Introduction
Osteosarcoma (OS) is an aggressive disease, and is
the most common type of bone tumor in children and adolescents
globally (1). The tumor predominantly
occurs near regions that produce osteoid. The prognosis of patients
with OS is poor due to its high rate of metastasis and
chemoresistance (2). Approximately
one-third of patients diagnosed with localized OS experience
relapse or progressive illness, and the mean survival time of
patients with OS recurrence is <1 year (3). Prior to the emergence of effective
chemotherapy, 2-year overall survival rates following surgical
resection and/or radiotherapy were 15–20% (4,5). Further
advances in therapeutic strategies for OS, including wide tumor
excision, neoadjuvant and adjuvant chemotherapy, and radiotherapy,
have resulted in significantly improved clinical outcomes and
long-term survival rates (2,6–9). However,
>30% of patients with OS survive for <5 years and succumb due
to pulmonary metastases following diagnosis (10,11).
Therefore, it is critical to identify OS effector molecules, novel
therapeutic strategies, and signaling pathways regulating OS growth
and metastasis. Furthermore, there is an ongoing need for a more
thorough understanding of the genetic and molecular mechanisms of
OS development and metastasis.
Emerging evidence in the research area of
epigenetics suggests that OS is caused by the accumulation of
genetic and epigenetic alterations (12). MicroRNAs (miRNAs) are highly
conserved, noncoding, small (22 nucleotides in length) RNAs that
have a critical role in regulating post-transcriptional gene
expression (13,14). Aberrant miRNA expression affects the
initiation and progression of carcinogenesis, and is associates
with abnormal cell proliferation, differentiation, growth,
apoptosis and invasion (15). Recent
advances in epigenetics have revealed that miRNA dysregulation
frequently occurs in various types of neoplasm, including gastric,
colon, lung and prostate cancer, as well as OS (16–20). The
focus of a vast number of studies on determining the expression of
miRNAs in clinical samples has identified an association between
miRNA function and oncogenesis or tumor suppressor genes, which was
dependent on the function of the miRNA target genes (21,22).
In recent decades, the majority of studies have
focused on miRNA expression in cell lines and tissues; these miRNAs
have also been identified in circulation as diagnostic and
prognostic biomarkers (23). Further
studies have shown that the circulating miRNA profile reflects
disease states, particularly in cancer (24,25).
However, the association between miRNA profiles in circulation and
in tumor tissues remains unclear. In our previous study (20), it was found that miR-17 levels were
marginally elevated in OS tissues and cell lines. The study also
observed that downregulation of miR-17 expression in OS cells,
which leads to increased phosphatase and tensin homolog (PTEN)
expression, resulted in the suppression of OS cell proliferation,
migration and invasion. However, expression of miR-17 in the
circulation has not been predicted or experimentally confirmed, and
the expression levels and patterns of miR-17 in the circulation of
patients with OS have not been well established. Therefore, the
present study detected miR-17 expression in OS cell lines and serum
samples. miR-17 expression was upregulated in the serum and
positively correlated with expression levels in the OS specimens.
These results suggest that miR-17 may be a promising prognostic
biomarker of OS.
Materials and methods
Cell lines, blood samples and OS
specimens
Human osteoblast cell line hFOB 1.19, and OS cell
lines U2OS, Saos-2, MG-63 and MNNG/HOS were purchased from The Cell
Bank of Type Culture Collection of Chinese Academy of Sciences,
Shanghai Institute of Cell Biology, Chinese Academy of Sciences
(Shanghai, China). Human osteosarcoma cell lines MNNG/HOS, U2OS,
Saos-2 and MG-63 were cultured in minimal essential medium,
Dulbecco's modified Eagle's medium (DMEM), RPMI 1640 and DMEM,
respectively, supplemented with 10% fetal bovine serum (FBS), 100
mg/ml streptomycin and 100 IU/ml penicillin, at 37°C with 5%
CO2. The human osteoblast hFOB 1.19 cell line was
maintained in DMEM/F12 medium supplemented with 10% FBS, 100 mg/ml
streptomycin and 100 IU/ml penicillin, at 37°C with 5%
CO2. The cells were serum-starved for 12 h prior to each
experiment.
The present study included 46 patients with OS and
46 matched normal tumor-free patients treated in the Department of
Orthopedics, Union Hospital, Tongji Medical College, Huazhong
University of Science and Technology (Wuhan, China) between March
2008 and March 2011. The patient cohort consisted of 27 males and
19 females, with an age at diagnosis that ranged from 9 to 48 years
and a mean age of 19.6 years. Inclusion criteria were a diagnosis
of osteosarcoma based on the results of imaging studies and
biopsies, while exclusion criteria included the presence of any
other type of neoplasm, liver and kidney dysfunction, and a
post-operative pathological diagnosis of osteosarcoma. Peripheral
blood samples (15 ml) were obtained from the OS patients and
matched normal tumor-free patients. The samples were centrifuged at
978 × g for 10 min at room temperature, serum was extracted
and stored immediately in liquid nitrogen until use.
OS and matched normal non-tumor tissues (n=46) were
collected from the patients with OS. All the tissues were resected
at the time of surgery (including amputation and limb-sparing
surgery) and immediately stored in liquid nitrogen until use. The
present study was approved by the Ethics Committee of Union
Hospital, Tongji Medical College, Huazhong University of Science
and Technology.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total miRNA was obtained from all cultured cells,
serum samples and tissues using RNeasy Mini and miRNeasy Mini kits
(Qiagen, Inc., Valencia, CA, USA). miR-17 expression was quantified
by performing RT-qPCR using TaqMan MicroRNA Assay kits (catalog no.
217004; Applied Biosystems; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) on a LightCycler 480 System II (Roche
Diagnostics, Rotkreuz, Switzerland). The primer sequences were as
follows: Forward, 5′-CCAGGACCAGAGGAAACCT-3′ and reverse,
5′-GCTAGCCTCTGGATTTGA-3′ for PTEN; U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′;
miR-17 forward, 5′-TGCTTACAGTGCAGGTAG-3′ and reverse,
5′-GAACATGTCTGCGTATCTC-3′; and forward, 5′-ATGTCGTGGAGTCTACTGGC-3′
and reverse, 5′-TGACCTTGCCCACAGCCTTG-3′ for GAPDH. The PCR
conditions were as follows: 50°C for 2 min, 95°C for 10 min, 95°C
for 30 sec and 60°C for 30 sec, for 40 cycles. PTEN expression was
detected using SYBR Premix Ex Taq II (Takara Bio, Inc., Otsu,
Japan). The RT-qPCR data was evaluated using the
2−ΔΔCt method (26) relative to GAPDH for mRNA or U6 small
nuclear RNA for miRNA. Each PCR experiment was conducted in
triplicate.
Statistical analysis
Data are presented as mean ± standard deviation and
were analyzed using SPSS software (version 16.0; SPSS, Inc.,
Chicago, IL, USA). The correlation between miR-17/PTEN expression
levels and prognosis was analyzed by Kaplan-Meier survival analysis
using GraphPad Prism 6 software (GraphPad Software, Inc., San
Diego, CA, USA). Analysis of variance (followed by Gabriel's
procedure or a two-tailed Student's t-test were used to examine the
data between >2 or 2 variables, respectively, and Spearman's
correlation analysis was performed to evaluate the association
between miR-17 and PTEN expression levels. P<0.05 was used to
indicate a statistically significant difference.
Results
Expression of miR-17 is increased in
OS cell lines, serum samples and tissues
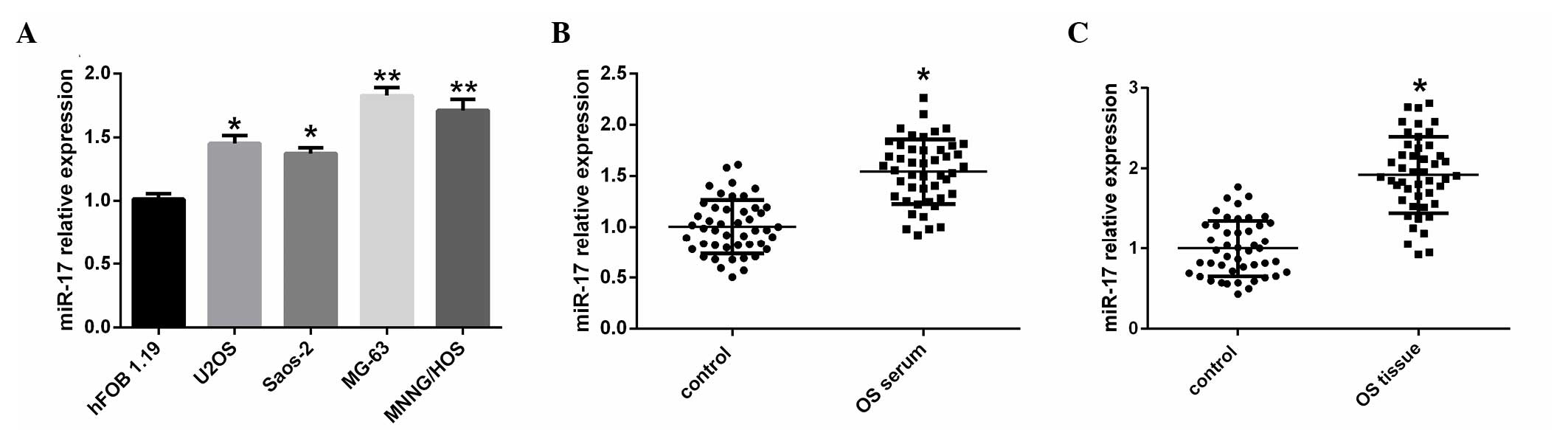
To investigate the potential significance of miR-17
in OS development and progression, the present study evaluated the
expression of miR-17 in a human osteoblast cell line, and OS cell
lines, serum samples and tissues using RT-qPCR. miR-17 expression
levels were significantly upregulated in all four OS cell lines,
U2OS, Saos-2 (P<0.05), MG-63 and MNNG/HOS (P<0.01), compared
with the hFOB 1.19 human osteoblast cell line (Fig. 1A). The expression of miR-17 was also
significantly increased in serum samples of patients with OS
compared with normal controls subjects (P<0.05; Fig. 1B). Furthermore, the expression of
miR-17 was significantly increased in OS tissues compared with
matched normal tissues (P<0.05; Fig.
1C).
Correlation between miR-17 and PTEN
expression in patients with OS
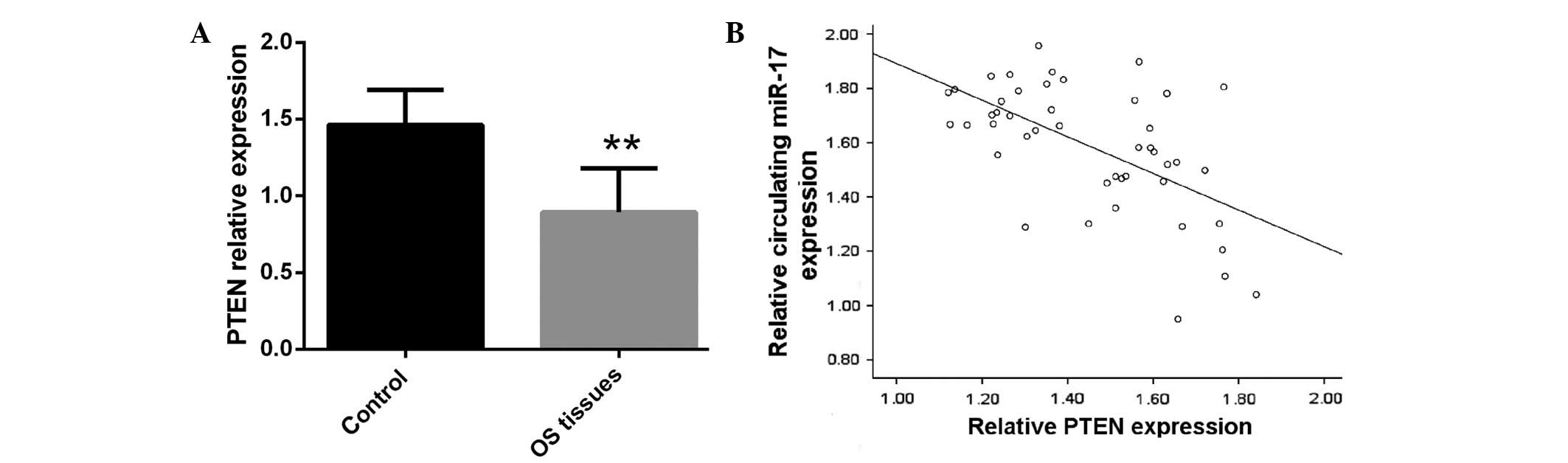
Previous studies demonstrated that PTEN is a target
gene of miR-17 and its expression appears to be inversely
correlated with miR-17 expression levels in OS tissues (27,28). In
the present study, PTEN expression was significantly decreased in
human OS tissues compared with in matched normal tissues
(P<0.01), indicating that downregulation of PTEN may have a
significant role in OS development and progression (Fig. 2A). To evaluate the association between
circulating miR-17 and PTEN expression levels, Spearman's
correlation analysis was performed, demonstrating that serum miR-17
expression is strongly inversely correlated with PTEN expression in
OS tissue (P=0.0002; Fig. 2B).
Upregulation of miR-17 expression and
downregulation of PTEN expression are correlated with poor survival
in patients with OS
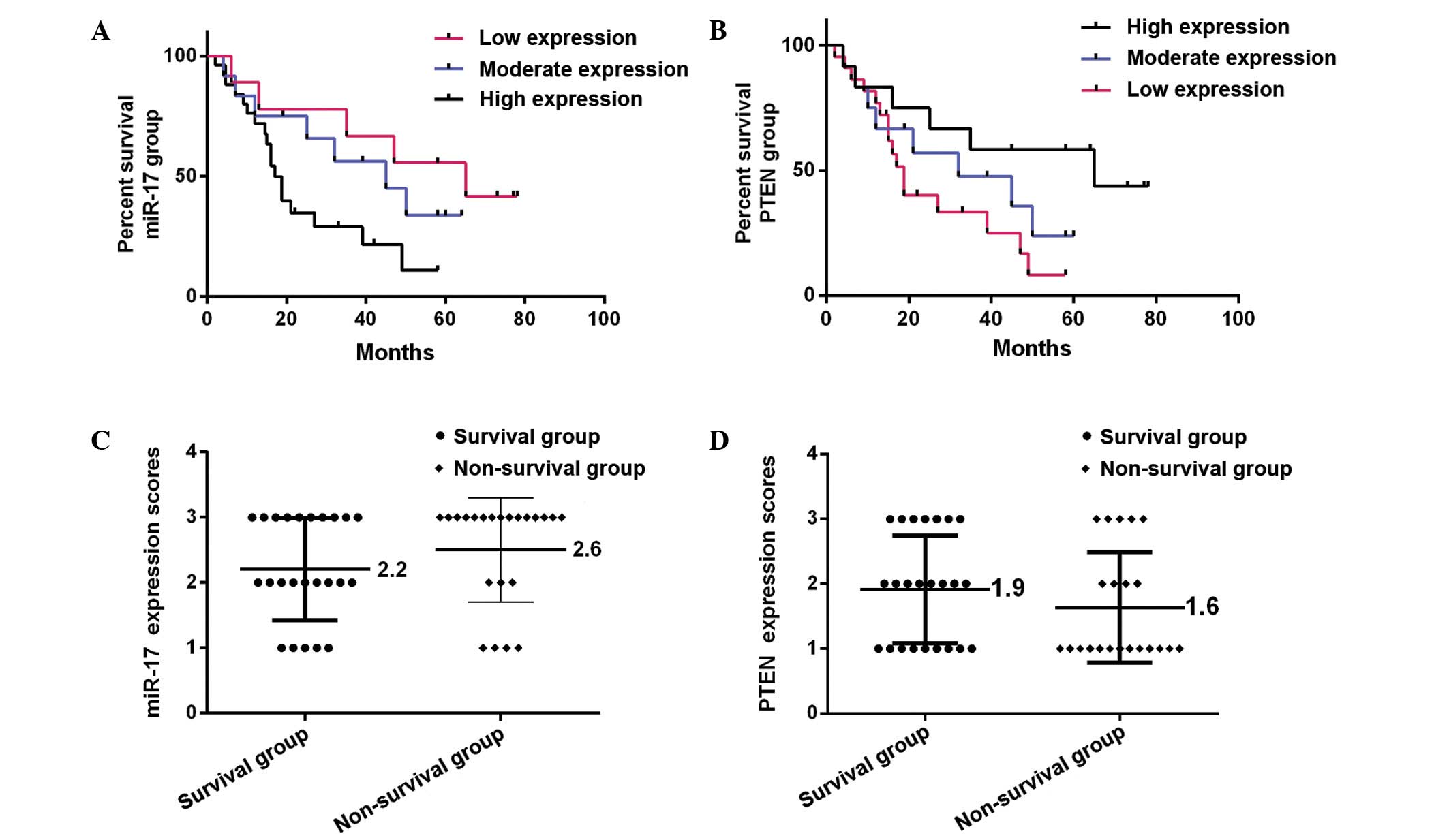
To further determine the potential roles of miR-17
in OS development and progression, the present study analyzed the
correlations between miR-17 expression in circulation and PTEN
expression in OS tissues, and clinical features of patients with
OS. The expression levels of miR-17 in serum and PTEN in OS tissues
were analyzed in the cohort of 46 patients with OS, and divided
into the following three groups: Low expression (score 1), moderate
expression (score 2) and high expression (score 3). The results
revealed that high expression of miR-17 in circulation was strongly
associated with poor prognosis in patients with OS, while
overexpression of PTEN in OS tissues was significantly correlated
with good prognosis in patients with OS (P<0.05; Table I). As indicated in Table I, the expression levels of miR-17 were
correlated with incidence at an early age (P>0.05).
 | Table I.Comparison of the clinical
characteristics of 46 OS patients with PTEN tissue expression and
circulating miR-17 expression. |
Table I.
Comparison of the clinical
characteristics of 46 OS patients with PTEN tissue expression and
circulating miR-17 expression.
|
| PTEN expression in OS
tissue | miR-17 expression in
serum |
|---|
|
|
|
|
|---|
| Characteristic | High | Moderate | Low | P-value | High | Moderate | Low | P-value |
|---|
| Total, n (%) | 12 (26.1) | 12 (26.1) | 22 (47.8) | 0.523 | 25 (54.3) | 12 (26.1) | 9 (19.6) | 0.546 |
| Age at diagnosis,
years |
|
|
Mean | 21.9 | 24.3 | 16.7 |
| 15.2 | 23.8 | 24.7 |
|
|
Range | 13–42 | 12–48 | 9–27 | 0.673 | 9–31 | 14–40 | 13–48 | 0.615 |
| Gender, n (%) |
|
|
Male | 7
(58.3) | 6
(50.0) | 14 (63.4) |
| 15 (60.0) | 7
(58.3) | 6 (66.7) |
|
|
Female | 5
(41.7) | 6
(50.0) | 8
(32.6) | 0.632 | 10 (40.0) | 5
(41.7) | 3 (33.3) | 0.754 |
| Prognosis, n
(%) |
|
|
Survived | 7
(58.3) | 8
(66.7) | 9
(40.9) | 0.045 | 10 (40.0) | 9
(75.0) | 5 (55.6) | 0.042 |
|
Deceased | 5
(41.7) | 4
(33.3) | 13 (59.1) | 0.032 | 15 (60.0) | 3
(25.0) | 4 (44.4) | 0.036 |
Furthermore, Kaplan-Meier analysis demonstrated that
the prognosis of patients with OS is significantly worse in those
with high serum miR-17 expression compared with those exhibiting
moderate and low miR-17 expression (P=0.0037; Fig. 3A). By contrast, patients with high
PTEN tissue expression exhibited a significantly better prognosis
than those with moderate and low PTEN expression (P=0.0187;
Fig. 3B). Furthermore, patients were
divided into two outcome groups: Survival group (survived for >5
years) and non-survival group (survived for <5 years). It was
observed that patients in the survival group (24 patients, 52.2%)
exhibited significantly lower miR-17 expression scores compared
with patients in the non-survival group (22 patients, 47.8%;
P=0.0026). By contrast, patients in the survival group had higher
PTEN expression scores than patients in the non-survival group
(P=0.0002). The mean miR-17 and PTEN expression score ranges in two
groups were 2.2–2.6 (Fig. 3C) and
1.9–1.6 (Fig. 3D), respectively.
Discussion
Emerging studies have indicated that miRNAs are
involved in several processes, including cell apoptosis,
proliferation, metastasis and differentiation, by regulating the
expression of multiple target genes (14,15).
Hence, exploring the profiles of miRNAs and their target genes
involved in tumorigenesis may promote understanding of the
underlying mechanisms of patients with human malignancies, and
provide valuable insight for the early diagnosis and treatment of
such diseases (20–22,24).
Results from previous studies have continually demonstrated that
the presence of miRNAs in the circulation and in bodily fluid may
function as promising diagnostic and prognostic biomarkers in
certain diseases, including human malignancies (23–25,29).
In the present study, the expression of miR-17 in
serum samples from patients with OS was compared with paired serum
samples of healthy control subjects. This analysis revealed that
the expression of miR-17 in circulation was significantly
upregulated in OS compared with the normal controls. The current
study also observed increased expression of miR-17 in OS tissues
compared with normal tissues. This is consistent with a previous
study, which identified that miR-17 expression is upregulated in OS
cell lines compared with a matched human osteoblast cell line
(20). Simiarly, Minami et al
demonstrated that the overexpression of miR-17 promotes cell growth
in synovial sarcoma (27). Our
results identified PTEN as a miR-17 target gene that affects OS
cell proliferation, colony formation, migration and invasion
(20). These results are consistent
with a previous report that PTEN is a direct target of miR-17 in
glioblastoma cells (30). Our
previous study demonstrated that downregulation of miR-17 leads to
the upregulation of PTEN expression in OS cells, which results in
the inhibition of OS cell growth and metastasis (20). Similarly, a previous study concerning
PTEN expression levels in OS tissues revealed a the positive
correlation between PTEN expression and the degree of
differentiation (31).
Initially identified in 1997, the PTEN protein is
considered an important tumor suppressor that inhibits tumor
development, invasion, angiogenesis and metastasis (32). PTEN predominantly functions as a tumor
suppressor gene through the phosphoinositide 3-kinase
(PI3K)/PTEN/AKT signaling pathway. It has been reported that the
PI3K/PTEN/AKT signaling pathway is frequently deregulated during
tumorigenesis (33). Considering the
importance of the PI3K/PTEN/AKT signaling pathway in regulating
numerous cellular behaviors, including cell proliferation and
survival, it is unsurprising that the PTEN gene undergoes
inactivation or loss of function in several types of human tumor
(34). As a tumor suppressor gene,
downregulation of PTEN in certain types of malignant cancer
activates AKT, promoting cell proliferation, survival, migration
and angiogenesis (35).
Immunohistochemical characterization of OS tumor tissues suggest
that PTEN expression is downregulated in a high percentage of OS
cases (36). Furthermore, the present
study results suggest that miR-17 modulates PTEN expression and
that miR-17 expression is downregulated in OS serum, which reveals
the involvement of miR-17 in the pathological process of OS.
In conclusion, the current results demonstrated that
downregulation of miR-17 decreases OS cell survival, and enhanced
miR-17 expression resulted in the downregulation of PTEN expression
in OS. Taken together, the current results suggest, for the first
time, that miR-17 in serum may function as a diagnostic biomarker
and prognostic factor in the progression of OS.
References
|
1
|
Marina N, Gebhardt M, Teot L and Gorlick
R: Biology and therapeutic advances for pediatric osteosarcoma.
Oncologist. 9:422–441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferguson WS and Goorin AM: Current
treatment of osteosarcoma. Cancer Investigation. 19:292–315. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schwartz CL, Gorlick R, Teot L, Krailo M,
Chen Z, Goorin A, Grier HE, Bernstein ML and Meyers P: Children's
Oncology Group: Multiple drug resistance in osteogenic sarcoma:
INT0133 from the children's oncology group. J Clin Oncol.
25:2057–2062. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marcove RC, Miké V, Hajek JV, Levin AG and
Hutter RV: Osteogenic sarcoma under the age of twenty-one. A review
of one hundred and forty-five operative cases. J Bone Joint Surg
Am. 52:411–423. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Friedman MA and Carter SK: The therapy of
osteogenic sarcoma: Current status and thoughts for the future. J
Surg Oncol. 4:482–510. 1972. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bernthal NM, Federman N, Eilber FR, Nelson
SD, Eckardt JJ, Eilber FC and Tap WD: Long-term results (>25
years) of a randomized, prospective clinical trial evaluating
chemotherapy in patients with high-grade, operable osteosarcoma.
Cancer. 118:5888–5893. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang J and Zhang W: New molecular insights
into osteosarcoma targeted therapy. Curr Opin Oncol. 25:398–406.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bruland OS and Pihl A: On the current
management of osteosarcoma. A critical evaluation and a proposal
for a modified treatment strategy. Eur J Cancer. 33:1725–1731.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Amankwah EK, Conley AP and Reed DR:
Epidemiology and therapies for metastatic sarcoma. Clin Epidemiol.
5:147–162. 2013.PubMed/NCBI
|
|
10
|
Rainusso N, Wang LL and Yustein JT: The
adolescent and young adult with cancer: State of the art-bone
tumors. Curr Oncol Rep. 15:296–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cote GM and Choy E: Role of epigenetic
modulation for the treatment of sarcoma. Curr Treat Options Oncol.
14:454–464. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zamore PD and Haley B: Ribo-gnome: The big
world of small RNAs. Science. 309:1519–1524. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cho WC: OncomiRs: The discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim JG, Kim TO, Bae JH, Shim JW, Kang MJ,
Yang K, Ting AH and Yi JM: Epigenetically regulated MIR941 and
MIR1247 target gastric cancer cell growth and migration.
Epigenetics. 9:1018–1030. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fang L, Li H, Wang L, Hu J, Jin T, Wang J
and Yang BB: MicroRNA-17-5p promotes chemotherapeutic drug
resistance and tumour metastasis of colorectal cancer by repressing
PTEN expression. Oncotarget. 5:2974–2987. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Okudela K, Tateishi Y, Umeda S, Mitsui H,
Suzuki T, Saito Y, Woo T, Tajiri M, Masuda M, Miyagi Y and Ohashi
K: Allelic imbalance in the miR-31 host gene locus in lung
cancer-its potential role in carcinogenesis. PLoS One.
9:e1005812014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jin M, Zhang T, Liu C, Badeaux MA, Liu B,
Liu R, Jeter C, Chen X, Vlassov AV and Tang DG: MicroRNA-128
suppresses prostate cancer by inhibiting BMI-1 to inhibit
tumor-initiating cells. Cancer Res. 74:4183–4195. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao Y, Luo LH, Li S and Yang C: miR-17
inhibitor suppressed osteosarcoma tumor growth and metastasis via
increasing PTEN expression. Biochem Biophys Res Commun.
444:230–234. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Browne G, Taipaleenmäki H, Stein GS, Stein
JL and Lian JB: MicroRNAs in the control of metastatic bone
disease. Trends Endocrinol Metab. 25:320–327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mulrane L, McGee SF, Gallagher WM and
O'Connor DP: miRNA dysregulation in breast cancer. Cancer Res.
73:6554–6562. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bihrer V, Friedrich-Rust M, Kronenberger
B, Forestier N, Haupenthal J, Shi Y, Peveling-Oberhag J, Radeke HH,
Sarrazin C, Herrmann E, et al: Serum miR-122 as a biomarker of
necroinflammation in patients with chronic hepatitis C virus
infection. Am J Gastroenterol. 106:1663–1669. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bryant RJ, Pawlowski T, Catto JW, Marsden
G, Vessella RL, Rhees B, Kuslich C, Visakorpi T and Hamdy FC:
Changes in circulating microRNA levels associated with prostate
cancer. Br J Cancer. 106:768–774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Joosse SA, Müller V, Steinbach B, Pantel K
and Schwarzenbach H: Circulating cell-free cancer-testis MAGE-A
RNA, BORIS RNA, let-7b and miR-202 in the blood of patients with
breast cancer and benign breast diseases. Br J Cancer. 111:909–917.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Minami Y, Kohsaka S, Tsuda M, Yachi K,
Hatori N, Tanino M, Kimura T, Nishihara H, Minami A, Iwasaki N and
Tanaka S: SS18-SSX-regulated miR-17 promotes tumor growth of
synovial sarcoma by inhibiting p21WAF1/CIP1. Cancer Sci.
105:1152–1159. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li X, Yang H, Tian Q, Liu Y and Weng Y:
Upregulation of microRNA-17-92 cluster associates with tumor
progression and prognosis in osteosarcoma. Neoplasma. 61:453–460.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zampetaki A, Kiechl S, Drozdov I, Willeit
P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E,
et al: Plasma microRNA profiling reveals loss of endothelial
mir-126 and other microRNAs in type 2 diabetes. Circ Res.
107:810–817. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li H and Yang BB: Stress response of
glioblastoma cells mediated by miR-17-5p targeting PTEN and the
passenger strand miR-17-3p targeting MDM2. Oncotarget. 3:1653–1668.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Y, Chen A, Guo F and Xia Y:
Expression and clinical significance of PTEN protein in
osteosarcoma. Chinese-German J Clin Oncol. 5:296–299. 2008.
View Article : Google Scholar
|
|
32
|
Li J, Yen C, Liaw D, Podsypanina K, Bose
S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al:
PTEN, a putative protein tyrosine phosphatase gene mutated in human
brain, breast and prostate cancer. Science. 275:1943–1947. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Di Cristofano A and Pandolfi PP: The
multiple roles of PTEN in tumor suppression. Cell. 100:387–390.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Naguib A and Trotman LC: PTEN plasticity:
How the taming of a lethal gene can go too far. Trends Cell Biol.
23:374–379. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Seront E, Pinto A, Bouzin C, Bertrand L,
Machiels JP and Feron O: PTEN deficiency is associated with reduced
sensitivity to mTOR inhibitor in human bladder cancer through the
unhampered feedback loop driving PI3K/Akt activation. Br J Cancer.
109:1586–1592. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Levine RA, Forest T and Smith C: Tumor
suppressor PTEN is mutated in canine osteosarcoma cell lines and
tumors. Vet Pathol. 39:372–378. 2002. View Article : Google Scholar : PubMed/NCBI
|