Introduction
Fas-associated death domain protein (FADD) is a
classical adaptor protein involved in the cytoplasmic transduction
of extracellular apoptotic signals by mediating the association of
Fas and caspase-8 (1,2). FADD deficiency largely leads to
compromised death receptor-mediated apoptosis (3,4). However,
previous studies have indicated that FADD is also required for
embryonic development and lymphocyte differentiation, which is
independent of apoptosis, suggesting that FADD interacts with
non-apoptotic cooperators in vivo (5–7).
Previously, the role of FADD in programmed necrosis has been
revealed (8–10). Inhibition of programmed necrosis by
inactivation of the receptor interacting protein 1 rescues
FADD-deficient embryos from death (8).
As a pro-apoptotic protein, FADD functions as a
tumor suppressor in certain types of cancer, including thymic
lymphoblastic lymphoma and thyroid adenoma/adenocarcinoma (11). Downregulation of FADD has also been
observed in the leukemic cells of acute myeloid leukemia patients
(12). By contrast, FADD is
overexpressed in certain types of cancer. FADD is located on
chromosome 11q13, a region that is frequently amplified in several
types of cancer, including lung, head and neck squamous cell
carcinoma, laryngeal and pharyngeal carcinomas, and breast and
ovarian carcinomas (13). FADD mRNA
and protein levels are known to be increased in these forms of
cancer (14–18). However, the role of FADD in these
diseases remains to be fully elucidated.
Pancreatic cancer is one of the most lethal types of
cancer worldwide and the early stages are difficult to diagnose,
thus hindering prompt surgery, chemotherapy and radiotherapy
(19). Additionally, pancreatic
cancer is highly resistant to traditional chemotherapy and
radiotherapy, which is primarily due to failure of inducing cell
cycle arrest and apoptosis in pancreatic cancer cells (20). Cyclin-dependent kinase inhibitor 1
(p21) is a potent cell cycle inhibitor at G1 and S phase, and is
usually suppressed in cancer cells (21). Therefore, identifying the mechanisms
for p21 repression in pancreatic cancer cell in response to drugs
may shed light on more efficient chemotherapy strategies.
Although the role of FADD in several types of cancer
has been revealed, its role in pancreatic cancer remains to be
elucidated. In the present study, it was demonstrated that FADD is
overexpressed in pancreatic cancer cells. Furthermore, its
expression level is correlated with pancreatic cancer cell drug
resistance. FADD RNA interference resulted in increased cell cycle
arrest and apoptosis in response to Adriamycin®
treatment in drug-resistant cells. The present data indicates that
FADD may have an important role in the development of drug
resistance in pancreatic cancer cells.
Materials and methods
Cell culture
Pancreatic cancer cells (Colo357, Capan-1 and
MIA-PaCa-2) were purchased from American Type Culture Collection
(Manassas, VA, USA) and cultured in Dulbecco's modified Eagle's
medium (DMEM) or RPMI-1640 (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS;
Invitrogen; Thermo Fisher Scientific, Inc.). Normal epithelial cell
line (HPDE6) was a gift from Dr. Zhangjun Cheng (Southeast
University, Nanjing, China) and cultured in RPMI-1640 with 10%
fetal bovine serum. All cells were maintained at 37°C with 5%
CO2 and a humidity of 95%.
Cell viability assay
Colo357, Capan-1 and MIA-PaCa-2 cells (2,000
cells/well) were plated into each well of multiple 96-well plates
and cultured for 24 h at 37°C in DMEM or RPMI-1640. Cells were then
treated with 0, 0.25, 0.5, 1, 2 or 4 µM of Adriamycin
(Sigma-Aldrich; Merck Millipore, Darmstadt, Germany). Cell
viability was determined by the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium (MTT) assay.
Cells were incubated with MTT for 2 h at 37°C. Following removal of
the media containing MTT solution, the remaining dark blue formazan
crystals in each well were dissolved in dimethyl sulfoxide. The
absorbance was determined with a microplate reader. Each experiment
was performed in triplicate.
RNA interference
FADD small interfering (siRNA) and negative control
siRNA (NC siRNA) were synthesized by GenePharma Co., Ltd.
(Shanghai, China). The FADD siRNA sequences used were as follows:
sense, 5′-CACAGAGAAGGAGAACGCA-3′; and antisense,
5′-UGCGUUCUCCUUCUCUGUG-3′ as described by Varfolomeev et al
(22). The NC siRNA sequences used
were as follows: sense, 5′-GGCAGCAACCGAGAAGAAA-3′ and antisense,
5′-UUUCUUCUCGGUUGCUGCC-3′, generated by siRNA Wizard v3.1
(InvivoGen, San Diego, CA, USA). The NC siRNA was a scrambled
sequence of the FADD siRNA, which had no match in the human genome
sequence. Colo-357 and Capan-1 cells (5×105) were plated
in a 6-well plate and cultured overnight at 37°C in RPMI-1640.
siRNA and Lipofectamine® 2000 Reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) were mixed and incubated with the
cells in serum-free Gibco™ Opti-MEM (Thermo Fisher Scientific,
Inc.) for 6 h at 37°C. Fresh RPMI-1640 with 10% FBS was then added
into the cells. For the effect of FADD on the proliferation of
Colo-357, the concentrations of FADD siRNA were 0, 10, 25, 50, 100
and 200 nM, and the concentrations of the total siRNA were made up
to 200 nM with NC siRNA for those transfections with a FADD siRNA
concentration <200 nM. For the effect of FADD on the drug
resistance of Colo-357 and Capan-1, the concentration of FADD siRNA
was 100 nM and cells were treated with Adriamycin following 48 h of
siRNA transfection.
Western blotting
Briefly, cells were collected and washed with
phosphate-buffered saline (PBS) and lysed in lysis buffer (20 mM
Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2 EDTA, 1 mM
EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM
beta-glycerophosphate, 1 mM Na3VO4, 1 µg/ml
leupeptin) for 30 min on ice, and subsequently centrifuged at
11,700 × g for 10 min at 4°C. The supernatants were
collected. Western blotting was performed, as described by Cheng
et al (23). Briefly, 50 µg of
protein from each sample were separated by using SDS-PAGE on a 12%
SDS gel and subsequently transferred onto PVDF membranes (Merck
Millipore). The membranes were then blocked in 5% non-fat milk in
Tris-buffered saline containing 0.1% Tween 20 for 1 h at room
temperature and probed with primary antibodies at a dilution of
1:500 overnight at 4°C (anti-FADD; cat. no. sc-5559; anti-p21; cat.
no. sc-6246; anti-GAPDH; cat no. sc-47724), then probed with
corresponding horseradish peroxidase-conjugated secondary
antibodies at a dilution of 1:5,000 (goat anti-mouse IgG-HRP; cat.
no. sc-2302; goat anti-rabbit IgG-HRP; cat no. sc-2301) (all Santa
Cruz Biotechnology, Inc., Dallas, TX, USA). Bands were visualized
with 20X LumiGLO® Reagent and 20X Peroxide (cat. no.
7003; Cell Signaling Technology, Inc., Danvers, MA, USA).
Cell cycle analysis
Following siRNA transfection and Adriamycin
treatment, Capan-1 cells were subjected to cell cycle analysis with
propidum iodide using standard flow cytometry protocols. Briefly,
cells were trypsinized, fixed with cold ethanol and stored
overnight at 4°C. Cells were subsequently pelleted and rinsed in
cold PBS containing RNase A and incubated for 30 min at 37°C.
Propidium iodide was added and incubated for an additional 30 min
at 37°C. Cells were analyzed using FACSCalibur™ (BD Biosciences,
Franklin Lakes, NJ, USA) for the fluorescence signals collected in
the FL-2 detector. A total of 100,000 events were collected. Cell
cycle profiles were analyzed using FlowJo software (version 10.0.8;
FlowJo, LLC, Ashland, OR, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA was extracted with TRIzol® reagent
(Thermo Fisher Scientific, Inc.) and cDNA was synthesized with
PrimeScript™ RT Master Mix (Takara Bio, Inc., Otsu, Japan)
according to the manufacturer's protocol. RT-qPCR was then
performed using the StepOne/StepOnePlus™ Real-time PCR system and
SYBR Green PCR Master Mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Cycling conditions were: 10 min of denaturation at 95°C and 40
cycles at 95°C for 15 sec and at 60°C for 1 min. The relative FADD
mRNA expression level was calculated using the 2−ΔΔCq
method (24), where ΔCq
was calculated as follows: Cq value of
FADD-Cq value of GAPDH. PCR primers for RT-qPCR were as
follows: FADD forward, ATTAATGCCTCTCCCGCACC and reverse,
TCTCTGCTTCGCTCCGATTC; GAPDH forward, CACCATCTTCCAGGAGCGAG and
reverse, GCAGGAGGCATTGCTGAT.
Statistical analysis
Data are expressed as the mean ± standard deviation,
and paired Student's t-test analysis was performed. Comparisons
within groups were conducted using a t-test with repeated measures.
P<0.05 was considered to indicate a statistically significant
difference. All statistical analyses were performed with Prism
software version 6.0 (GraphPad Software Inc., La Jolla, CA,
USA).
Results
FADD is overexpressed in pancreatic
cancer cells
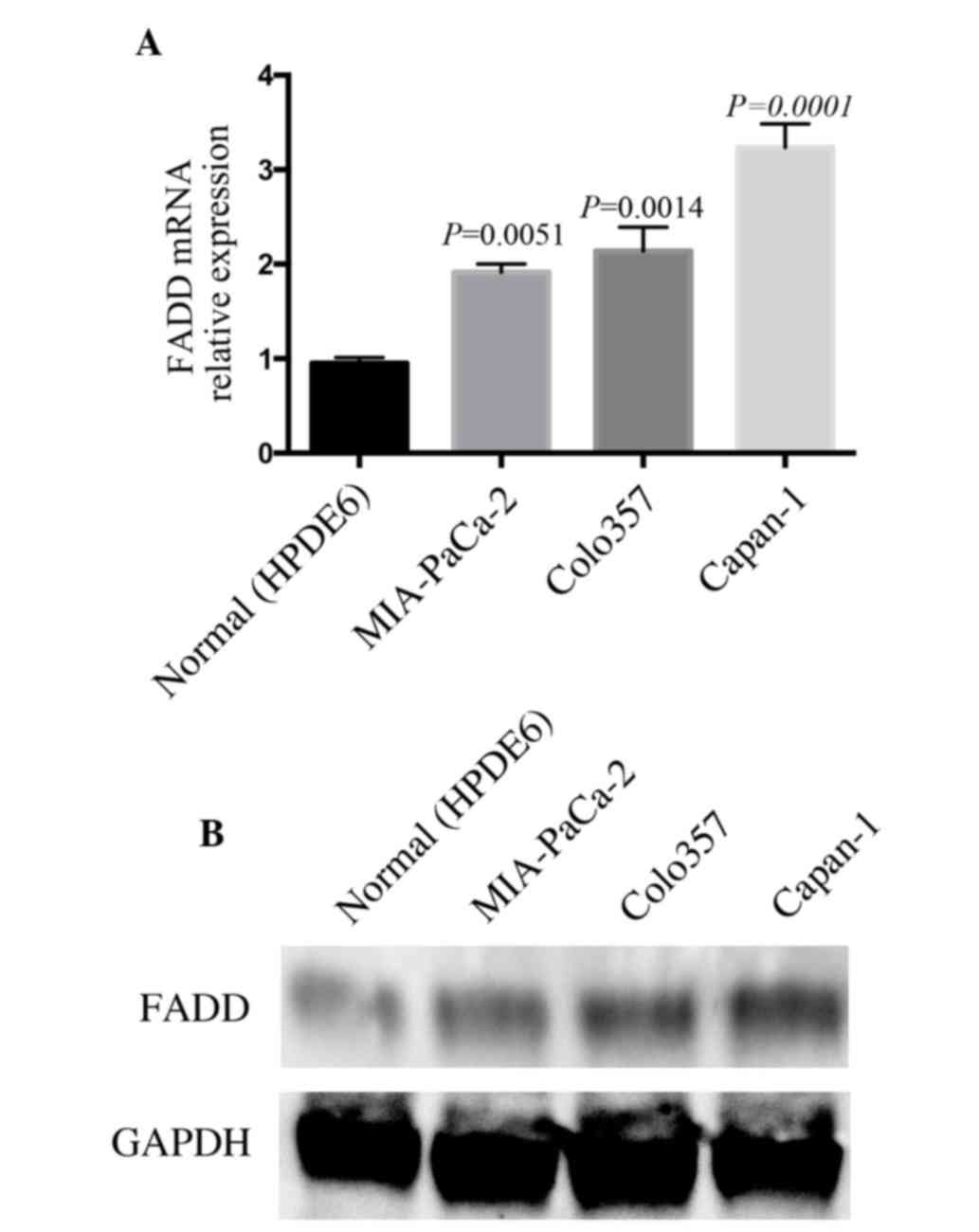
To determine the expression of FADD in pancreatic
cancer cells, FADD mRNA levels were examined in the wild-type
epithelial cell line (HPDE6) and cancerous pancreatic cells. FADD
mRNA expression was elevated significantly in pancreatic cancer
cells compared to in HPDE6 cell, with P-values of 0.0051, 0.0014
and 0.0001 in MIA-PaCa-2, Colo357 and Capan-1, respectively
(Fig. 1A). To confirm the changes in
the FADD mRNA level, the protein levels were subsequently
determined using western blotting. FADD protein expression was also
increased in pancreatic cancer cells (Fig. 1B). Increased levels of FADD in
pancreatic cancer cells were in conflict to its classic apoptotic
role, suggesting a non-apoptotic functions of FADD in these
cells.
FADD is required for pancreatic cancer
cell proliferation
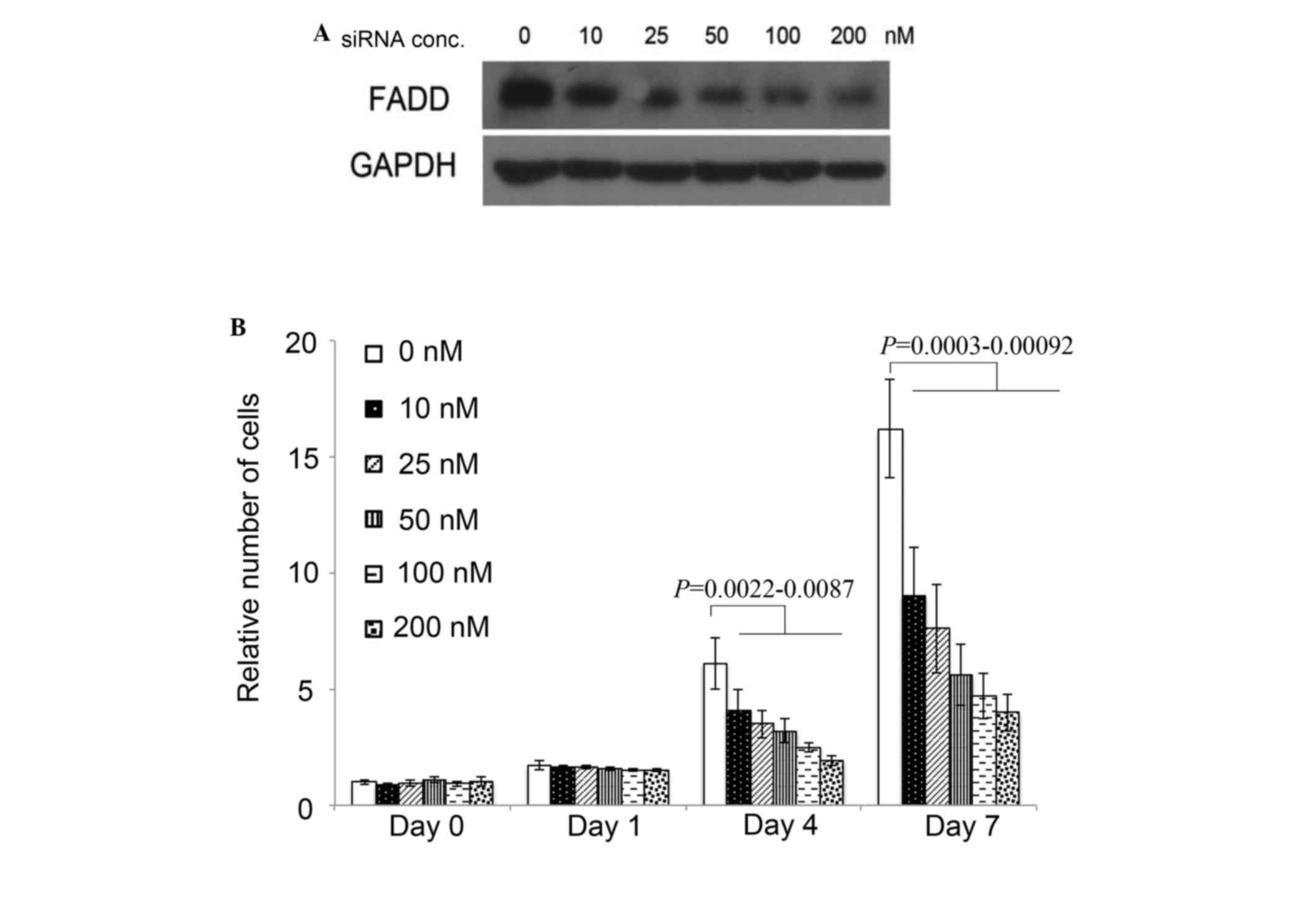
As FADD is overexpressed in pancreatic cancer cells,
its effect on pancreatic cancer cell proliferation was examined.
Cell proliferation was inhibited significantly by FADD RNAi at 4
days subsequent to FADD siRNA transfection compared to NC siRNA
transfection, indicated by P-values of 0.0087, 0.0061, 0.0051, and
0.0022 corresponding to the FADD siRNA concentrations of 10, 25,
50, 100 and 200 nM, respectively. A total of 7 days following
transfection, FADD knockdown inhibited Colo357 proliferation by
>50% with concentrations of FADD siRNA of 25 nM or higher, ,
with P-values of 0.00092, 0.00045, 0.00063 and 0.0003 corresponding
to FADD siRNA concentrations of 10, 25, 50, 100 and 200 nM,
respectively (Fig. 2).
FADD is correlated with drug
resistance in pancreatic cancer cells
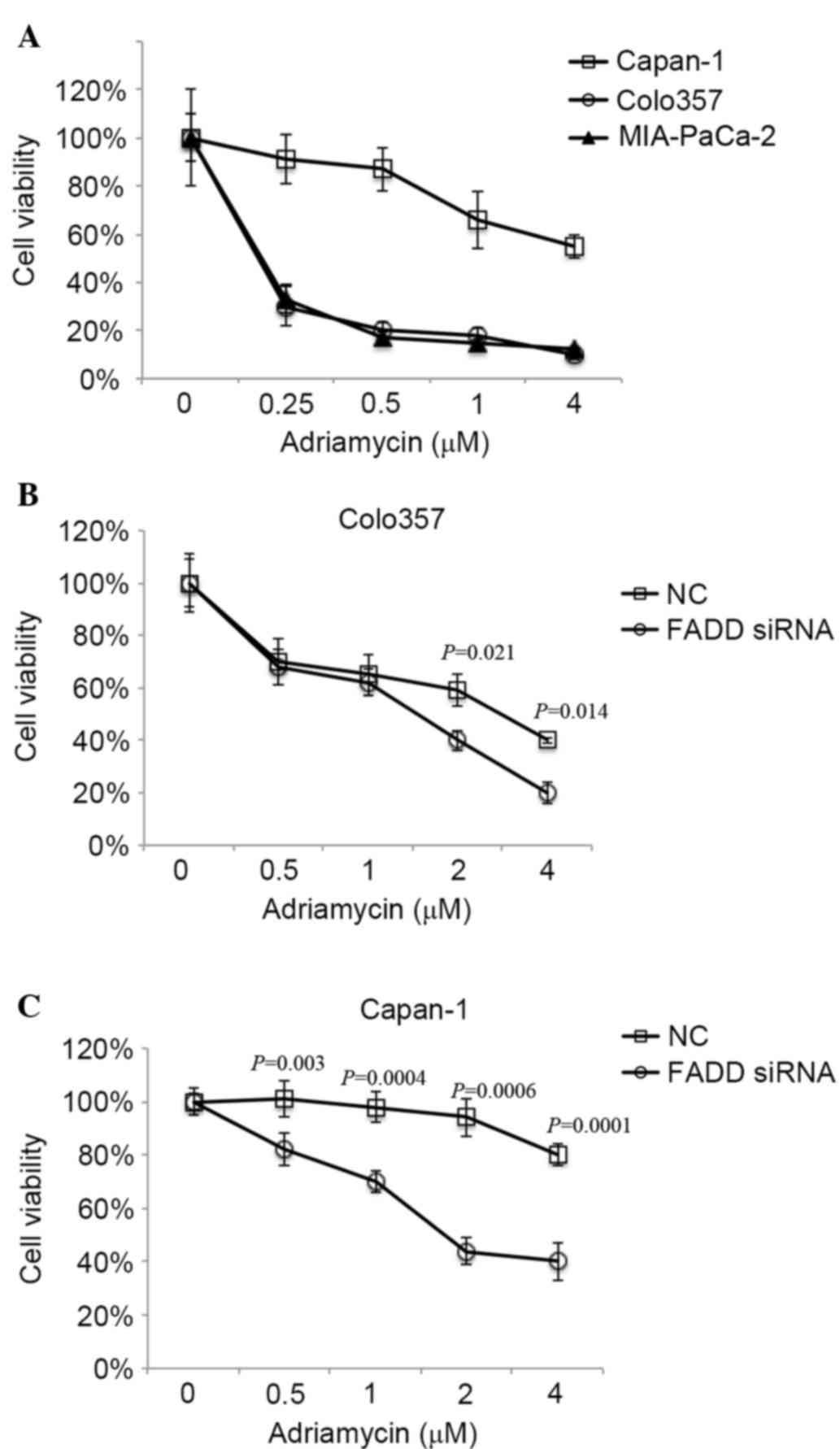
It has been demonstrated that FADD is overexpressed
in 3 pancreatic cancer cell lines to varying degrees (Fig. 1). It has previously been demonstrated
that Capan-1 cells exhibit increased resistance to anti-cancer
drugs compared with other cancer cell lines (25). As FADD is highly expressed in Capan-1,
the effect of FADD on drug resistance was analyzed. The Adriamycin
sensitivity of the cell lines was initially measured. Capan-1
exhibited increased resistance to Adriamycin compared with Colo357
and MIA-PaCa2 (Fig. 3A). Furthermore,
FADD knockdown sensitized Colo357 to Adriamycin significantly, with
P-values of 0.021 and 0.014 corresponding to Adriamycin
concentrations of 2 and 4 µM, respectively (Fig. 3B). Additionally, FADD RNAi increased
the sensitivity of Capan-1 cells to Adriamycin significantly, with
P-values of 0.003, 0.0004, 0.0006 and 0.0001 corresponding to
Adriamycin concentrations of 0.5, 1, 2 and 4 µM, respectively
(Fig. 3C). The present data indicate
a promoting role for FADD in the development of drug resistance in
pancreatic cancer cells.
FADD protects pancreatic cancer cells
from drug-induced apoptosis
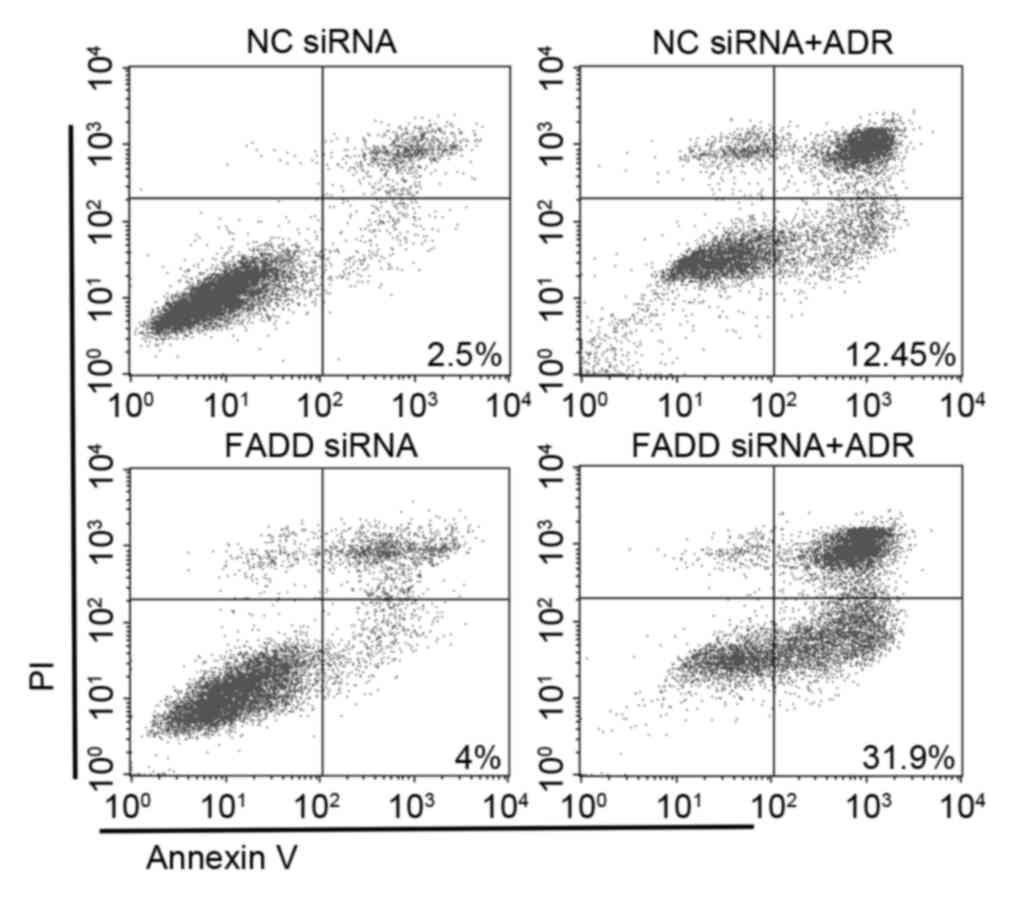
Adriamycin-induced apoptosis in FADD knockdown cells
was examined. While FADD RNAi alone had no effect on apoptosis, 2
µM Adriamycin treatment was able to induce a moderate level of
apoptosis. FADD RNAi dramatically promoted Adriamycin-induced
apoptosis (Annexin V+/PI-) (Fig. 4).
The present data indicates that FADD is critical to the development
of drug resistance in Capan-1 cells and protects them from
apoptosis.
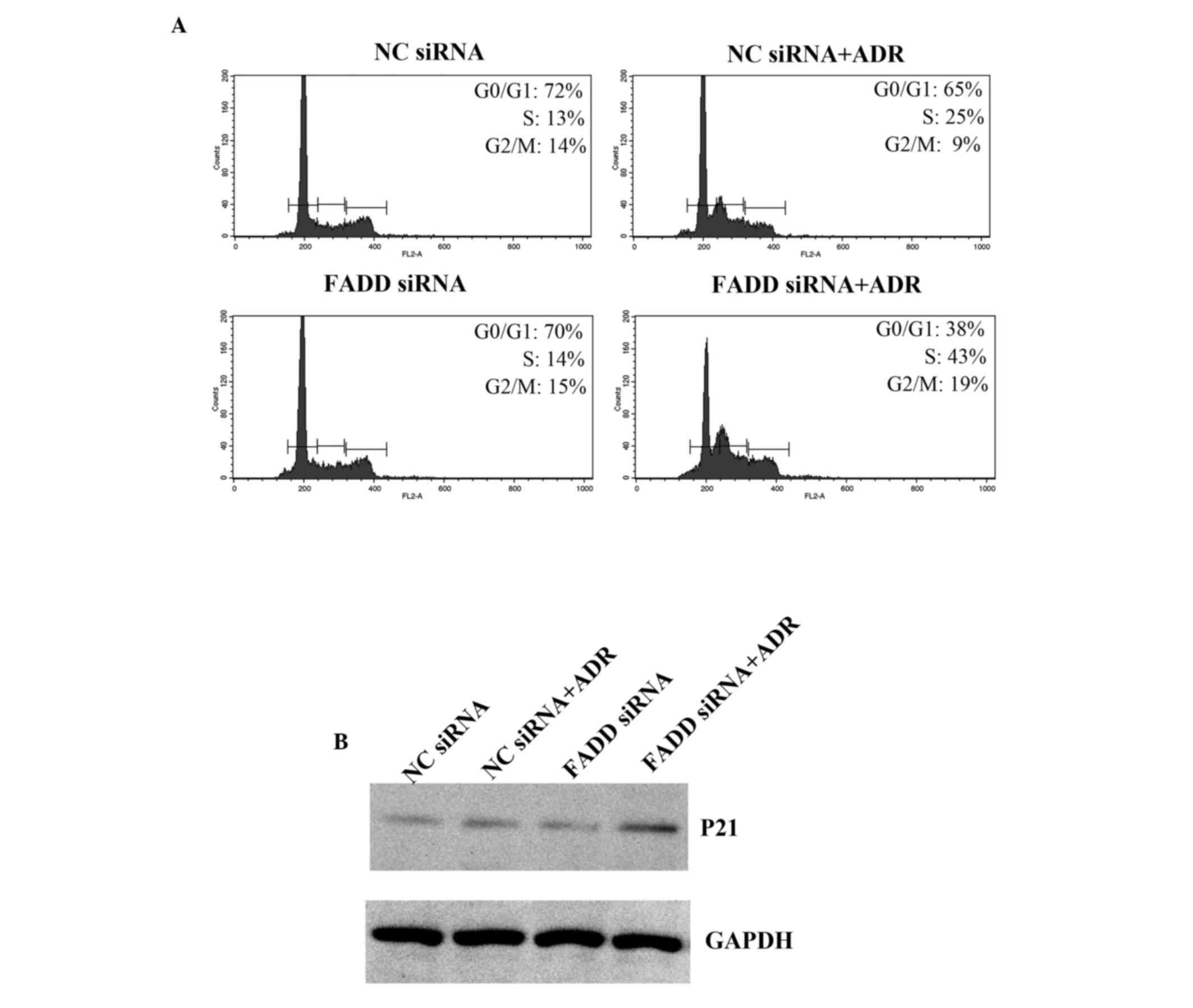
FADD regulates the cell cycle
It has been shown that Adriamycin induces cell cycle
arrest, which subsequently leads to apoptosis (21). Cell cycle phase distribution in FADD
knockdown Capan-1 cells was examined. Adriamycin treatment resulted
in cell cycle arrest (Fig. 5A). FADD
RNAi promoted increased cell cycle arrest in the S phase when
combined with Adriamycin treatment. p21 levels were also
significantly elevated in FADD knockdown Capan-1 cells following
Adriamycin treatment (Fig. 5B). Thus
the present data indicate that FADD partially inhibits Adriamycin
induced cell cycle arrest.
Discussion
FADD was initially identified as an adaptor protein
that couples Fas receptor and procaspase-8 to form a death-inducing
signaling complex. Procaspase-8 is subsequently cleaved leading to
its activation and the initiation of apoptosis (2). It is therefore reasonable to hypothesize
that FADD has a repressive role in cancer cell proliferation and
survival. This concept was supported by the fact that FADD is
deficient in certain types of cancer (11,12).
However, multiple studies have revealed that FADD is overexpressed
in many other forms of cancer (14–17).
Consistent with previous observations, the present study indicated
that FADD is overexpressed in pancreatic cancer cells. Knockdown of
FADD impairs pancreatic cancer cell proliferation, suggesting that
FADD is required for cell proliferation in the context of cancer.
It has been shown that FADD translocates into the nucleus during
mitosis to promote cell cycle progression, implying that FADD has
apoptotic and non-apoptotic roles depending upon its cellular
localization (26,27). Its nuclear localization promotes cell
cycle progression and simultaneously deprives proliferative cells
of apoptotic stimuli (18). This
model proposes a mechanism for the FADD-mediated coupling of cancer
cell proliferation and resistance to apoptosis, however further
research is required (18).
In addition to facilitating proliferation, the
present study also demonstrated that FADD induces drug resistance
in pancreatic cancer cells, indicating that FADD is required for
cancer cell survival in response to certain apoptotic stimuli.
Although FADD is required for death receptor-mediated apoptosis, it
is not required in Adriamycin-induced fibroblast apoptosis
(5). As demonstrated by the present
study, FADD deficiency results in apoptosis in response to
Adriamycin in pancreatic cancer cells. This discrepancy largely
depends on cellular context. Accelerated cell cycle machinery in
cancer cells may be more sensitive to stimuli that cause cell cycle
arrest. However, the complete mechanisms remain to be fully
elucidated.
In conclusion, the present data demonstrated
critical roles for FADD in pancreatic cancer cell proliferation and
the development of resistance to Adriamycin. The results indicate
more complex regulatory mechanisms for pancreatic cancer cell
proliferation and survival. The data also reveal new strategies in
the development of more efficient pancreatic cancer treatments.
Acknowledgements
The authors are grateful for grants from the
National Key Basic Research Program of the Ministry of Science and
Technology (grant nos. 2012CB967004, 2014CB744501 and
2011CB933502), the Chinese National Nature Sciences Foundation
(grant nos. 81121062, 30425009, 30330530 and 30270291), the Jiangsu
Provincial Nature Science Foundation (grant nos. BE2013630,
BZ2012050 and BK2011573) and the Bureau of Science and Technology
of Changzhou (grant nos. CM20122003, CZ20120004, CZ20130011 and
CE20135013s).
References
|
1
|
Zhang J and Winoto A: A mouse
Fas-associated protein with homology to the human Mort1/FADD
protein is essential for Fas-induced apoptosis. Mol Cell Biol.
16:2756–2763. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chinnaiyan AM, O'Rourke K, Tewari M and
Dixit VM: FADD, a novel death domain-containing protein, interacts
with the death domain of Fas and initiates apoptosis. Cell.
81:505–512. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kuang AA, Diehl GE, Zhang J and Winoto A:
FADD is required for DR4- and DR5-mediated apoptosis: Lack of
trail-induced apoptosis in FADD-deficient mouse embryonic
fibroblasts. J Biol Chem. 275:25065–25068. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chinnaiyan AM, Tepper CG, Seldin MF,
O'Rourke K, Kischkel FC, Hellbardt S, Krammer PH, Peter ME and
Dixit VM: FADD/MORT1 is a common mediator of CD95 (Fas/APO-1) and
tumor necrosis factor receptor-induced apoptosis. J Biol Chem.
271:4961–4965. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yeh WC, de la Pompa JL, McCurrach ME, Shu
HB, Elia AJ, Shahinian A, Ng M, Wakeham A, Khoo W, Mitchell K, et
al: FADD: Essential for embryo development and signaling from some,
but not all, inducers of apoptosis. Science. 279:1954–1958. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kabra NH, Kang C, Hsing LC, Zhang J and
Winoto A: T cell-specific FADD-deficient mice: FADD is required for
early T cell development. Proc Natl Acad Sci USA. 98:6307–6312.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang J, Cado D, Chen A, Kabra NH and
Winoto A: Fas-mediated apoptosis and activation-induced T-cell
proliferation are defective in mice lacking FADD/Mort1. Nature.
392:296–300. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang H, Zhou X, McQuade T, Li J, Chan FK
and Zhang J: Functional complementation between FADD and RIP1 in
embryos and lymphocytes. Nature. 471:373–376. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Osborn SL, Diehl G, Han SJ, Xue L, Kurd N,
Hsieh K, Cado D, Robey EA and Winoto A: Fas-associated death domain
(FADD) is a negative regulator of T-cell receptor-mediated
necroptosis. Proc Natl Acad Sci USA. 107:13034–13039. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Welz PS, Wullaert A, Vlantis K, Kondylis
V, Fernández-Majada V, Ermolaeva M, Kirsch P, Sterner-Kock A, van
Loo G and Pasparakis M: FADD prevents RIP3-mediated epithelial cell
necrosis and chronic intestinal inflammation. Nature. 477:330–334.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Newton K, Harris AW and Strasser A:
FADD/MORT1 regulates the pre-TCR checkpoint and can function as a
tumour suppressor. EMBO J. 19:931–941. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tourneur L, Delluc S, Lévy V, Valensi F,
Radford-Weiss I, Legrand O, Vargaftig J, Boix C, Macintyre EA,
Varet B, et al: Absence or low expression of fas-associated protein
with death domain in acute myeloid leukemia cells predicts
resistance to chemotherapy and poor outcome. Cancer Res.
64:8101–8108. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fantl V, Smith R, Brookes S, Dickson C and
Peters G: Chromosome 11q13 abnormalities in human breast cancer.
Cancer Surv. 18:77–94. 1993.PubMed/NCBI
|
|
14
|
Gibcus JH, Menkema L, Mastik MF, Hermsen
MA, de Bock GH, van Velthuysen ML, Takes RP, Kok K, Marcos CA
Alvarez, van der Laan BF, et al: Amplicon mapping and expression
profiling identify the Fas-associated death domain gene as a new
driver in the 11q13.3 amplicon in laryngeal/pharyngeal cancer. Clin
Cancer Res. 13:6257–6266. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Prapinjumrune C, Morita K, Kuribayashi Y,
Hanabata Y, Shi Q, Nakajima Y, Inazawa J and Omura K: DNA
amplification and expression of FADD in oral squamous cell
carcinoma. J Oral Pathol Med. 39:525–532. 2010.PubMed/NCBI
|
|
16
|
Rasamny JJ, Allak A, Krook KA, Jo VY,
Policarpio-Nicolas ML, Sumner HM, Moskaluk CA, Frierson HF Jr and
Jameson MJ: Cyclin D1 and FADD as biomarkers in head and neck
squamous cell carcinoma. Otolaryngol Head Neck Surg. 146:923–931.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kwek SS, Roy R, Zhou H, Climent J,
Martinez-Climent JA, Fridlyand J and Albertson DG: Co-amplified
genes at 8p12 and 11q13 in breast tumors cooperate with two major
pathways in oncogenesis. Oncogene. 28:1892–1903. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen G, Bhojani MS, Heaford AC, Chang DC,
Laxman B, Thomas DG, Griffin LB, Yu J, Coppola JM, Giordano TJ, et
al: Phosphorylated FADD induces NF-kappaB, perturbs cell cycle, and
is associated with poor outcome in lung adenocarcinomas. Proc Natl
Acad Sci USA. 102:12507–12512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Andrán-Sandberg A: Pancreatic cancer:
Chemotherapy and radiotherapy. N Am J Med Sci. 3:1–12. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
King KL and Cidlowski JA: Cell cycle
regulation and apoptosis. Annu Rev Physiol. 60:601–617. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Varfolomeev E, Maecker H, Sharp D,
Lawrence D, Renz M, Vucic D and Ashkenazi A: Molecular determinants
of kinase pathway activation by Apo2 ligand/tumor necrosis
factor-related apoptosis-inducing ligand. J Biol Chem.
280:40599–40608. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cheng W, Wang L, Zhang R, Du P, Yang B,
Zhuang H, Tang B, Yao C, Yu M, Wang Y, et al: Regulation of protein
kinase C inactivation by Fas-associated protein with death domain.
J Biol Chem. 287:26126–26135. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Arlt A, Vorndamm J, Breitenbroich M,
Fölsch UR, Kalthoff H, Schmidt WE and Schäfer H: Inhibition of
NF-kappaB sensitizes human pancreatic carcinoma cells to apoptosis
induced by etoposide (VP16) or doxorubicin. Oncogene. 20:859–868.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Alappat EC, Feig C, Boyerinas B, Volkland
J, Samuels M, Murmann AE, Thorburn A, Kidd VJ, Slaughter CA, Osborn
SL, et al: Phosphorylation of FADD at serine 194 by CKIalpha
regulates its nonapoptotic activities. Mol Cell. 19:321–332. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cheng W, Wang L, Yang B, Zhang R, Yao C,
He L, Liu Z, Du P, Hammache K, Wen J, et al: Self-renewal and
differentiation of muscle satellite cells are regulated by the
Fas-associated death domain. J Biol Chem. 289:5040–5050. 2014.
View Article : Google Scholar : PubMed/NCBI
|