positively
correlated with metastasis in human liver cancer tissues and cell
lines
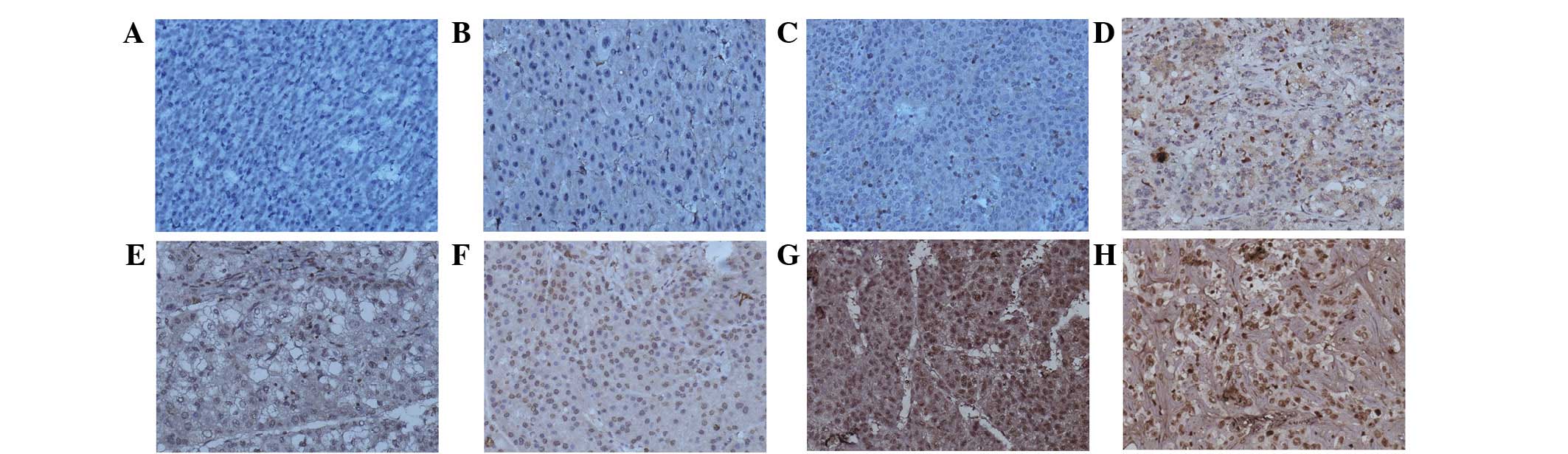
As shown in Fig. 1,
SATB1 was predominantly localized in the nucleus with a low level
of expression in the perinuclear cytoplasm, in accordance with that
reported previously (19). Absent or
extremely weak SATB1 expression was identified in 50/60 (83.33%)
carcinoma-adjacent tissues (Fig. 1A).
Positive SATB1 expression was observed in 40/60 (66.67%)
hepatocellular carcinoma tissues, which ranged from weak to strong
in intensity (Fig. 1C-H). SATB1
expression was significantly higher in hepatocellular carcinoma
tissues compared with carcinoma-adjacent tissues (P<0.0001). In
addition, the percentage of positive SATB1 expression in
hepatocellular carcinoma samples with tumors ≥5 cm, moderately and
poorly-differentiated tumors, hemorrhage and/or necrosis, invasion
and/or metastases and TNM stages III–IV were 83.33, 82.50, 84.62,
85.71 and 78.95%, respectively (Table
I), suggesting that SATB1 expression is associated with tumor
size, differentiation degree, hemorrhage and/or necrosis, invasion
and/or metastases and TNM stage (P<0.05; Table I).
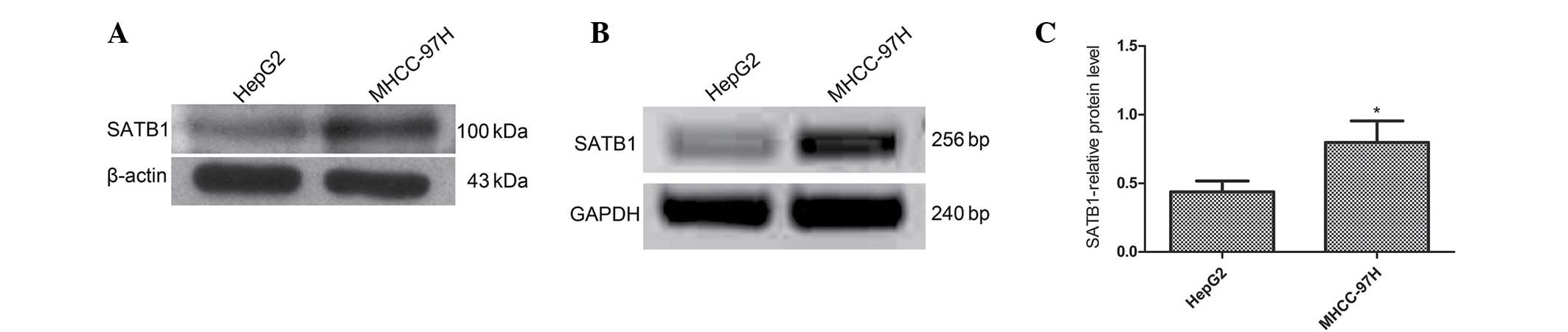
SATB1 mRNA and protein expression levels were also
analyzed in two different liver cancer cell lines, MHCC-97H (high
metastatic potential) and HepG2 (low metastatic potential)
(30,31). SATB1 mRNA and protein expression was
higher in the MHCC-97H cell line than the HepG2 cell line (P=0.002
and P=0.001, respectively). It was also observed that SATB1
expression is correlated with metastasis in human liver cancer
tissues and cell lines (P<0.05; Fig.
2).
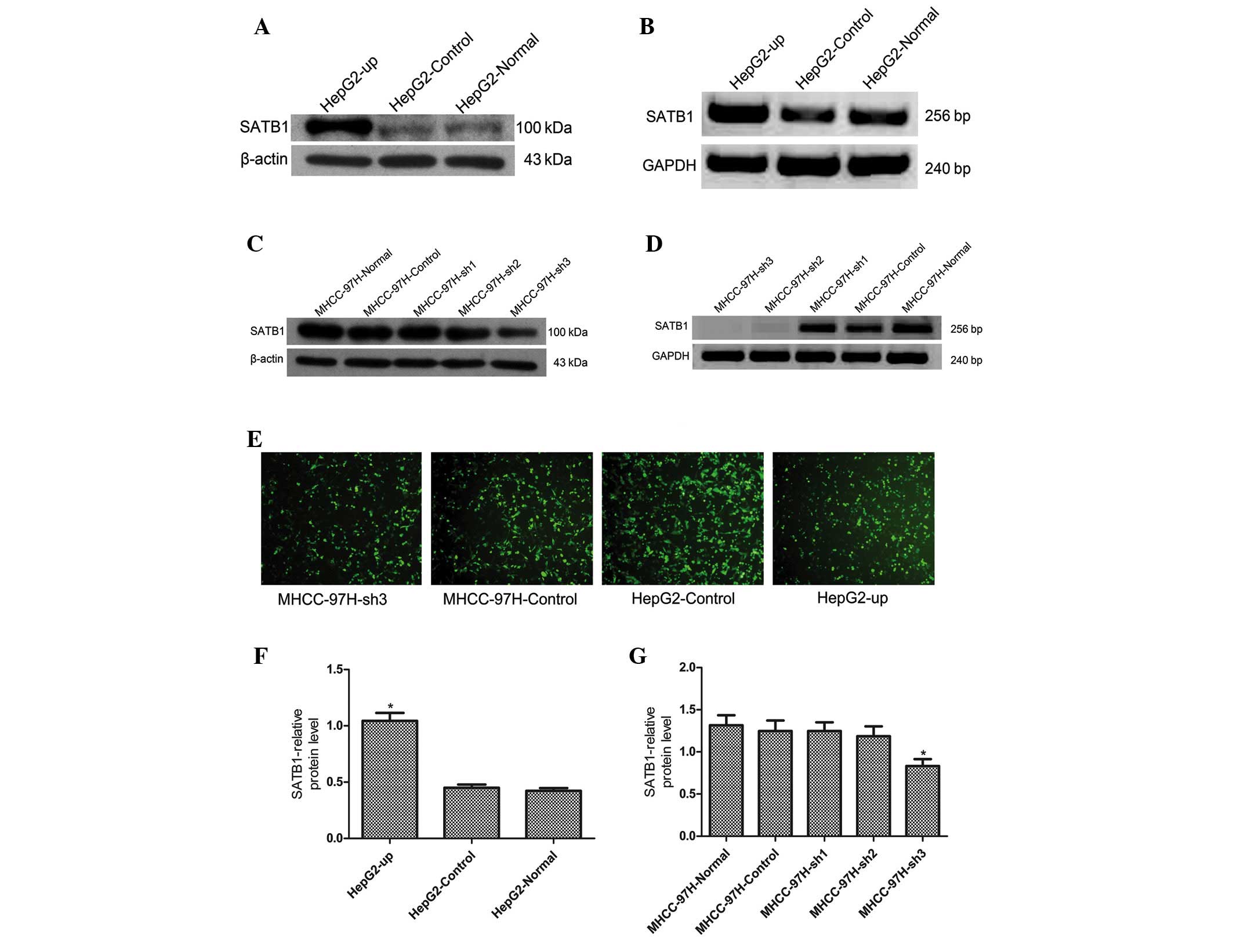
Silencing and upregulation of SATB1
expression in human liver cancer cell lines
The MHCC-97H (high SATB1 expression) and HepG2 (low
SATB1 expression) cell lines were selected to investigate the
effects of SATB1 gene silencing and overexpression, respectively,
in order to further investigate the association between SATB1
expression and liver cancer metastasis (Fig. 3). SATB1 mRNA and protein levels were
increased significantly in HepG2 cells following transfection with
the HepG2-up plasmid, which exhibited a transfection efficiency of
80%, however, no significant differences in SATB1 mRNA or protein
expression levels were identified in the cells transfected with the
control CON107 plasmid, which exhibited a transfection efficiency
of 90% (Fig. 3A, B and F; P<0.05).
MHCC-97H cells were transfected with 3 silencing plasmids, sh1, sh2
and sh3, or the corresponding control CON036 plasmid. The results
revealed that SATB1 protein levels were significantly lower in the
sh3-transfection group, which exhibited a transfection efficiency
of 70% compared with the sh1, sh2 and control CON036-transfected
groups, which exhibited a transfection efficiency of 80% (Fig. 3C). No significant differences in SATB1
protein expression were identified in the sh1, sh2 and CON036
groups compared with normal MHCC-97H cells (Fig. 3C and G; P<0.05). SATB1 mRNA
expression was significantly lower in the sh2 and sh3-transfected
groups compared with the normal MHCC-97H (Fig. 3D; P<0.05), while no significant
differences in SATB1 mRNA expression were identified in the sh1 and
CON036-transfection groups compared with normal MHCC-97H cells.
Therefore, the sh3 silencing plasmid was selected to transfect
MHCC-97H cells for subsequent experiments.
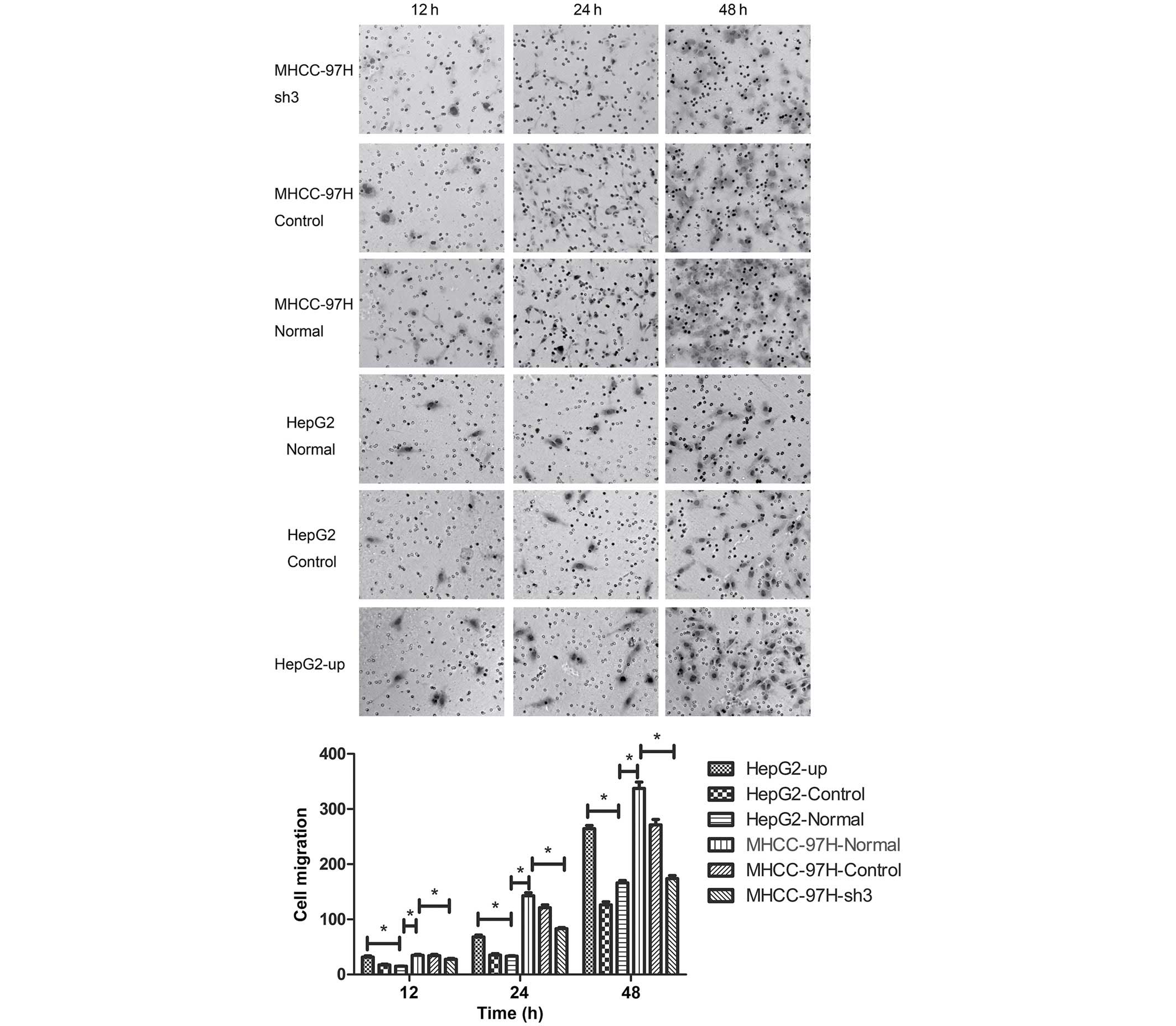
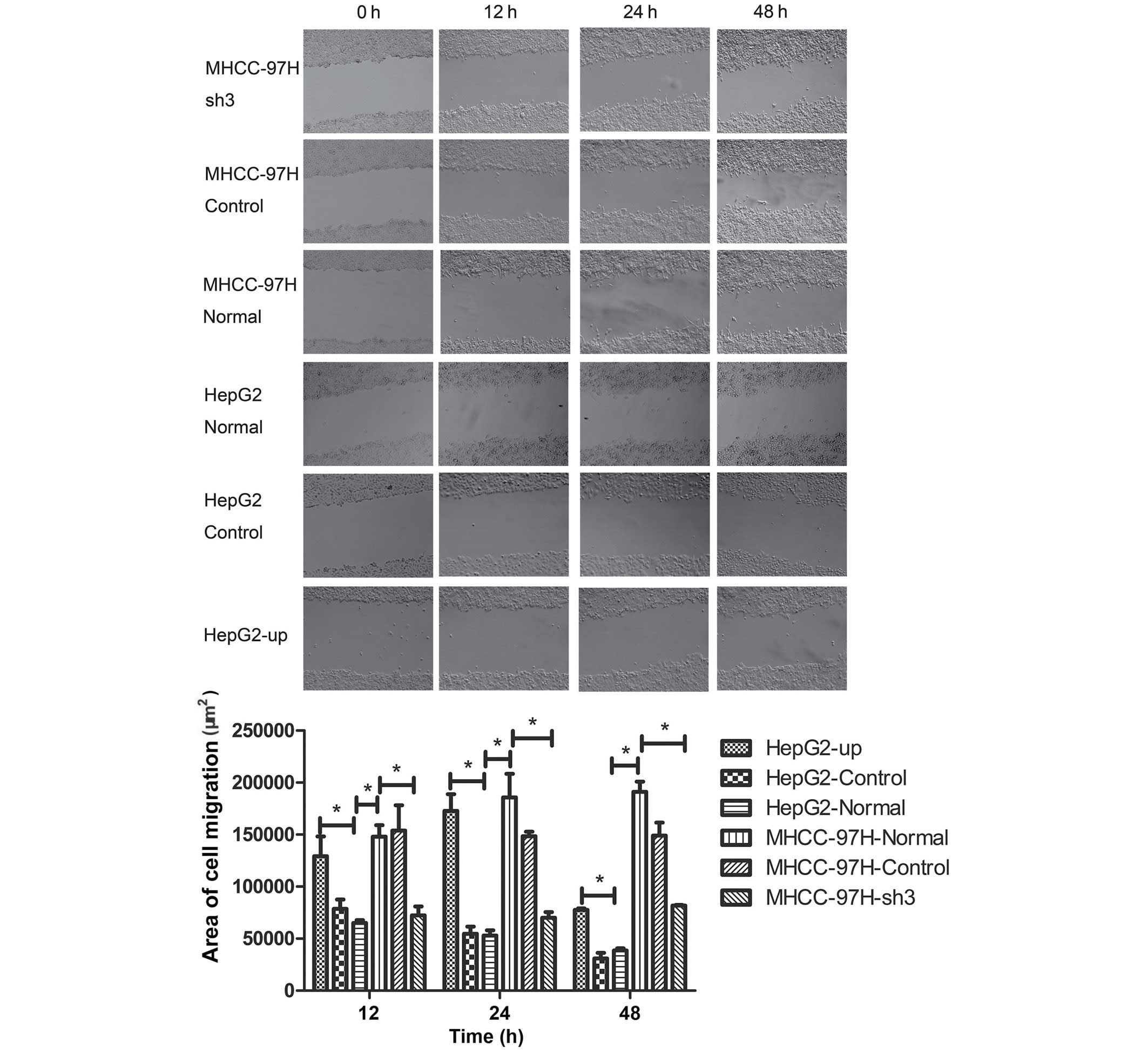
Metastasis of liver cancer MHCC-97H
and HepG2 cell lines is positively correlated with SATB1
expression
As our previous experiments had indicated that SATB1
expression was associated with the metastasis of liver cancer, the
biological behavior of MHCC-97H and HepG2 cells was assessed using
Transwell migration and wound-healing assays. The number of cells
adhering to the lower surface of the membrane in the MHCC-97H liver
cancer cell line was significantly higher compared than the HepG2
cell line (P<0.05; Fig. 4). The
number of cells adhering to the lower surface of the membrane
decreased significantly in the MHCC-97H cell line following
transfection with the sh3 plasmid (Fig.
4; P<0.05). By contrast, the number of cells adhering to the
lower surface of the membrane in the HepG2 cell line increased
significantly compared with the normal group following transfection
with the HepG2-up plasmid (Fig. 4;
P<0.05). No significant differences in migration capability were
identified between the control groups CON036 and CON107 and the
corresponding normal groups (Fig. 4;
P>0.05). Wound-healing assays were performed and the area of
cell migration of the scratch area was used to determine the
migration of cancer cell lines. The area of cell migration was
greater in the MHCC-97H liver cancer cell line than in the HepG2
cell line (Fig. 5; P<0.05). The
area of cell migration of the scratch area decreased significantly
compared with the normal group after the sh3 plasmid was
transfected into the MHCC-97H liver cancer cell line (Fig. 5; P<0.05). The area of cell
migration of the scratch area increased significantly in the HepG2
liver cancer cell line following transfection with the HepG2-up
plasmid when compared with the normal group (Fig. 5; P<0.05). No significant
differences in the area of cell migration were identified between
the control groups CON036 and CON107 and the corresponding normal
groups (Fig. 5; P<0.05).
Discussion
The results of the present study revealed that SATB1
mRNA expression was higher in MHCC-97H cells (high metastatic
potential) than HepG2 cells (low metastatic potential) and that the
metastatic potential of liver cancer cells was correlates with
increased SATB1 expression. Han et al (13) reported that SATB1 expression may used
as an independent factor of poor prognosis and demonstrated that
SATB1 correlated with poor prognosis, even in breast cancer
patients that exhibited low immunohistochemical expression levels.
Furthermore, Tu et al (26)
demonstrated that SATB1 promotes growth and metastasis of liver
cancer in vitro and in vivo via promotion of cell
cycle progression, apoptosis inhibition and induction of
epithelial-mesenchymal transition. However, in the present study,
the association between SATB1 expression and liver cancer
metastasis was demonstrated by both SATB1 up- and downregulation in
biological behavior assays. The results revealed that SATB1
expression positively correlates with the metastasis of liver
cancer, in accordance with previously reported data (16–26). At
present, an increasing number of patients with liver cancer are
diagnosed at an early stage as a result of the advent of precision
instruments, including ultrasonic imaging and the development of
fine-needle puncture. In addition, histopathological classification
and immunohistochemical results provided by specific antibodies are
easier to obtain than results following surgery, which is the
typical treatment route in cases of liver cancer. The surgical
approach requires the expertise of a clinical pathologist to
improve the diagnostic level possible with a limited tissue sample,
thus improving the utility of the diagnosis for clinical treatment.
In the present study, a correlation was identified between SATB1
expression and liver cancer metastasis. Furthermore, SATB1
expression was associated with tumor size, differentiation degree,
hemorrhage and/or necrosis, invasion and/or metastases and TNM
stage, which may are often considered as prognostic factors during
clinicopathological diagnosis. Therefore, future studies using a
larger sample size with adequate clinical follow-up data are
required. The aim of the present study was to investigate the
association between SATB1 expression and metastasis of liver
cancer. The results indicate that SATB1 expression may present a
prognostic factor for liver cancer, and thus these findings may
lead to improved guidance for clinical treatment. The results
indicate that SATB1 may present a novel target for liver cancer
treatment.
References
|
1
|
Parkin DM: Global cancer statistics in the
year 2000. Lancet Oncol. 2:533–543. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li G, Yang D, Li X, Zhong K, Liu X, Bi M,
Liu Y, Liao X and Lin L: Expression of SATB1 in hepatocellular
carcinoma cell lines with different invasive capacities. Nan Fang
Yi Ke Da Xue Xue Bao. 32:986–994. 2012.(In Chinese). PubMed/NCBI
|
|
3
|
Xu L, Deng HX, Xia JH, Yang Y, Fan CH,
Hung WY and Siddque T: Assignment of SATB1 to human chromosome band
3p23 by in situ hybridization. Cytogenet Cell Genet. 77:205–206.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dickinson LA, Joh T, Kohwi Y and
Kohwi-Shigematsu T: A tissue-specific MAR/SAR DNA-binding protein
with unusual binding site recognition. Cell. 70:631–645. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wen J, Huang S, Rogers H, Dickinson LA,
Kohwi-Shigematsu T and Noguchi CT: SATB1 family protein expressed
during early erythroid differentiation modifies globin gene
expression. Blood. 105:3330–3339. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kumar PP, Purbey PK, Ravi DS, Mitra D and
Galande S: Displacement of SATB1-bound histone deacetylase 1
corepressor by the human immunodeficiency virus type 1
transactivator induces expression of interleukin-2 and its receptor
in T cells. Mol Cell Biol. 25:1620–1633. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yasui D, Miyano M, Cai S, Varga-Weisz P
and Kohwi-Shigematsu T: SATB1 targets chromatin remodelling to
regulate genes over long distances. Nature. 419:641–645. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Z, Yang X, Chu X, Zhang J, Zhou H,
Shen Y and Long J: The structural basis for the oligomerization of
the N-terminal domain of SATB1. Nucleic Acids Res. 40:4193–4202.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kumar PP, Bischof O, Purbey PK, Notani D,
Urlaub H, Dejean A and Galande S: Functional interaction between
PML and SATB1 regulates chromatin-loop architecture and
transcription of the MHC class I locus. Nat Cell Biol. 9:45–56.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Galande S, Purbey PK, Notani D and Kumar
PP: The third dimension of gene regulation: Organization of dynamic
chromatin loopscape by SATB1. Curr Opin Genet Dev. 17:408–414.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Beyer M, Thabet Y, Müller RU, Sadlon T,
Classen S, Lahl K, Basu S, Zhou X, Bailey-Bucktrout SL, Krebs W, et
al: Repression of the genome organizer SATB1 in regulatory T cells
is required for suppressive function and inhibition of effector
differentiation. Nat Immunol. 12:898–907. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Notani D, Gottimukkala KP, Jayani RS,
Limaye AS, Damle MV, Mehta S, Purbey PK, Joseph J and Galande S:
Global regulator SATB1 recruits beta-catenin and regulates T(H)2
differentiation in Wnt-dependent manner. PLoS Biol. 8:e10002962010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Han HJ, Russo J, Kohwi Y and
Kohwi-Shigematsu T: SATB1 reprogrammes gene expression to promote
breast tumour growth and metastasis. Nature. 452:187–193. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hanker LC, Karn T, Mavrova-Risteska L,
Ruckhäberle E, Gaetje R, Holtrich U, Kaufmann M, Rody A and
Wiegratz I: SATB1 gene expression and breast cancer prognosis.
Breast. 20:309–313. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li QQ, Chen ZQ, Cao XX, Xu JD, Xu JW, Chen
YY, Wang WJ, Chen Q, Tang F, Liu XP and Xu ZD: Involvement of
NF-κB/miR-448 regulatory feedback loop in chemotherapy-induced
epithelial-mesenchymal transition of breast cancer cells. Cell
Death Differ. 18:16–25. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nodin B, Hedner C, Uhlén M and Jirström K:
Expression of the global regulator SATB1 is an independent factor
of poor prognosis in high grade epithelial ovarian cancer. J
Ovarian Res. 5:242012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun F, Lu X, Li H, Peng Z, Wu K, Wang G
and Tong Q: Special AT-rich sequence binding protein 1 regulates
the multidrug resistance and invasion of human gastric cancer
cells. Oncol Lett. 4:156–162. 2012.PubMed/NCBI
|
|
18
|
Nodin B, Johannesson H, Wangefjord S,
O'Connor DP, Lindquist KE, Uhlén M, Jirström K and Eberhard J:
Molecular correlates and prognostic significance of SATB1
expression in colorectal cancer. Diagn Pathol. 7:1152012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang J, Zhang B, Zhang X, Sun Y, Wei X,
McNutt MA, Lu S, Liu Y, Zhang D, Wang M, et al: SATB1 expression is
associated with biologic behavior in colorectal carcinoma in vitro
and in vivo. PloS One. 8:e479022013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meng WJ, Yan H, Zhou B, Zhang W, Kong XH,
Wang R, Zhan L, Li Y, Zhou ZG and Sun XF: Correlation of SATB1
overexpression with the progression of human rectal cancer. Int J
Colorectal Dis. 27:143–150. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chu SH, Ma YB, Feng DF, Zhang H, Zhu ZA,
Li ZQ and Jiang PC: Upregulation of SATB1 is associated with the
development and progression of glioma. J Transl Med. 10:1492012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Y, van den Berg A, Veenstra R, Rutgers
B, Nolte I, van Imhoff G, Visser L and Diepstra A: PML nuclear
bodies and SATB1 are associated with HLA class I expression in EBV+
Hodgkin lymphoma. PloS One. 8:e729302013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang B, Zhou H, Wang X and Liu Z:
Silencing SATB1 with siRNA inhibits the proliferation and invasion
of small cell lung cancer cells. Cancer Cell Int. 13:82013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shukla S, Sharma H, Abbas A, MacLennan GT,
Fu P, Danielpour D and Gupta S: Upregulation of SATB1 is associated
with prostate cancer aggressiveness and disease progression. PloS
One. 8:e535272013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mao L, Yang C, Wang J, Li W, Wen R, Chen J
and Zheng J: SATB1 is overexpressed in metastatic prostate cancer
and promotes prostate cancer cell growth and invasion. J Transl
Med. 11:1112013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tu W, Luo M, Wang Z, Yan W, Xia Y, Deng H,
He J, Han P and Tian D: Upregulation of SATB1 promotes tumor growth
and metastasis in liver cancer. Liver Int. 32:1064–1078. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bosman FT, Carneiro F, Hruban RH and
Theise ND: WHO Classification of Tumours of the Digestive System.
4th. World Health Organization; pp. 4172010
|
|
28
|
Liang CC, Park AY and Guan JL: In vitro
scratch assay: A convenient and inexpensive method for analysis of
cell migration in vitro. Nat Protoc. 2:329–333. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Obermajer N, Doljak B and Kos J:
Cytokeratin 8 ectoplasmic domain binds urokinase-type plasminogen
activator to breast tumor cells and modulates their adhesion,
growth and invasiveness. Mol Cancer. 8:882009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cheng J, Huo DH, Kuang DM, Yang J, Zheng L
and Zhuang SM: Human macrophages promote the motility and
invasiveness of osteopontin-knockdown tumor cells. Cancer Res.
67:5141–5147. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Y, Tang Y, Ye L, Liu B, Liu K, Chen J
and Xue Q: Establishment of a hepatocellular carcinoma cell line
with unique metastatic characteristics through in vivo selection
and screening for metastasis-related genes through cDNA microarray.
J Cancer Res Clin Oncol. 129:43–51. 2003.PubMed/NCBI
|