Introduction
Lung cancer accounts for 70–80% of cancer-associated
mortalities worldwide (1). Lung
cancers are classified according to histological type (2) in clinical practice. For therapeutic
purposes, there are two broad classes: Small cell lung carcinoma
(SCLC) and non-SCLC (3).
Adenocarcinoma (ADC) and squamous-cell carcinoma (SCC) are the two
major subtypes of NSCLC, and account for 40 and 30% of lung cancer
cases, respectively (2). In SCLC, the
malignant cancer cells contain dense neurosecretory granules
(vesicles containing neuroendocrine hormones), which render SCLC an
endocrine/paraneoplastic-associated tumor, and tumor malignance is
attributed to this hormone production (4). Although complete surgical resection can
be particularly important in the treatment of lung cancer, only a
limited number of patients undergo curative surgery, as lung cancer
is most commonly diagnosed at an advanced stage, at which point
patients are unable to undergo a surgical procedure (5). Lung cancer is extremely aggressive and
often results in a poor prognosis. An improved understanding of
lung cancer is urgently required to identify novel drug targets and
effective therapeutic strategies for lung cancer patients.
Tumor suppressor candidate 3 (TUSC3), originally
termed N33, has been known to be responsible for autosomal
recessive mental retardation for several years (6–8). Only
recently was TUSC3 identified as a potential tumor suppressor gene
when it was found deleted in several cancer types, including
prostate (9) and liver (10) cancer. However, the manner in which
TUSC3 functions as a tumor suppressor remains unclear. TUSC3 is
expressed in the majority of non-lymphoid human tissues, including
the prostate, lungs, liver and colon tissue (11). The protein is localized in the
endoplasmic reticulum and encodes a subunit of the endoplasmic
reticulum-bound oligosaccharyl transferase (OST) complex, which is
primarily responsible for protein N-linked glycosylation (12). While several studies have focused on
the correlation between TUSC3 expression and tumor development
(13–16), no data are currently available
regarding the expression of TUSC3 in lung cancer. In the present
study, the expression of TUSC3 in lung cancer, and the association
between TUSC3 expression and the clinicopathological parameters of
the disease were investigated.
Materials and methods
Tissue samples
Tissue microarray slides were purchased from Xi'an
Alena Biotechnology Ltd., Co., (Xi'an, China). The slides included
35 SCLC specimens, 80 SCC specimens, 80 ADC specimens and 37 normal
lung tissue specimens. The detailed clinicopathological
characteristics of the patients with lung cancer are listed in
Table I. All patients were clinically
staged [tumor-node-metastasis (TNM) staging] according to the
seventh edition of the American Joint Committee on Cancer system
for lung cancer (2). The degrees of
pathological differentiation were defined as follows: 1,
Well-differentiated carcinoma; 2, moderately-differentiated
carcinoma; 3, poorly-differentiated carcinoma; and 4,
undifferentiated carcinoma. The degree of differentiation for the
tumors in each of the patients was evaluated by two pathologists.
Written informed consent was obtained from all patients, and the
study was approved by the Ethical Committe of Shandong Provincial
Qianfoshan Hospital (Shandong University, Jinan, China).
 | Table I.Basic characteristics of the
patients. |
Table I.
Basic characteristics of the
patients.
| Characteristic | Number (%) | Positive, n | Negative, n | Positive rate, % | χ2 | P-valuea |
|---|
| Age,
yearsb |
|
|
|
| 0.691 | 0.406 |
|
<56 | 90 (46.2) | 39 | 61 | 43.3 |
|
|
| ≥56 | 105 (53.8) | 50 | 65 | 47.6 |
|
|
| Gender |
|
|
|
| 0.239 | 0.625 |
| Male | 144 (73.8) | 63 | 93 | 43.8 |
|
|
|
Female | 51 (26.2) | 26 | 33 | 51.0 |
|
|
| Histological
type |
|
|
|
| 11.304 | 0.001 |
|
SCLC | 35 (17.9) | 7 | 28 | 20.0 |
|
|
|
NSCLC | 160 (82.1) | 82 | 78 | 51.3 |
|
|
| TNM
stagingc |
|
|
|
| 4.903 | 0.027 |
|
I+II | 76 (39.0) | 39 | 37 | 51.3 |
|
|
|
III+IV | 119 (61.0) | 42 | 77 | 35.3 |
|
|
| LNM |
|
|
|
| 6.459 | 0.011 |
|
Negative | 65 (33.3) | 38 | 27 | 58.5 |
|
|
|
Positive | 130 (66.7) | 51 | 79 | 39.2 |
|
|
| Degree of
differentiationd |
|
|
|
| 21.817 | 0.000 |
|
1–2 | 144 (73.8) | 80 | 64 | 55.6 |
|
|
|
3–4 | 51 (26.2) | 9 | 42 | 17.6 |
|
|
Immunohistochemistry (IHC) assay
IHC staining was performed directly on the 5-µm
tissue slides. Briefly, following incubation for 2 h at 56°C, the
slides were dewaxed with xylene and rehydrated using a graded
alcohol series (100, 90, 70 and 50% ethanol; 5 min each).
Endogenous peroxidase activity was blocked with 3%
H2O2 for 15 min. For antigen retrieval, the
sections were incubated in sodium citrate buffer (0.01 M, pH 6.0)
for 20 min in a household microwave oven (600 W). Next, the slides
were incubated with 10% normal goat serum to block non-specific
binding sites. Thereafter, the slides were incubated with the TUSC3
goat polyclonal antibody (1:100 final dilution; catalog no.
sc-98191; Santa Cruz, USA) overnight at 4°C. Subsequent to washing
the slides with phosphate-buffered saline, the bio-labeled
secondary antibody, rabbit anti-goat IgG (catalog no. ZDR 5308;
ZSGB-Bio, Beijing, China), was applied at a 1:200 dilution for 40
min at 37°C. The sections were then stained with diaminobenzidine.
Finally, the sections were counterstained with hematoxylin and
eosin, dehydrated with graded alcohol and mounted using neutral
gum. Stained cell scoring using a digital pathology system was
performed by Aperio ImageScope (Aperio Technologies, Inc., Vista,
CA, USA).
Immunoreactivity was observed in the cytoplasm of
the cells and the scoring was based on cytoplasmic staining.
Immunoreactivity for TUSC3 expression was independently evaluated
by three pathologists from the Shandong Provincial Qianfoshan
Hospital, and categorized according to the immunoreactive score
(IRS) as follows: IRS = staining intensity (SI) × percentage of
positively stained cells (PP). SI was determined as 0 (negative), 1
(weak), 2 (moderate) or 3 (strong). PP was scored as 0 (negative),
1 (<25% of the cells), 2 (25–50% of the cells), 3 (50–75% of the
cells) or 4 (>75% of the cells). A final score was then
calculated by multiplying these two scores. Additionally, all the
specimens were divided into two groups, showing negative or
positive expression, using an IRS of 6 as the cut-off value.
Statistical analysis
The SPSS 13.0 software (SPSS, Inc., Chicago, IL,
USA) was used for the statistical analyses. Levels of TUSC3
expression were compared using a rank sum test. Comparisons of the
positive rates between two groups were performed using Fisher's
exact test, and Spearman's correlation method was used to evaluate
the association of scores. The significance of the correlation
between clinical pathological parameters (age, gender, grading,
T-stage, N-stage and differentiation degree) and IRS of TUSC3 was
determined using Fisher's exact test. All reported P-values were
two-sided, and P<0.05 was considered to indicate a statistically
significant difference.
Results
Patient characteristics
The basic characteristics of the patients are shown
in Table I. The median age of the
healthy controls was 53 years (range, 26–76 years) and that of lung
cancer patients was 56 years (range, 16–76 years). No significant
difference was observed in gender or age between the normal
controls and patients. Regarding the TNM staging, a significant
decrease in TUSC3 expression could be observed in the patients with
stage III+IV disease compared with those with stage I+II disease
(P=0.027; Table I). When lymph node
metastasis (LNM) was considered, analysis revealed a marked
decrease in TUSC3 expression in the patients who were
LNM+ compared with patients who were LNM−
(P=0.011, Table I). Additionally, the
positive rate of TUSC3 expression in the patients with a
differentiation degree of 1–2 was significantly higher than that in
the patients with a differentiation degree of 3–4 (P<0.001;
Table I). The representative IHC
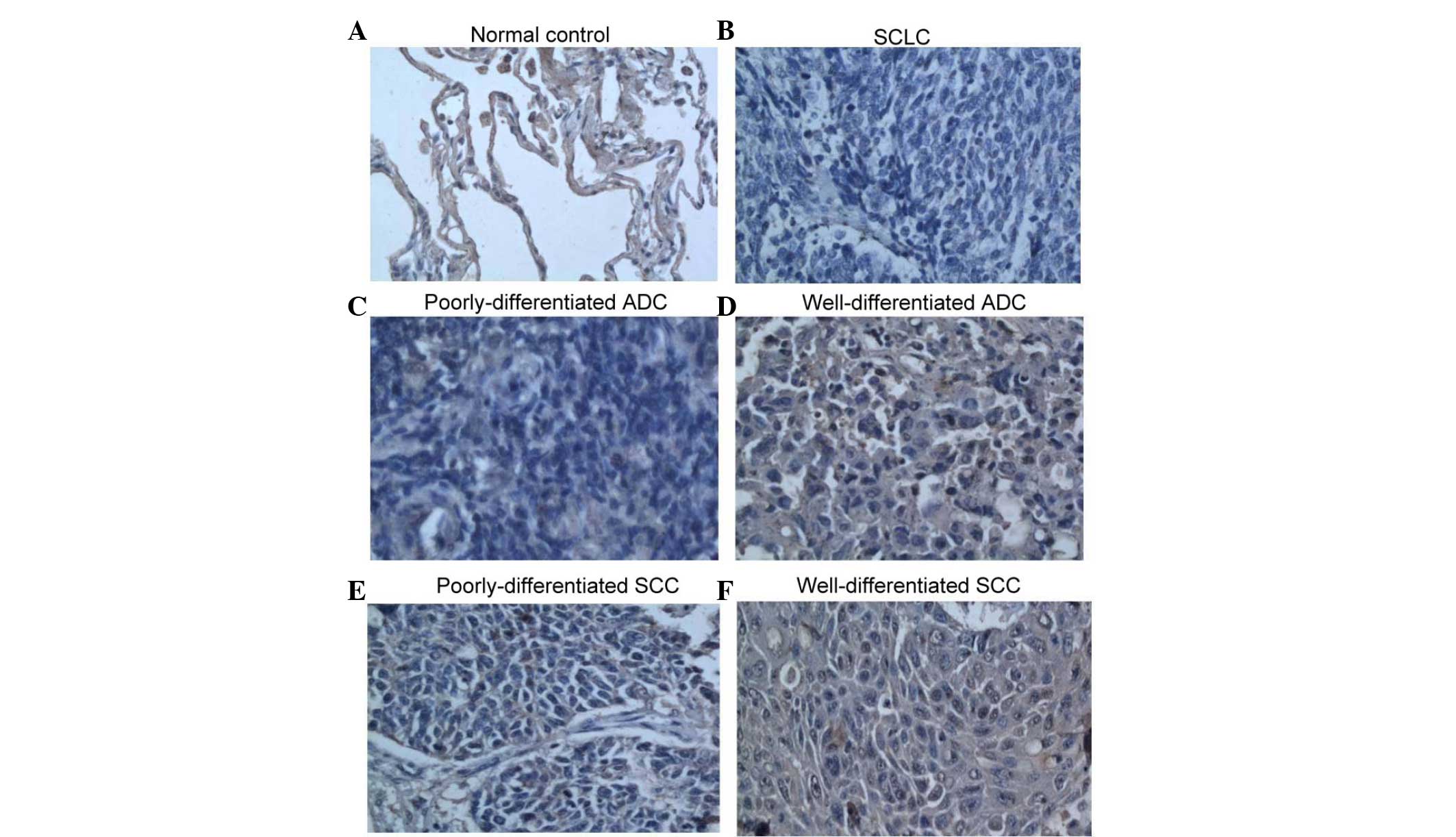
assay is shown in Fig. 1.
General expression levels in normal
controls and cancer patients
The positive rates of TUSC3 expression in the normal
controls and lung cancer patients were 59.5% (22/37) and 45.6%
(89/195), respectively (Table II).
There was no significant difference between the normal controls and
lung cancer patients with regard to the TUSC3 expression rate
(χ2=0.238, P=0.123; Table
II). When the patients were grouped according to clinical
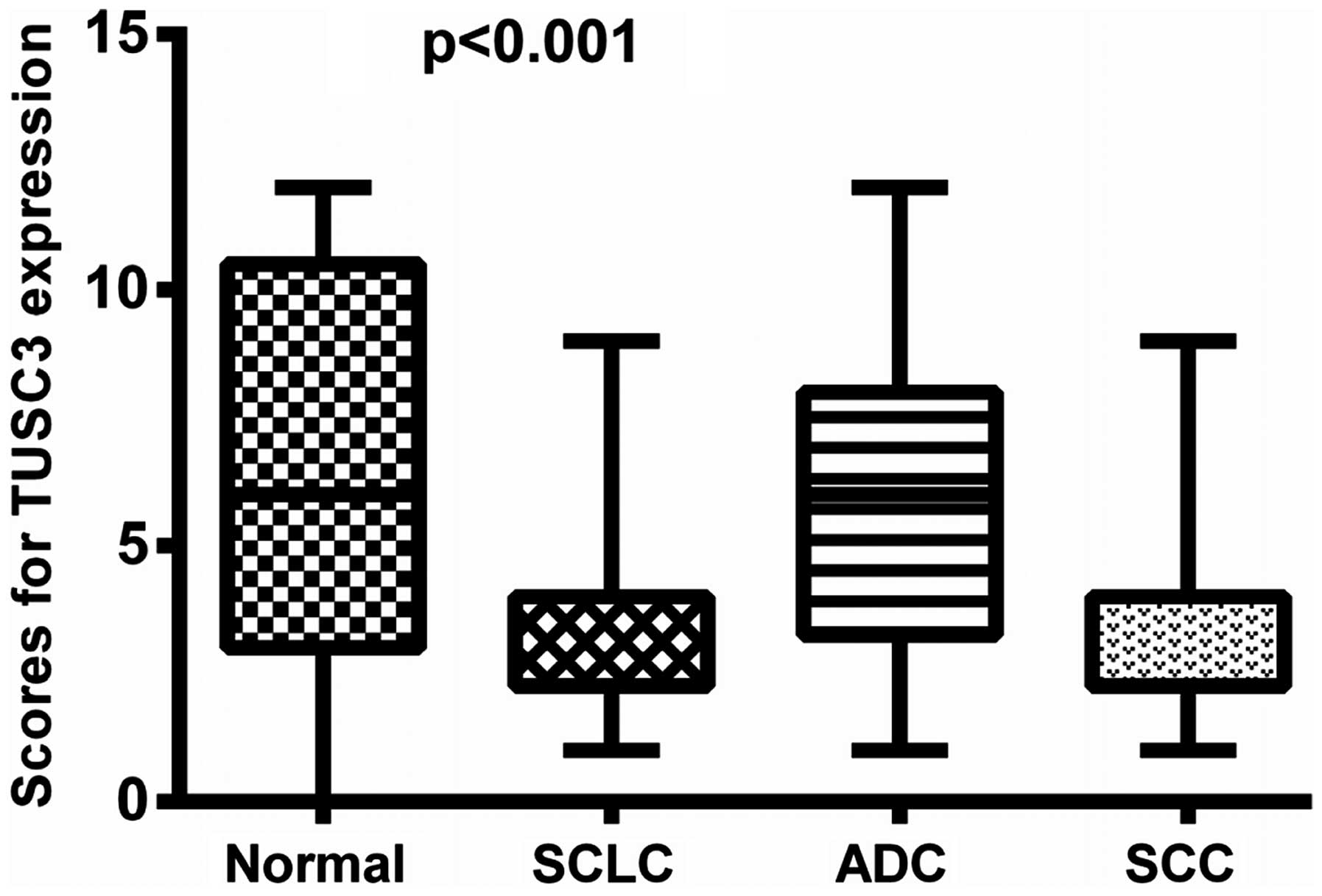
classification, i.e., the SCLC, SCC, ADC and normal control group,
differences in TUSC3 expression were identified among multiple
groups (normal vs. SCLC, P=0.0005; normal vs. ADC, P=0.3250; normal
vs. SCC, P=0.0546; SCLC vs. ADC, P=0.0012; SCLC vs. SCC, P=0.0636;
SCC vs. ADC, P=0.6490) (rank sum; Fig.2). Moreover, the positive rate of TUSC3
expression in the SCLC patients was significantly lower than that
in the normal controls (χ2=11.642, P=0.001; Table III). However, the positive rate of
TUSC3 expression showed no significant difference between the
normal controls and the ADC patients (χ2=2.499, P=0.114;
Table IV), or between the controls
and the SCC patients (χ2=0.255, P=0.614; Table V). Similarly, no significant
difference in the positive rate of TUSC3 was observed between the
ADC and SCC groups (χ2=3.602, P=0.058; Table VI).
 | Table II.Comparison of TUSC3 expression
between normal controls and lung cancer patients. |
Table II.
Comparison of TUSC3 expression
between normal controls and lung cancer patients.
|
|
| TUSC3 expression,
n |
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| Patient type | Number | + | − | Positive rate,
% | χ2 |
P-valuea |
|---|
| Normal control | 37 | 22 | 15 | 59.5 | 0.238 | 0.123 |
| Lung
cancerb | 195 | 89 | 106 | 45.6 |
|
|
 | Table III.Comparison of TUSC3 expression
between normal controls and SCLC patients. |
Table III.
Comparison of TUSC3 expression
between normal controls and SCLC patients.
|
|
| TUSC3 expression,
n |
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| Patient type | Number | + | − | Positive rate,
% | χ2 |
P-valuea |
|---|
| Normal | 37 | 22 | 15 | 59.5 | 11.642 | 0.001 |
| SCLC | 35 | 7 | 28 | 20.0 |
|
|
 | Table IV.Comparison of TUSC3 expression
between normal controls and ADC patients. |
Table IV.
Comparison of TUSC3 expression
between normal controls and ADC patients.
|
|
| TUSC3 expression,
n |
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| Patient type | Number | + | − | Positive rate,
% | χ2 |
P-valuea |
|---|
| Normal | 37 | 22 | 15 | 59.5 | 2.499 | 0.114 |
| ADC | 80 | 47 | 33 | 58.8 |
|
|
 | Table V.Comparison of TUSC3 expression
between normal controls and SCC patients. |
Table V.
Comparison of TUSC3 expression
between normal controls and SCC patients.
|
|
| TUSC3 expression,
n |
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| Patient type | Number | + | − | Positive rate,
% | χ2 |
P-valuea |
|---|
| Normal | 37 | 22 | 15 | 59.5 | 0.255 | 0.614 |
| SCC | 80 | 35 | 45 | 43.8 |
|
|
 | Table VI.Comparison of TUSC3 expression
between ADC and SCC patients. |
Table VI.
Comparison of TUSC3 expression
between ADC and SCC patients.
|
|
| TUSC3 expression,
n |
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| Patient type | Number | + | − | Positive rate,
% | χ2 |
P-valuea |
|---|
| ADC | 80 | 47 | 33 | 58.8 | 3.602 | 0.058 |
| SCC | 80 | 35 | 45 | 43.8 |
|
|
Association between TUSC3 expression
and the TNM staging of lung cancer patients
As shown in Fig. 3,
there were significant differences in TUSC3 expression among the
patients with different TNM staging (stages I–IV) in the SCLC
(P=0.0006; Fig. 3A) and ADC
(P<0.0001; Fig. 3D) patients, but
not in the SCC patients (P=0.216; Fig.
3H). Additionally, significant differences in TUSC3 expression
were identified among the patients with different N stages in the
SCLC (P=0.0012; Fig. 3C), ADC
(P<0.0001; Fig. 3F) and SCC
patients (P<0.0001; Fig. 3J).
However, no such differences were found among the patients with
different T stages in the SCLC (P=0.208; Fig. 3B), ADC (P=0.208; Fig. 3E) and SCC patients (P=0.641; Fig. 3I).
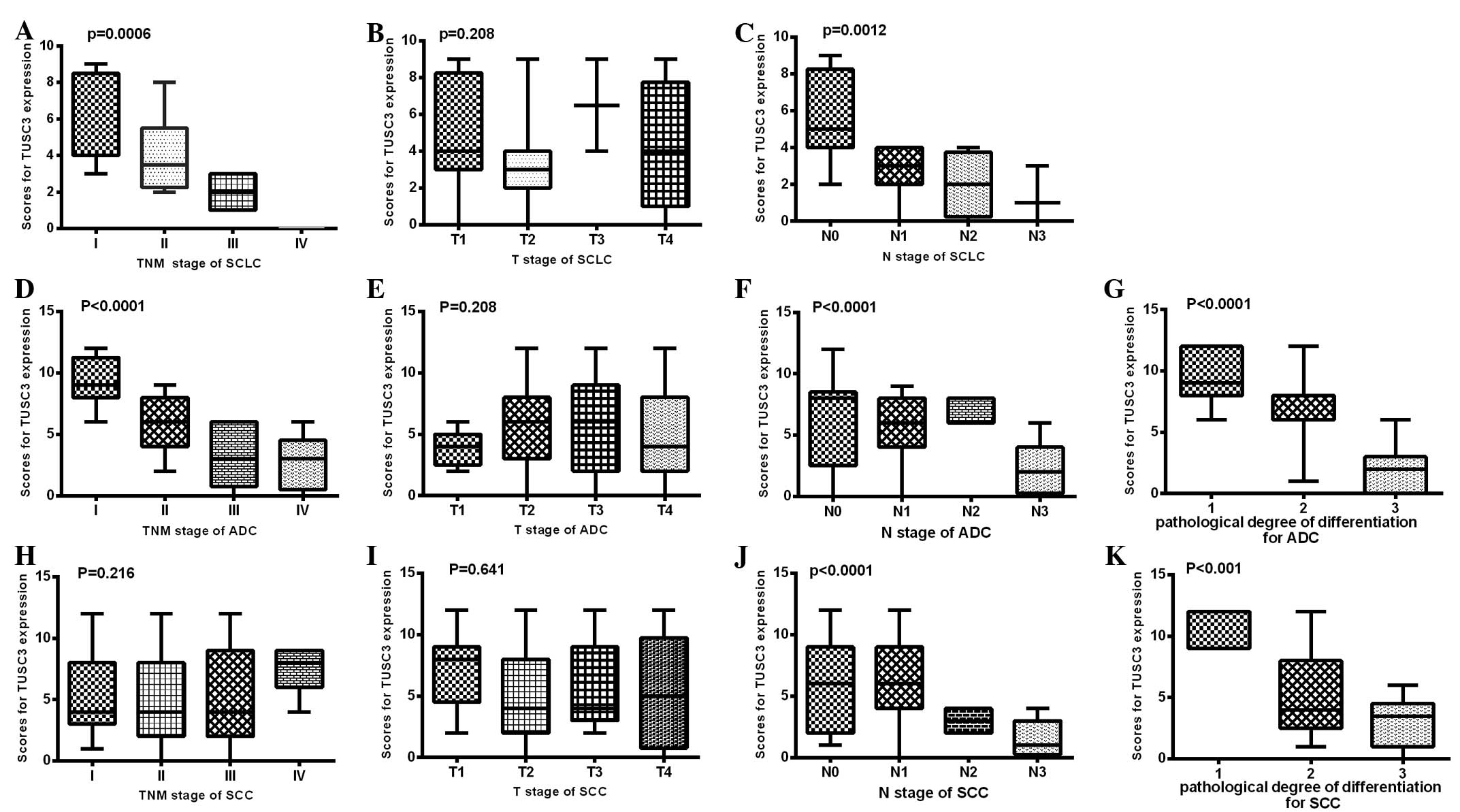 | Figure 3.TUSC3 expression was compared among
different clinical TNM stages and pathological degrees of
differentiation in three types of lung cancer patients. In (A-C)
SCLC, (D-G) ADC and (H-K) SCC patients, TUSC3 expression was
compared among different (A, D and E) clinical TNM stages, (B, E
and I) T stages, (C, F and J) N stages, and (G and K) pathological
degrees of differentiation. The rank sum test was used to analyze
the differences between groups. SCLC, small cell lung cancer; ADC,
adenocarcinoma; SCC, squamous cell carcinoma; TUSC3, tumor
suppressor candidate 3. |
Further analysis showed that TUSC3 expression was
negatively correlated with clinical TNM staging in the SCLC
(P<0.0001; rs=0.463; Fig. 4A) and
ADC (P<0.0001; rs=0.5245; Fig. 4D)
patients, but not in the SCC patients (P=0.173; rs=0.024; Fig. 4H). Additionally, TUSC3 expression was
negatively correlated with N stage in the SCLC (P<0.0001;
rs=0.3835; Fig. 4C), ADC (P=0.0002;
rs=0.167; Fig. 4F) and SCC (P=0.002;
rs=0.119; Fig. 4J) patients. However,
there were no correlations between TUSC3 expression and T stage in
the SCLC (P=0.857; rs=0.00099; Fig.
4B), ADC (P=0.903; rs=0.00019; Fig.
4E) or SCC (P=0.678; rs=0.002; Fig.
4I) patients.
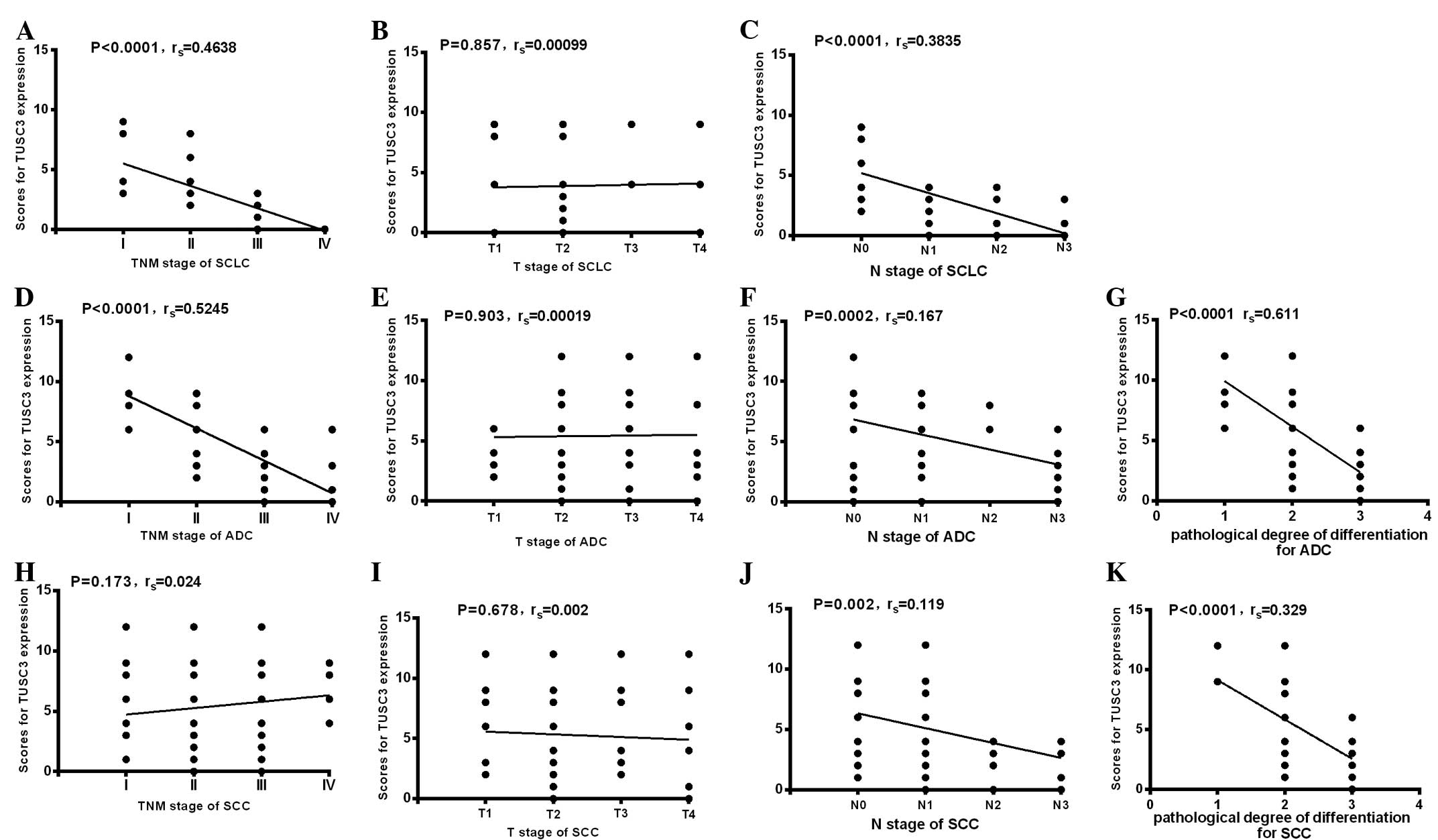 | Figure 4.Correlation between TUSC3 expression
and different clinical TNM stages or pathological degrees of
differentiation in three types of lung cancer patients. In (A-C)
SCLC, (D-G) ADC and (H-K) SCC patients, correlations between TUSC3
expression and (A, D and H) clinical TNM, (B, E and I) T stage, (C,
F and J) N stage and (G and K) degree of differentiation were
analyzed. Spearman's correlation method was used to evaluate the
association of scores. SCLC, small cell lung cancer; ADC,
adenocarcinoma; SCC, squamous cell carcinoma; TUSC3, tumor
suppressor candidate 3. |
Association between TUSC3 expression
and the pathological differentiation of lung cancer patients
The analysis revealed significant differences
between TUSC3 expression and pathological differentiation in ADC
(P<0.0001; Fig. 3G) and SCC
(P<0.001; Fig. 3K) patients.
Moreover, correlation analysis showed that there were negative
correlations between TUSC3 expression and pathological
differentiation in the ADC (P<0.0001; rs=0.611; Fig. 4G) and SCC (P<0.0001; rs=0.3289;
Fig. 4K) patients. As all SCLC
patients presented with a pathological degree of differentiation of
4 (undifferentiated), the correlation between TUSC3 expression and
pathological degree of differentiation was not analyzed in the SCLC
patients.
Discussion
Lung cancer presents with a high mortality rate due
mainly to its late diagnosis (17,18). In
total, >75% of lung cancer cases are diagnosed once the disease
has become locally advanced or metastatic, resulting in a current
5-year survival rate of <15% (17). Therefore, there is an urgent
requirement for reliable predictors and indicators of diagnosis and
prognosis for lung cancer.
TUSC3, a subunit of the human endoplasmic
reticulum-bound OST complex, has been recognized as a candidate
tumor suppressor gene involved in the tumor development of multiple
tumors. Loss of TUSC3 slows glycoprotein folding, and induces
proliferation, migration and invasion of cancerous cells during
tumor progression (13,16). The clinical significance of TUSC3
expression levels have been determined in several human tumors.
Guervós et al (13) found that
TUSC3 plays a role in metastasis in larynx and pharynx squamous
cell carcinomas, and that the loss of TUSC3 is negatively
correlated with LNM and survival rate. Pils et al (14) found that TUSC3 loss may facilitate
tumor growth. Reconstitution of TUSC3 in vitro decreases
proliferation and the binding of cancer cells to the extracellular
matrix. Therefore, TUSC3 represents a potential predictive factor
for survival. Khalid et al (15) found that TUSC3 is involved in
testicular spermatogenesis, and that it acts in the normal
development of the prostate and in the suppression of tumors. Horak
et al (16) found that TUSC3
expression is frequently lost in prostate cancer cell lines,
leading to the increased proliferation, migration and invasion of
cancer cells. However, the significance of TUSC3 expressions in
lung cancer patients has not yet been reported. To the best of our
knowledge, the present study is the first to analyze the
association between TUSC3 expression and the clinicopathological
parameters of lung cancer.
The study showed there was no significant
differences between normal controls and lung cancer patients in
terms of TUSC3 expression rate (χ2=0.238, P=0.123;
Table II). The results may be due to
the heterogeneity of different types of lung cancer. Therefore, the
TUSC3 expression rate was analyzed in SCLC, ADC and SCC patients,
respectively. The analysis showed decreased expression of TUSC3 in
the SCLC patients compared with the normal controls (P=0.001;
Table III). However, no difference
in TUSC3 expression was identified between the normal controls and
the ADC patients (χ2=2.499, P=0.114; Table IV), or between the controls and the
SCC patients (χ2=0.255, P=0.614; Table V). When ADC and SCC were considered
together as NSCLC according to the histological type, TUSC3
expression in the SCLC patients was significantly lower than that
in the NSCLC patients (P=0.001; Table
I). The aforementioned results indicated that decreased TUSC3
expression may play a more significant role in the tumorigenesis of
SCLC than in that of ADC and SCC. However, a larger sample size
will be used in an upcoming study of lung cancer patients, with a
focus on ADC and SCC patients.
Additionally, the association between TUSC3
expression and the pathological degree of differentiation was
analyzed. The TUSC3 expression in the patients with a pathological
differentiation degree of 1–2 was significantly higher than that in
the patients with a differentiation degree of 3–4 (P<0.001;
Table I). Further analysis showed
that TUSC3 expression levels were negatively correlated with the
pathological degree of differentiation in ADC (P<0.001;
rs=0.611; Fig. 4G) and SCC
(P<0.001; rs=0.3289; Fig. 4K)
patients. The correlation between TUSC3 expression and pathological
degree of differentiation could not be analyzed in the SCLC
patients, as all SCLC patients presented with an differentiation
degree of 4 (undifferentiated). Together, these results indicated
that decreased TUSC3 expression levels may be associated with a
poorly-differentiated tumor grade.
The correlations between TUSC3 expression and
clinical TNM staging were evaluated. Significant differences in
TUSC3 expression were identified among patients with different TNM
stages (stage I–IV) in the SCLC (P=0.0006; Fig. 3A) and ADC (P<0.0001; Fig. 3D) patients, but not in the SCC
patients (P=0.216; Fig. 3H). Further
correlation analysis also confirmed these results (Fig. 4A, D and H). It is known that TNM
staging is an index to reflect tumor progression in clinical
practice (2). TUSC3 expression may
similarly be a useful predictor of the progression of SCLC and ADC.
However, this conclusion cannot be obtained in SCC patients. Our
further study will verify the aforementioned results using a larger
sample size.
Notably, in the present study, the analysis revealed
a marked decrease in TUSC3 expression in patients who were
LNM+ compared with the expression level in patients who
were LNM− (P=0.011; Table
I). Furthermore, significant differences in TUSC3 expression
were identified among the different N stages (LNM status) in SCLC
(P=0.0012; Fig. 3C), ADC
(P<0.0001; Fig. 3F) and SCC
(P<0.0001; Fig. 3J) patients.
Similarly, correlation analysis also identified a negative
correlation between TUSC3 expression and LNM in all the three
pathological types of lung cancer tested (Fig. 4C, F and J). The results suggested that
lower TUSC3 expression may indicate a higher probability of LNM in
lung cancer patients. These results have significance in clinical
practice. For an individual patient, a combined analysis of TUSC3
expression and the clinical variables will assist in predicting the
incidence of LNM.
In conclusion, the present findings provide the
first evidence that a loss or reduction of TUSC3 may be associated
with a poorly-differentiated grade of lung cancer. Notably, TUSC3
may be a novel predictor of LNM in lung cancer patients.
Acknowledgements
This study was supported by grants from the Natural
Science Foundation of Shandong Province (grant nos. ZR2011HQ010 and
ZR2015HM077), the Technology Development Plan of Shandong Province
(grant no. 2015WSB04012) and the National Natural Science
Foundation of China (grant no. 30901712).
Glossary
Abbreviations
Abbreviations:
|
TUSC3
|
tumor suppressor candidate 3
|
|
IHC
|
immunohistochemistry
|
|
SCLC
|
small cell lung cancer
|
|
SCC
|
squamous lung cancer
|
|
ADC
|
adenocarcinoma lung cancer
|
|
NSCLC
|
non-SCLC
|
References
|
1
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. N Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ettinger DS, Akerley W, Borghaei H, Chang
AC, Cheney RT, Chirieac LR, D'Amico TA, Demmy TL, Govindan R,
Grannis FW Jr, et al: Non-small cell lung cancer. Version 2.2013. J
Natl Compr Canc Netw. 11:645–653; quiz 653. 2013.PubMed/NCBI
|
|
3
|
Kumar V, Abbas AK and Aster JC: Robbins
basic pathology (9th). Elsevier. Saunders, PA: 505. 2013.
|
|
4
|
Rosti G, Bevilacqua G, Bidoli P, Portalone
L, Santo A and Genestreti G: Small cell lung cancer. Ann Oncol.
17:(Suppl 2). ii5–ii10. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chute JP, Chen T, Feigal E, Simon R and
Johnson BE: Twenty years of phase III trails for patients with
extensive-stage small-cell lung cancer: Perceptible progress. J
Clin Oncol. 17:1794–1801. 1999.PubMed/NCBI
|
|
6
|
Loddo S, Parisi V, Doccini V, Filippi T,
Bernardini L, Brovedani P, Ricci F, Novelli A and Battaglia A:
Homozygous deletion in TUSC3 causing syndromic intellectual
disability: A new patient. Am J Med Genet A 161A. 2084–2087. 2013.
View Article : Google Scholar
|
|
7
|
Garshasbi M, Kahrizi K, Hosseini M, Vahid
L Nouri, Falah M, Hemmati S, Hu H, Tzschach A, Ropers HH, Najmabadi
H and Kuss AW: A novel nonsense mutation in TUSC3 is responsible
for non-syndromic autosomal recessive mental retardation in a
consanguineous Iranian family. Am J Med Genet A 155A. 1976–1980.
2011. View Article : Google Scholar
|
|
8
|
Garshasbi M, Hadavi V, Habibi H, Kahrizi
K, Kariminejad R, Behjati F, Tzschach A, Najmabadi H, Ropers HH and
Kuss AW: A defect in the TUSC3 gene is associated with autosomal
recessive mental retardation. Am J Hum Genet. 82:1158–1164. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bova GS, MacGrogan D, Levy A, Pin SS,
Bookstein R and Isaacs WB: Physical mapping of chromosome 8p22
markers and their homozygous deletion in a metastatic prostate
cancer. Genomics. 35:46–54. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
MacGrogan D, Levy A, Bova GS, Isaacs WB
and Bookstein R: Structure and methylation-associated silencing of
a gene within a homozygously deleted region of human chromosome
band 8p22. Genomics. 35:55–65. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Levy A, Dang UC and Bookstein R:
High-density screen of human tumor cell lines for homozygous
deletions of loci on chromosome arm 8p. Genes Chromosomes Cancer.
24:42–47. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mohorko E, Owen RL, Malojčić G, Brozzo MS,
Aebi M and Glockshuber R: Structural basis of substrate specificity
of human oligosaccharyl transferase subunit N33/Tusc3 and Its role
in regulating protein N-glycosylation. Structure. 22:590–601. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guervós MA, Marcos CA, Hermsen M, Nuño AS,
Suárez C and Llorente JL: Deletions of N33, STK11 and TP53 are
involved in the development of lymph node metastasis in larynx and
pharynx carcinomas. Cell Oncol. 29:327–334. 2007.PubMed/NCBI
|
|
14
|
Pils D, Horak P, Gleiss A, Sax C, Fabjani
G, Moebus VJ, Zielinski C, Reinthaller A, Zeillinger R and Krainer
M: Five genes from chromosomal band 8p22 are significantly
down-regulated in ovarian carcinoma: N33 and EFA6R have a potential
impact on overall survival. Cancer. 104:2417–2429. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Khalid AM, Asano A, Hosaka YZ, Takeuchi T
and Yamano Y: Tumor suppressor candidate TUSC3 expression during
rat testis maturation. Biosci Biotechnol Biochem. 77:2019–2024.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Horak P, Tomasich E, Vaňhara P,
Kratochvílová K, Anees M, Marhold M, Lemberger CE, Gerschpacher M,
Horvat R, Sibilia M, et al: TUSC3 loss alters the ER stress
response and accelerates prostate cancer growth in vivo. Sci Rep.
4:37392014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
National Lung Screening Trial Research
Team, ; Aberle DR, Berg CD, Black WC, Church TR, Fagerstrom RM,
Galen B, Gareen IF, Gatsonis C, Goldin J, et al: The national lung
screening trial: Overview and study design. Radiology. 258:243–253.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|