Introduction
Cyclophilins (Cyps) are a group of cellular proteins
known as immunophilins, which display the enzymatic activity of
peptidyl-prolyl isomerases (PPIases) (1). These enzymes catalyze the cis to trans
conversion of proline-containing peptides (1,2). CypA is a
key member of the Cyp family, and was first shown to mediate the
immunosuppressive function of cyclosporine (Cs) A through the
formation of a CsA-CypA complex (3).
As a multifunctional chaperone, CypA has been demonstrated to
participate in a range of cellular functions, including protein
folding, trafficking, immunomodulation and cell signaling (4). Previously, CypA was shown to participate
in numerous important processes, including exacerbation of
oxidative stress (5), inflammation
(5), pathogenesis of vascular
diseases (6), human immunodeficiency
virus infection (7) and rheumatoid
arthritis (8). Furthermore,
overexpression of CypA often correlates with severity of cancers,
including pancreatic and hepatic cancer, suggesting its role in
tumor malignancies (3).
Human hepatocellular carcinoma (HCC) is one of the
most common malignances in the world, with a particular high
prevalence in Asian and African countries (9). HCC is the third-leading cause of
cancer-associated mortalities, and the survival rate is 30–40% at 5
years post-surgery (10). Infection
with hepatitis B and C viruses is considered to account for >80%
of primary liver cancers (11,12). As an
essential host factor, CypA is critical for virus replication, and
may facilitate hepatotropic viral infections (13). In addition, CypA was initially noticed
to be upregulated in HCC, and may promote HCC metastasis through
the upregulation of matrix metalloproteinase (MMP)3 and MMP9
(14). To date, CypA has been
considered as a potential therapeutic target for molecular cancer
therapy, due to its important role in tumor formation and
metastasis (3). However, the
biological functions of CypA in HCC are far from being
understood.
The present study screened human fetal liver
complementary DNA (cDNA) for proteins interacting with CypA using
the yeast two-hybrid system. A nuclear protein,
serine/arginine-rich (SR)-25, was isolated as a novel protein that
is distinct from the CypA-binding proteins previously described in
the literature. Binding assays and co-immunoprecipitation were used
to confirm the physical association between CypA and SR-25.
Furthermore, the messenger RNA (mRNA) levels of SR-25 and CypA in
24 HCC cases were also evaluated in the present study.
Materials and methods
Isolation of interacting proteins
The Matchmaker LexA Two-Hybrid System (Clontech
Laboratories, Inc., Mountain View, CA, USA) was used to isolate the
CypA-interacting proteins following the manufacturer's protocol.
Saccharomyces cerevisiae EGY48 was transformed first with
the p8op-LacZ reporter plasmid, and then with pLexA-CypA. A single
colony grown in selective synthetic defined (SD) medium lacking
uracil and histidine (Ura−His−) was
transformed with the pB42AD activation domain plasmid (Clontech,
Inc.) of the human fetal liver cDNA library provided in the
Matchmaker LexA Two-Hybrid System. Interacting plasmids were
selected using
SD/galactose/raffinose/Ura−His−leucine−
medium and verified by sequencing.
Plasmid construction
For expression of CypA in the Escherichia
coli strain BL21 (Novagen, Inc., Madison, WI, USA), human CypA
cDNA (GenBank accession number NM_021130) was inserted in frame
into the pGEX6P-1 vector (GE Healthcare Life Sciences, Chalfont,
UK). The CypA PPIase mutation CypAm (R55A and F60A) was also
constructed as previously described (15).
To investigate their subcellular localization, human
CypA and SR-25 (GenBank accession number NM_016638) cDNAs were
introduced into the pCMV-Myc (Clontech Laboratories, Inc.) and
pCMV-Flag (Clontech Laboratories, Inc.) vectors, respectively.
Cell culture and transfection
Hep3B cells (American Type Culture Collection,
Manassas, VA, USA) were grown in RPMI 1640 medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
fetal calf serum (Thermo Fisher Scientific, Inc.). Cells
(3.5×105) were seeded in 60-mm dishes. Upon overnight
growth, cells were 80% confluent, and were transfected with 3 µg of
plasmid constructs using Lipofectamine Reagent (Thermo Fisher
Scientific, Inc.) in serum-free medium. After 5 h of incubation,
the medium was replaced with fresh complete medium, and cells were
cultured for an additional 48 h prior to collection.
Glutathione S-transferase (GST)-fusion
protein pull-down experiments
Hep3B cells that expressed Flag-SR-25 were harvested
and lysed in 500 µl lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM
NaCl, 5 mM EDTA, 0.5% NP-40, 1 mM PMSF and 10 µg/ml each aprotinin
and leupeptin). Cell lysates were centrifuged at 10,000 × g for 10
min at 4 °C. The expression and purification of GST fusion proteins
was conducted following the protocol provided by the manufacturer
of Glutathione Sepharose 4B (GE Healthcare Life Sciences). Purified
GST, GST-CypA or GST-CypAm (R55A and F60A) proteins were covalently
attached to the 50% slurry of Glutathione Sepharose 4B beads, and
then incubated with the whole-cell lysates of cells expressing
Flag-SR-25 at 4°C for 3 h. The beads were washed three times with
lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.5%
NP-40, 1 mM PMSF and 10 µg/ml each aprotinin and leupeptin), and
the bound proteins were analyzed by western blotting using an
anti-Flag monoclonal antibody (mAb; 1:1,000; F1804; Sigma-Aldrich;
Merck Millipore, Darmstadt, Germany) and an anti-GST mAb (1:1,000;
sc-33613; Santa Cruz Biotechnology, Inc., Dallas, TX, USA).
Immunoprecipitation
Cells lysates were pre-clarified with Protein A/G
Plus Agarose (Thermo Fisher Scientific, Inc.) by rotating at 4°C
for 30 min. Upon separation from the beads by centrifugation (at
1,000 × g 2 min and 4°C), the lysates were then immunoprecipitated
with ANTI-FLAG M2 Affinity Agarose Gel (Sigma-Aldrich) for 3 h at
4°C. The beads were washed four times with the aforementioned cell
lysis buffer and finally analyzed by western blotting using
anti-Flag mAb and anti-GST mAb.
Western blotting
Samples were separated by 10% SDS-PAGE, followed by
transfer to polyvinylidene difluoride membranes. Upon blocking with
PBS containing 5% bovine serum albumin (BSA; Sigma-Aldrich) and
0.1% Tween 20, the membrane was incubated with appropriate primary
antibodies at room temperature for 2 h, followed by incubation with
a peroxidase-conjugated goat anti-rabbit IgG or
peroxidase-conjugated goat anti-mouse IgG (both 1:5,000; ZB-2301;
ZB-2305; ZSGB-BIO, Beijing, China) at room temperature for 1 h. The
signals were detected using Western Blotting Luminol Reagent (Santa
Cruz Biotechnology, Inc.). The primary antibodies were anti-Flag
mAb (1:1,000; F1804; Sigma-Aldrich), anti-GST antibody (1:1,000;
sc-33613; Santa Cruz Biotechnology, Inc.) and anti-Myc antibody
(1:1,000; A7470; Sigma-Aldrich).
Immunofluorescence analysis
Hep3B cells grown on coverslips were fixed in 4%
paraformaldehyde for 10 min and washed once with TBS. Fixed cells
were permeabilized with 0.2% Triton X-100 for 5 min, washed three
times with TBS and incubated for 5 min in 0.1% sodium borohydride
(freshly prepared in TBS) to quench endogenous fluorescence. Upon
blocking with blocking buffer [1% horse serum (Thermo Fisher
Scientific, Inc.) 1% BSA, 0.02% NaN3 and 1X PBS] for 1
h, the cells were incubated with primary rabbit anti-Myc mAb
(1:500; A7470; Sigma-Aldrich) and mouse anti-Flag mAb (1:500;
F1804; Sigma Aldrich) at 4°C overnight. The coverslips were then
rinsed three times in PBS and reacted with fluorescein
isothiocyanate-conjugated goat anti-mouse IgG (green; 1:200;
ZF0312; ZSGB-BIO) and rhodamine-conjugated goat anti-rabbit IgG
(red; 1:200; ZF0316; ZSGB-BIO) secondary antibodies in the dark for
1 h. Subsequently, cells were counterstained with 1 µg/µl DAPI
(Shanghai Yeasen Biotechnology Co., Ltd., Shanghai, China) at 37°C
for 20 min and mounted on glass slides prior to visualization.
Images were recorded using a DC500 camera (Leica Microsystems,
Inc., Buffalo Grove, IL, USA) on a microscope equipped with DMRA2
fluorescence optics (Leica Microsystems, Inc.).
Tumor samples
A total of 24 pairs of primary HCC tissue samples
and adjacent tumor-free tissue samples were obtained at Yantai
Yuhuangding Hospital (Yantai, China). The samples were obtained
from patients during cytoreductive surgery between January 2013 and
May 2015. No patient was administered radiotherapy or chemotherapy
prior to surgery. All experimental procedures were approved by the
Ethics Committee of Yantai Yuhuangding Hospital. Informed consent
was obtained from all patients prior to the procedure.
Semi-quantitative reverse
transcription-polymerase chain reaction (RT-PCR) analysis
Total RNA was isolated from cultured cells or tissue
samples by a single-step isolation method using TRIzol reagent
(Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. Total RNA (2 µg) was reverse transcribed with ReverTra
Ace-α-® (Toyobo Co., Ltd., Osaka, Japan). PCR was
performed under the following conditions: A 5-min initial
denaturation step at 94°C, followed by 29 cycles of denaturation at
94°C for 30 sec, annealing at different temperatures for 30 sec and
extension at 72°C for 30 sec, and a final extension step at 72°C
for 10 min. The forward and reverse primers were selected to span
several introns to avoid genomic DNA amplification. Amplimer
contamination was controlled with a complete PCR reaction mixture
without cDNA. The primer sequences were as follows: CypA forward
(F), 5′-TACGGGTCCTGGCATCTT-3′ and reverse (R),
5′-CAGTCAGCAATGGTGATCTTCT-3′; SR-25 F, 5′-CCTCCTCTTCTTCCAGTTCTTC-3′
and R, 5′-ATTCGGGACTTCTGCTCATC-3′; β-macroglobulin (MG) F,
5′-ATGAGTATGCCTGCCGTGTGAAC-3′ and R, 5′-TGTGGAGCAACCTGCTCAGATAC-3′;
and GAPDH F, 5′-TGTGTCCGTCGTGGATCTGA-3′ and R,
5′-TTGCTGTTGAAGTCGCAGGAG-3′.
The semi-quantitative RT-PCR results were scanned
with Syngene G:BOX EF2 (Syngene, Frederick, MD, USA) and analyzed
using GeneTools image analysis software version 4.02 (Syngene),
according to Li et al (16).
The CypA and SR-25 mRNA level in cancer and normal tissues was
calculated using the dosage ratio (DR) of the ethidium bromide
intensity of the SR-25/β-MG and CypA/β-MG bands in a 1.5% agarose
gel.
Statistical analysis
The correlation between the scoring of SR-25 and
CypA expression levels was analyzed by the exact permutation test
for Spearman correlation coefficient. P<0.05 was considered to
indicate a statistical significant difference. All statistical
analyses were performed using SPSS version 19 (IBM SPSS, Armonk,
NY, USA).
Results
Screening of proteins that interact
with CypA
To identify the proteins that interact with CypA, a
total of 2×107 transformants were screened from a human
fetal liver cDNA library, and the initial screen revealed 43 cDNA
clones. By restriction mapping, nine of these candidate clones with
the same length of 1.2 kb were selected for further study. DNA
sequence analysis revealed that this set of cDNA molecules encoded
the human SR-25 protein. The binding specificity between CypA and
SR-25 was analyzed in yeast using pLexA-CypA and pB42-SR25 combined
with different control constructs, as shown in Table I. All yeast transformants in negative
control experiments either did not grow or became blue on
appropriate SD minimal medium, indicating a specific interaction
between CypA and SR-25 in yeast.
 | Table I.Interaction of CypA and SR-25 in the
yeast two-hybrid system. |
Table I.
Interaction of CypA and SR-25 in the
yeast two-hybrid system.
| Experiments | PADH1-BD
fusion | PGAL1-AD
fusion | SD minimal
medium | Colony
growth/color |
|---|
| Interaction
test | PlexA-CypA | pB42-SR-25 | SD-4a | Blue |
| Negative
control | PlexA-CypA | pB42 | SD-4a | No growth |
| Negative
control | PlexA-CypJ | pB42 | SD-4a | No growth |
| Negative
control | PlexA | pB42-SR-25 | SD-3b | White |
| Negative
control | PlexA | pB42 | SD-3b | White |
| Negative
control | PlexA | − | SD-2c | White |
| Negative
control | − | pB42 | SD-2c | White |
Interaction of SR-25 with CypA in
vitro
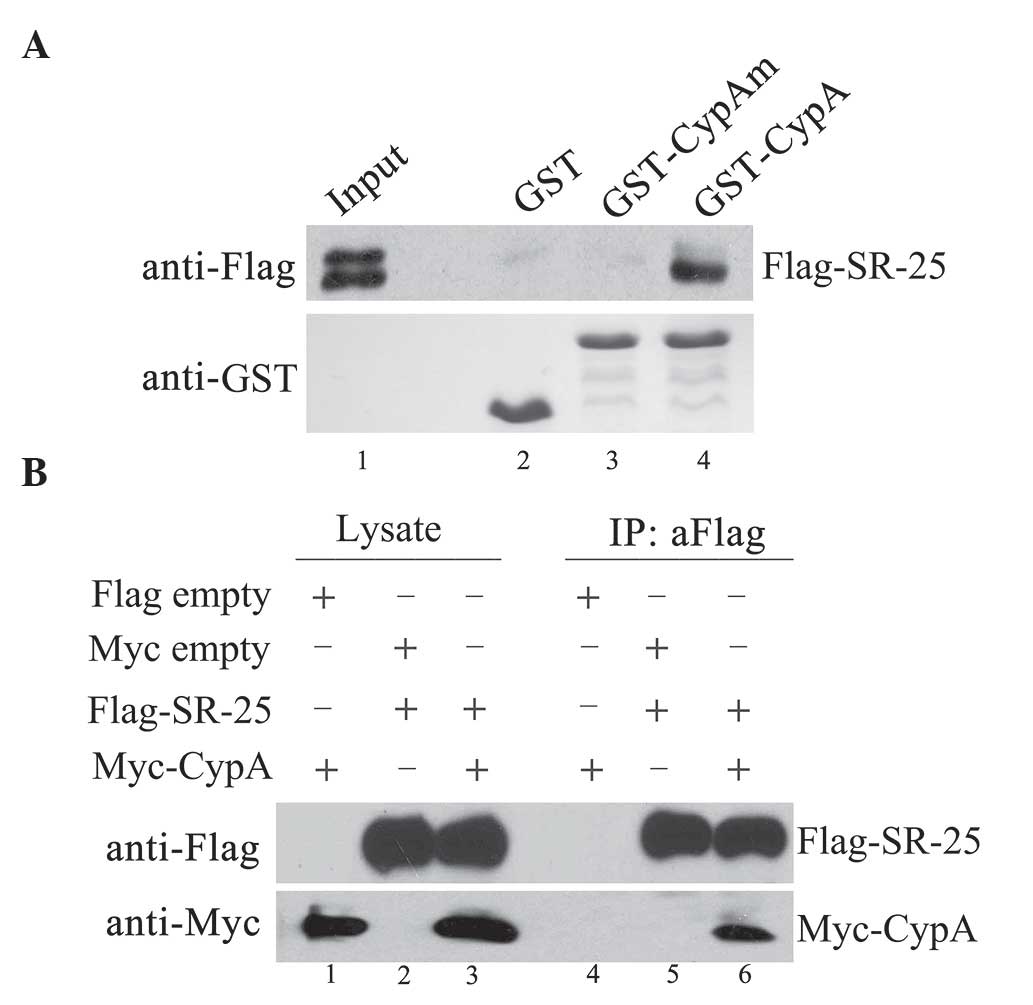
Using a GST-CypA fusion protein, in vitro GST
pull-down assays were performed. The results indicated that
Flag-SR-25 protein associated with GST-CypA fusion protein
(Fig. 1A, lane 4), but not with
control GST proteins (Fig. 1A, lane
2). In addition, the PPIase mutation of CypA (CypAm, R55A and F60A)
affected the interaction of CypA with SR-25, as no band was
detected when Flag-SR-25 was incubated with CypAm (Fig. 1A, lane 3).
Furthermore, the interaction of SR-25 with CypA in
Hep3B cells was also detected by immunoprecipitation experiments.
The results revealed that, when Flag-SR-25 was immunoprecipitated,
Myc-CypA co-precipitated with SR-25 (Fig.
1B, lane 6).
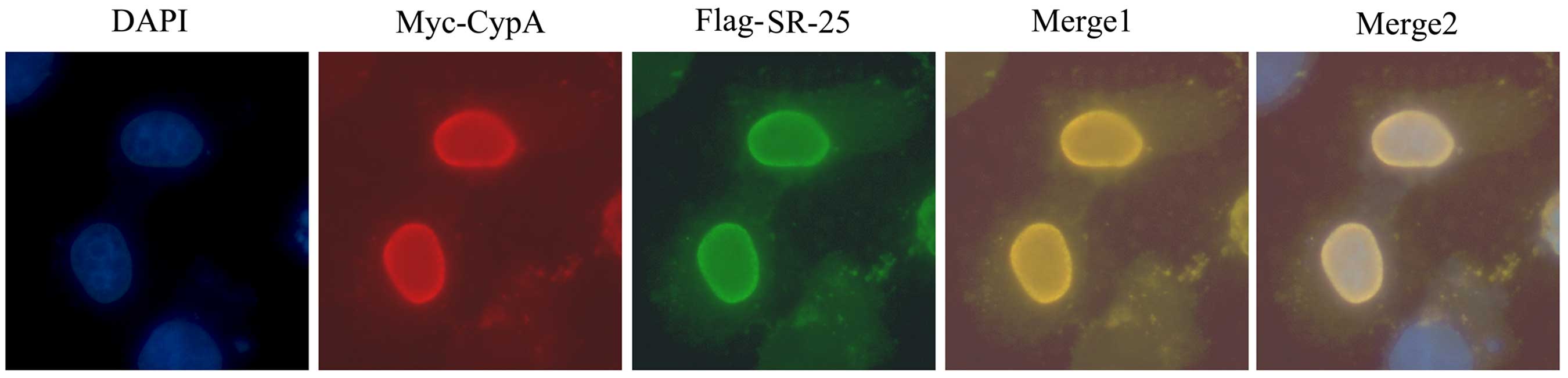
CypA and SR-25 co-localize at the cell
nucleus
Immunofluorescence microscopic examination revealed
that SR-25 and CypA co-localized mainly in the nuclei of
co-transfected Hep3B cells in a diffused pattern (Fig. 2), which supported the hypothesis that
SR-25 and CypA physically interact with each other.
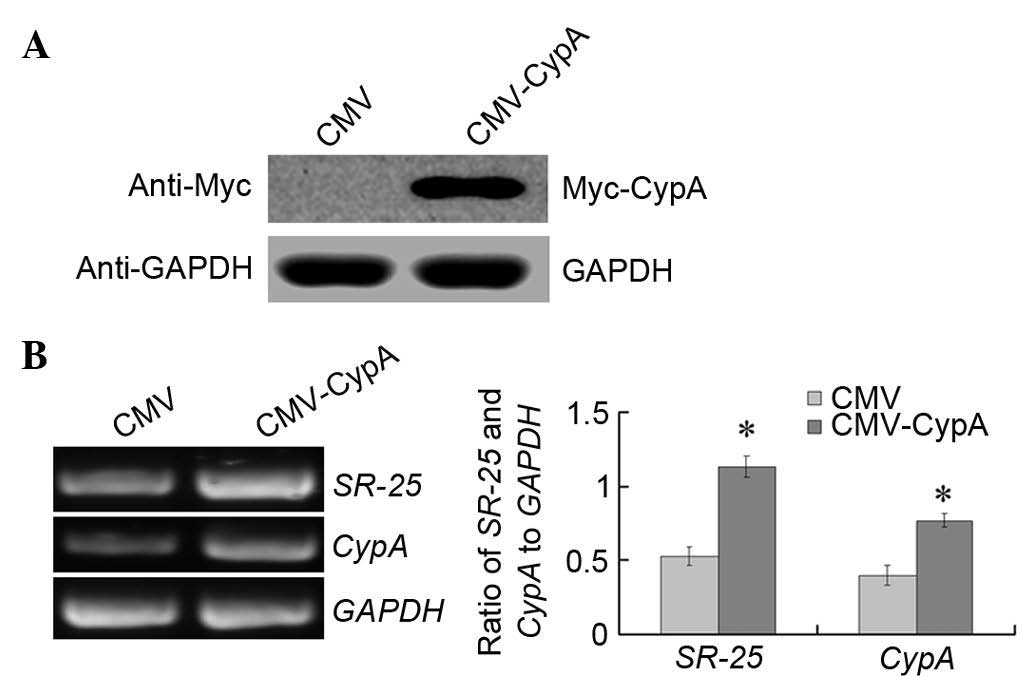
SR-25 is upregulated by CypA
overexpression in vitro
To detect if the expression of SR-25 was enhanced by
CypA overexpression, CypA was overexpressed in Hep3B cells
transiently transfected with the pCMV-CypA-Myc plasmid for 36 h.
Overexpression of CypA was confirmed at the protein and mRNA level
(Fig. 3). Semi-quantitative RT-PCR
analysis was conducted to determine the expression of SR-25. The
results indicated that the expression of SR-25 was upregulated by
>2-fold upon transient transfection of the cells with the
CypA-expression vector (Fig. 3B,
right panel) (P<0.001).
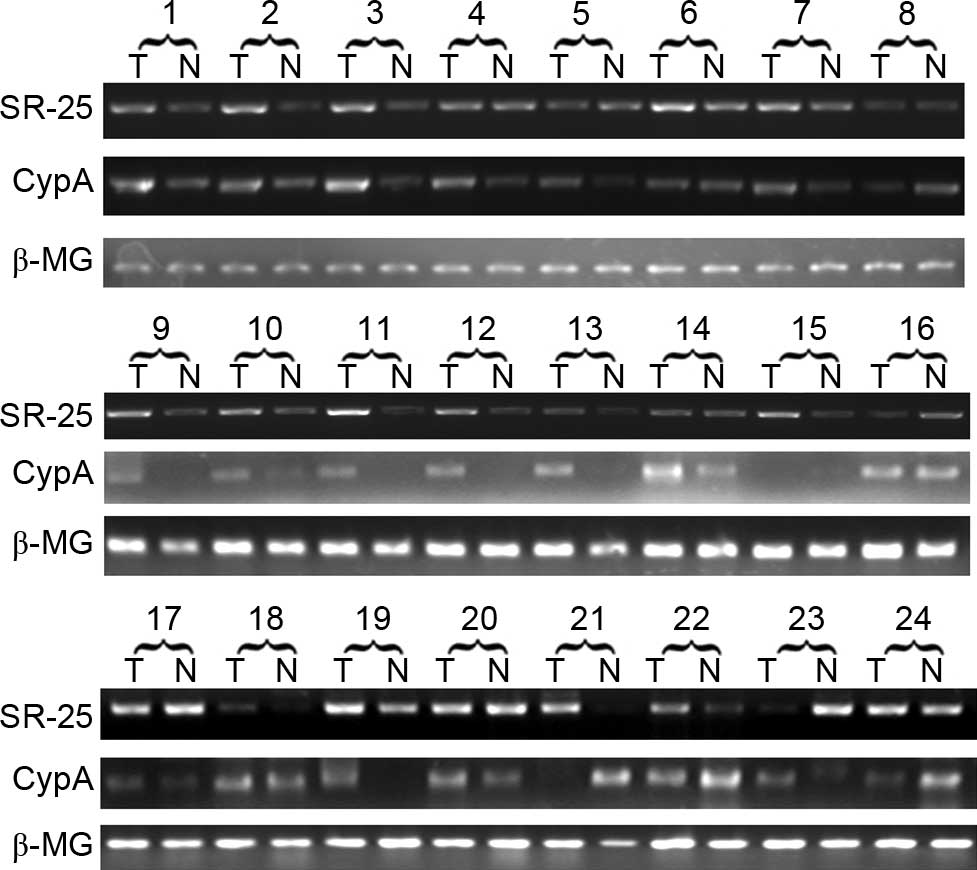
CypA and SR-25 exhibit elevated
expression in HCC tissues
The expression of CypA and SR-25 in 24 pairs of
HCC/adjacent non-cancerous tissues was determined and compared at
the mRNA level via semi-quantitative RT-PCR. The results revealed
that CypA and SR-25 shared a similar expression pattern in HCC
(Fig. 4). The DR of SR-25 and CypA in
each case was calculated, and the results are presented in Table II. SR-25 was upregulated in 33.3% of
the 24 HCC cases (8/24), while CypA was upregulated in 50% of cases
(12/24).
 | Table II.Characteristics of HCC patients. |
Table II.
Characteristics of HCC patients.
| N | Age (years) | Gender | Family history | AFP (ng/ml) | HbsAg (+/−) | Tumor (n) | Size | HCC Edmondson
grade | Fibrous capsule
(+/−) | Cancer embolus
(+/−) | SR-25a | CypAa |
|---|
| 1 | 34 | M | + | − | + | Sol | S | II | + | − | → | ↑ |
| 2 | 66 | M | + | − | + | Sol | S | II–III | + | − | → | → |
| 3 | 71 | M | − | − | + | Sol | S | I | − | − | ↑ | ↑ |
| 4 | 55 | F | − | − | + | Sol | S | I | − | − | → | ↑ |
| 5 | 63 | M | + | − | + | Sol | S | II–III | + | − | → | ↑ |
| 6 | 49 | M | − | + | + | Sol | L | III | − | − | → | → |
| 7 | 52 | M | − | − | + | Sol | S | III | − | − | ↑ | ↑ |
| 8 | 61 | F | + | + | + | Sol | S | II | + | − | → | → |
| 9 | 58 | M | − | + | + | Sol | S | II–III | − | − | → | ↑ |
| 10 | 57 | M | + | + | + | Sol | L | III | + | − | → | ↑ |
| 11 | 62 | M | + | + | + | Sol | L | III | − | − | ↑ | ↑ |
| 12 | 54 | F | − | + | + | Sol | S | II | + | − | ↑ | ↑ |
| 13 | 50 | M | + | + | + | Sol | S | III | + | − | → | ↑ |
| 14 | 48 | M | + | − | + | Sol | S | III | − | − | → | ↑ |
| 15 | 38 | F | + | − | + | Sol | L | II | + | − | ↑ | → |
| 16 | 35 | M | − | − | + | Sol | S | II | + | − | → | → |
| 17 | 45 | M | + | + | + | Sol | S | III | + | − | → | → |
| 18 | 53 | M | − | − | + | Sol | L | II–III | + | − | ↑ | → |
| 19 | 40 | M | − | + | + | Sol | S | II | + | − | ↑ | ↑ |
| 20 | 65 | F | − | − | + | Sol | S | II | + | − | → | → |
| 21 | 51 | M | − | + | + | Sol | S | III | + | − | ↑ | ↓ |
| 22 | 82 | M | − | + | + | Sol | L | III | + | − | → | ↓ |
| 23 | 50 | M | − | + | + | Sol | L | III | + | + | ↑ | → |
| 24 | 53 | M | + | − | + | Sol | S | II | + | − | → | ↓ |
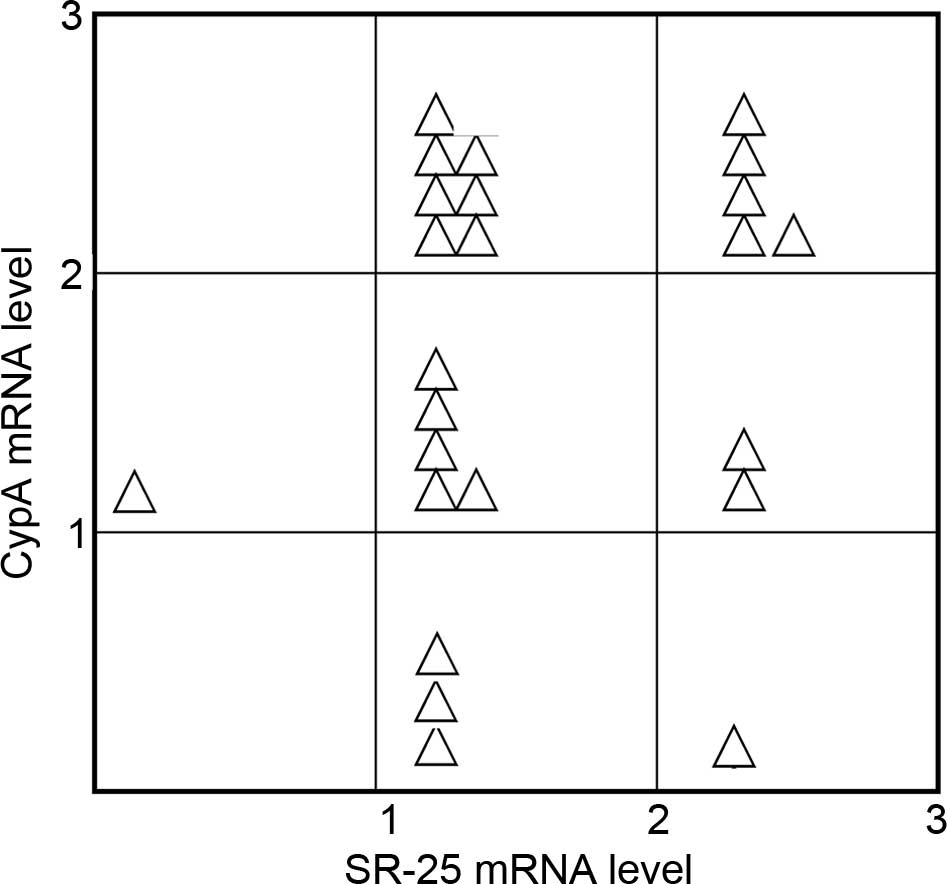
Statistical analysis demonstrated that there was a
significant positive correlation between SR-25 and CypA (Fig. 5). The Spearman correlation coefficient
between SR-25 and CypA was 0.4747 (P<0.05).
Discussion
Previous studies demonstrated that CypA is one of
the most abundant proteins in the cytoplasm, which accounts for
0.1% of total cytosolic proteins (17,18).
However, several previous immunolocalization studies reported that
CypA may also be present in the nucleus, and that CypA was
implicated in several cellular processes through interacting with
known nuclear proteins (19,20). Using the yeast two-hybrid system, the
present study isolated a novel CypA-binding nuclear protein, SR-25.
SR-25 is a novel member of a highly conserved family of splicing
factors, the SR proteins (21,22). SR
proteins are major modulators of alternative splicing, and usually
contain a SR domain that is required for protein-protein
interactions during splicing (23–25). It
has been suggested that SR proteins may be important in the
expression of specific disease phenotypes via alternative splicing
of disease-causing genes or mutation-triggered alternative splicing
events (26–28). Based on the presence of numerous
nuclear localizing signals and their similarity to RNA splicing
proteins, SR-25 was considered to contribute to RNA splicing
(29). In the present study, both the
binding assay and the co-immunoprecipitation assay confirmed the
physical association between CypA and SR-25. Furthermore, our study
revealed that the expression of SR-25 may be induced by CypA. It
may be speculated that the interaction between CypA and SR-25
proteins may be involved in potential carcinogenic functions of
CypA in HCC.
Various PPIases have been reported to interact with
transcription factors and to affect their activity (30,31). It
can be speculated that the PPIase activity may be involved in the
interaction between CypA and SR-25. SR proteins are
phosphoproteins, and their phosphorylation status can affect their
ability to interact with splicing complexes (21,32).
SR-25, as other SR proteins, is also rich in potential
phosphorylation residues and motifs (29). Previous studies demonstrated that a
specific conformation of the SR proteins is required for their
protein-protein interactions or for their
phosphorylation/dephosphorylation during the splicing cycle
(32). However, the highly repetitive
sequence composition and the presence of multiple proline residues
in the SR domains of SR proteins indicate that they are rather
unstructured (33). Therefore, SR
proteins require certain chaperones to mediate their conformational
changes in the spliceosome (33). As
a multifunctional chaperone, CypA could act specifically to alter
protein conformations (4). In
addition, the present study demonstrated that the disruption of the
PPIase domain affected the interaction of CypA with SR-25,
indicating that this interaction may depend on the PPIase
domain.
The present study revealed that CypA could induce
the expression of SR-25 when CypA was overexpressed in Hep3B cells.
Furthermore, the mRNA levels of CypA and SR-25 in HCC indicated
that the expression of CypA exhibited a significant correlation
with that of SR-25 in HCC tissue. A previous study indicated that
SR-25 is one of the mediators in the Ras-related C3 botulinum toxin
substrate (Rac)1 signaling pathway (34). Upregulated SR-25 in dominant negative
mutant of Rac1 (Rac1N17) cells reduces the apoptosis sensitivity
toward paclitaxel of melanoma cells, suggesting a role in the
regulation of apoptosis (34). As
CypA expression was also observed to be upregulated in
paclitaxel-resistant cancer cells (35), it can be speculated that the
interaction of CypA with SR-25 may participate in the regulation of
apoptosis in HCC. However, whether there is a signaling loop that
involves CypA, SR-25 and Rac1 in the regulation of apoptosis
requires further investigation.
In conclusion, for the first time, the present study
revealed a novel CypA-binding protein, SR-25. The present study
revealed that CypA could induce the expression of SR-25 in Hep3B
cells, and that this interaction may depend on the PPIase domain of
CypA. These results suggested that the interaction between CypA and
SR-25 proteins may participate in potential carcinogenic functions
of CypA in HCC. Additionally, there was a significant correlation
between the expression of CypA and that of SR-25 in HCC. Whether
there is a signaling loop that involves CypA, SR-25 and Rac1 in the
regulation of apoptosis in HCC requires further study. In addition,
the potential therapeutic value of these two proteins for HCC is
worth further investigation.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (Beijing, China; project
numbers 30801338 and 81071758), the Shandong Province Young and
Middle-Aged Scientists Research Awards Fund (Jinan, China; project
numbers BS2009SW052, 2008BSA02012 and ZR2015HQ031) and the Yantai
Science and Technology Program (Yantai, China; project numbers
2012085, 2009155-3 and 2015WS024).
References
|
1
|
Davis TL, Walker JR, Campagna-Slater V,
Finerty PJ, Paramanathan R, Bernstein G, MacKenzie F, Tempel W,
Ouyang H, Lee WH, et al: Structural and biochemical
characterization of the human cyclophilin family of peptidyl-prolyl
isomerases. PLoS Biol. 8:e10004392010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Naoumov NV: Cyclophilin inhibition as
potential therapy for liver diseases. J Hepatol. 61:1166–1174.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee J: Cyclophilin A as a new therapeutic
target for hepatitis C virus-induced hepatocellular carcinoma.
Korean J Physiol Pharmacol. 17:375–383. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nigro P, Pompilio G and Capogrossi MC:
Cyclophilin A: A key player for human disease. Cell Death Dis.
4:e8882013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Satoh K, Nigro P, Matoba T, O'Dell MR, Cui
Z, Shi X, Mohan A, Yan C, Abe J, Illig KA and Berk BC: Cyclophilin
A enhances vascular oxidative stress and the development of
angiotensin II-induced aortic aneurysms. Nat Med. 15:649–656. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Satoh K, Matoba T, Suzuki J, O'Dell MR,
Nigro P, Cui Z, Mohan A, Pan S, Li L, Jin ZG, et al: Cyclophilin A
mediates vascular remodeling by promoting inflammation and vascular
smooth muscle cell proliferation. Circulation. 117:3088–3098. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sokolskaja E and Luban J: Cyclophilin,
TRIM5, and innate immunity to HIV-1. Curr Opin Microbiol.
9:404–408. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang Y, Lu N, Zhou J, Chen ZN and Zhu P:
Cyclophilin A up-regulates MMP-9 expression and adhesion of
monocytes/macrophages via CD147 signalling pathway in rheumatoid
arthritis. Rheumatology (Oxford). 47:1299–1310. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hussain SA, Ferry DR, El-Gazzaz G, Mirza
DF, James ND, McMaster P and Kerr DJ: Hepatocellular carcinoma. Ann
Oncol. 12:161–172. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Aravalli RN, Steer CJ and Cressman EN:
Molecular mechanisms of hepatocellular carcinoma. Hepatology.
48:2047–2063. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
El-Serag HB: Epidemiology of viral
hepatitis and hepatocellular carcinoma. Gastroenterology.
142:1264–1273.e1. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bouchard MJ and Navas-Martin S: Hepatitis
B and C virus hepatocarcinogenesis: Lessons learned and future
challenges. Cancer Lett. 305:123–143. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Towers GJ, Hatziioannou T, Cowan S, Goff
SP, Luban J and Bieniasz PD: Cyclophilin A modulates the
sensitivity of HIV-1 to host restriction factors. Nat Med.
9:1138–1143. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang M, Dai C, Zhu H, Chen S, Wu Y, Li Q,
Zeng X, Wang W, Zuo J, Zhou M, et al: Cyclophilin A promotes human
hepatocellular carcinoma cell metastasis via regulation of MMP3 and
MMP9. Mol Cell Biochem. 357:387–395. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen S, Zhang M, Ma H, Saiyin H, Shen S,
Xi J, Wan B and Yu L: Oligo-microarray analysis reveals the role of
cyclophilin A in drug resistance. Cancer Chemother Pharmacol.
61:459–469. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li S, Li N, Zhu P, Wang Y, Tian Y and Wang
X: Decreased β-catenin expression in first-trimester villi and
decidua of patients with recurrent spontaneous abortion. J Obstet
Gynaecol Res. 41:904–911. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qu X, Wang C, Zhang J, Qie G and Zhou J:
The roles of CD147 and/or cyclophilin A in kidney diseases.
Mediators Inflamm. 2014:7286732014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Seizer P, Gawaz M and May AE: Cyclophilin
A and EMMPRIN (CD147) in cardiovascular diseases. Cardiovasc Res.
102:17–23. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Arevalo-Rodriguez M and Heitman J:
Cyclophilin A is localized to the nucleus and controls meiosis in
Saccharomyces cerevisiae. Eukaryot Cell. 4:17–29. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chiu R, Rey O, Zheng JQ, Twiss JL, Song J,
Pang S and Yokoyama KK: Effects of altered expression and
localization of cyclophilin A on differentiation of p19 embryonic
carcinoma cells. Cell Mol Neurobiol. 23:929–943. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lin S and Fu XD: SR proteins and related
factors in alternative splicing. Adv Exp Med Biol. 623:107–122.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Long JC and Caceres JF: The SR protein
family of splicing factors: Master regulators of gene expression.
Biochem J. 417:15–27. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hertel KJ and Graveley BR: RS domains
contact the pre-mRNA throughout spliceosome assembly. Trends
Biochem Sci. 30:115–118. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shen H, Kan JL and Green MR:
Arginine-serine-rich domains bound at splicing enhancers contact
the branchpoint to promote prespliceosome assembly. Mol Cell.
13:367–376. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bradley T, Cook ME and Blanchette M: SR
proteins control a complex network of RNA-processing events. RNA.
21:75–92. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guil S, Gattoni R, Carrascal M, Abián J,
Stévenin J and Bach-Elias M: Roles of hnRNP A1, SR proteins, and
p68 helicase in c-H-ras alternative splicing regulation. Mol Cell
Biol. 23:2927–2941. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ghigna C, Giordano S, Shen H, Benvenuto F,
Castiglioni F, Comoglio PM, Green MR, Riva S and Biamonti G: Cell
motility is controlled by SF2/ASF through alternative splicing of
the Ron protooncogene. Mol Cell. 20:881–890. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cartegni L, Hastings ML, Calarco JA, de
Stanchina E and Krainer AR: Determinants of exon 7 splicing in the
spinal muscular atrophy genes, SMN1 and SMN2. Am J Hum Genet.
78:63–77. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sasahara K, Yamaoka T, Moritani M, Tanaka
M, Iwahana H, Yoshimoto K, Miyagawa J, Kuroda Y and Itakura M:
Molecular cloning and expression analysis of a putative nuclear
protein, SR-25. Biochem Biophys Res Commun. 269:444–450. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bauer K, Kretzschmar AK, Cvijic H, Blumert
C, Löffler D, Brocke-Heidrich K, Schiene-Fischer C, Fischer G, Sinz
A, Clevenger CV and Horn F: Cyclophilins contribute to Stat3
signaling and survival of multiple myeloma cells. Oncogene.
28:2784–2795. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Horowitz DS, Lee EJ, Mabon SA and Misteli
T: A cyclophilin functions in pre-mRNA splicing. EMBO J.
21:470–480. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Keshwani MM, Aubol BE, Fattet L, Ma CT,
Qiu J, Jennings PA, Fu XD and Adams JA: Conserved proline-directed
phosphorylation regulates SR protein conformation and splicing
function. Biochem J. 466:311–322. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lorkovic ZJ, Lopato S, Pexa M, Lehner R
and Barta A: Interactions of Arabidopsis RS domain containing
cyclophilins with SR proteins and U1 and U11 small nuclear
ribonucleoprotein-specific proteins suggest their involvement in
pre-mRNA Splicing. J Biol Chem. 279:33890–33898. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee SC, Sim N, Clement MV, Yadav SK and
Pervaiz S: Dominant negative Rac1 attenuates paclitaxel-induced
apoptosis in human melanoma cells through upregulation of heat
shock protein 27: A functional proteomic analysis. Proteomics.
7:4112–4122. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li Z, Min W and Gou J: Knockdown of
cyclophilin A reverses paclitaxel resistance in human endometrial
cancer cells via suppression of MAPK kinase pathways. Cancer
Chemother Pharmacol. 72:1001–1011. 2013. View Article : Google Scholar : PubMed/NCBI
|