Introduction
Osteosarcoma (OS) is the most common type of primary
malignancy of the bones and joints, and accounts for ~2.4% of all
malignancies in child and adolescent patients, and ~20% of all
types of primary bone cancer (1,2). The
estimated incidence of OS is four to five cases per million
worldwide, with a peak incidence at 15–19 years old (3). Currently, the main standard therapeutic
methods for OS include local control of the primary lesion by
surgery and the use of combinational chemotherapy (4). OS cells are characteristically
aggressive, with capabilities of rapid growth and early metastasis.
Lymph node and/or distant metastasis is developed in >30% of
patients with locally advanced OS (5,6). Although
progress in therapeutic treatments has occurred, prognosis remains
poor. The 5-year overall survival rate for locally advanced
patients is 60–70%, whereas for patients who present with
metastatic disease it is <30% (7,8).
Understanding the molecular mechanisms underlying the rapid growth
and early metastasis of OS and investigating novel therapeutic
regimens to prevent metastasis during the early stages is,
therefore, important.
microRNAs (miRNAs/miRs) are a group of endogenous,
non-protein-coding and short RNAs (18–25 nucleotides) with highly
conserved sequences in plants, animals and DNA viruses (9). Several studies have demonstrated that
miRNAs regulate mRNA expression in tumor and normal cells, by
binding to sites in the 3′ untranslated regions (3′UTR) of mRNAs in
a base-pairing manner, resulting in the degradation of mRNAs or
translational inhibition at the post-transcription level (10–12). It
has been estimated that miRNAs regulate more than two-thirds of
human genes (13). Abnormal
expression of miRNAs has been reported in various diseases,
particularly in cancer (14).
Numerous studies have suggested that the abnormal expression of
miRNAs in cancer serves a crucial function in several physiological
and pathological processes, including cell growth, differentiation,
the cell cycle, apoptosis, survival, migration and invasion
(15,16). miRNAs may act as tumor suppressors or
oncogenes in the initiation and development of various types of
human malignancies, depending on the roles of the target mRNAs
(17). Therefore, an investigation
into miRNAs may reveal the prognostic value and therapeutic
potential of miRNAs in OS.
The present study aimed to investigate the
expression, functions and molecular mechanisms of miR-150 in OS
carcinogenesis and progression. In the present study, the miR-150
expression levels in OS tissues and cell lines were analyzed,
followed by functional studies of miR-150 in human OS cell lines.
The results of the present study revealed that miR-150 was
significantly downregulated in OS tissues and cell lines. Low
expression levels of miR-150 were associated with clinical stage
and distant metastasis in patients with OS. In addition, miR-150
inhibited OS cell growth, migration and invasion. Additionally,
zinc finger E-box binding homeobox 1 (ZEB1) was identified as a
direct target of miR-150. Therefore, miR-150 was determined to be
an antioncogenic regulator in OS via the direct targeting of ZEB1.
These findings indicated a novel molecular mechanism underlying the
pathogenic process in OS carcinogenesis and progression, and may
facilitate the development of novel targeted therapeutic regimens
for patients with OS.
Materials and methods
Clinical specimens
The current study was approved by the Ethical Review
Committee of Tianjin Hospital (Tianjin, China). In addition,
written informed consent and clinicopathological information was
obtained from each patient with OS involved in the present study. A
total of 67 pairs of OS tissues and matched normal adjacent tissues
(NATs) were obtained from patients (39 male and 28 female; age
range, 16–65 years) who underwent surgical resection at Tianjin
Hospital between June 2013 and January 2015. All the patients with
OS had not received any therapeutic treatments prior to surgery.
Specimens had been histologically and clinically diagnosed
following surgery. Tissues were snap-frozen in liquid nitrogen and
stored at −80°C until use.
Cell culture
The HOS, U2OS, MG-63 and SAOS-2 human OS cell lines
and the human normal osteoblastic hFOB 1.19 cell line were
purchased from the American Type Culture Collection (Manassas, VA,
USA). The HEK293T cell line was obtained from the Chinese Center
for Type Culture Collection (Wuhan, China). All cell lines were
maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher, Scientific, Inc.),
100 U/ml penicillin and 100 U/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.). All cell lines were cultured at 37°C in a
humidified atmosphere containing 5% CO2.
Cell transfection
miR-150 mimics and negative controls (NC) were
obtained from GenePharma Co., Ltd. (Shanghai, China). ZEB1 small
interfering RNA (siRNA) and negative control (NC) siRNA were
purchased from Guangzhou RiboBio (Guangzhou, China). When the
growth of the cells reached the exponential phase they were plated
into 6-well plates at a density of 7.5×105 per well and
maintained in DMEM containing 10% FBS without antibiotics. The
cells were transfected with miR-150 mimics, NC, ZEB1 siRNA or NC
siRNA using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues and cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. cDNA was synthesized
using a PrimeScript RT Reagent kit (Takara Bio, Inc., Otsu, Japan).
RT-qPCR was carried out to evaluate miR-150 expression with a SYBR
Premix Ex Taq™ kit (Takara Biotechnology Co., Ltd.,
Dalian, China), and U6 small nuclear RNA was used as an internal
control. The thermocycling conditions for qPCR of miR-150 and U6
were as follows: 95°C for 30 sec; 40 cycles of 95°C for 5 sec; 60°C
for 30 sec. ZEB1 mRNA expression was analyzed using SYBR Green PCR
Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.),
and GADPH was used as the internal reference gene. The
thermocycling conditions for qPCR of ZEB1 and GADPH were as
follows: 95°C for 10 min; 40 cycles of 95°C for 15 sec; 60°C for 1
min. RT-qPCR was performed on an Applied Biosystems 7500 Real-time
PCR detection system (ABI; Thermo Fisher Scientific, Inc.).
Relative expression was calculated using the 2−ΔΔCq
method (18).
MTT assay
An MTT assay (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) was performed to assess the OS cell viability. After 24 h
of transfection at 37°C, the cells were collected and seeded into
96-well plates at a density of 3,000 cells/well. Cells were
cultured in a cell culture box at 37°C with 5% CO2 for
1, 2, 3 and 4 days. A total of 20 µl MTT solution (5 mg/ml) was
added into each well and incubated for a further 4 h at 37°C. Then,
the cells were lysed in 150 µl dimethyl sulfoxide for 10 min at
37°C. The absorbance was measured using a microplate reader
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) at a 490 nm
wavelength. Each sample was evaluated in triplicate.
Transwell migration and invasion
assays
Cell migration and invasion assays were used to
evaluate OS cell motility ability using Transwell chambers (8 µm
pore size; Costar, Cambridge, MA, USA). For the cell invasion
assay, the Transwell chamber was coated with 50 µg Matrigel (BD
Biosciences, San Jose, CA, USA), according to the manufacturer's
protocol. After 48 h of transfection, cells were collected,
1×105 cells were resuspended in 200 µl DMEM without FBS
and were then added into the upper chamber, while the lower chamber
was filled with 500 µl DMEM supplemented with 20% FBS. After 24 h
of incubation, the cells were fixed with 100% methanol for 10 min
and stained with 0.5% crystal violet for 20 min. Subsequently,
cells that had not migrated or invaded to the lower membrane were
carefully removed with cotton swabs. The cells in >5 randomly
selected fields (magnification, ×100) were counted under an
inverted microscope (CKX41; Olympus Corporation, Tokyo, Japan). All
experiments were repeated at least three times.
miR-150 targets prediction
The target genes of miR-150 were predicted using the
following TargetScan (http://www.targetscan.org/index.html), PicTar
(http://pictar.mdc-berlin.de/) and
miRanda (http://www.microrna.org).
Western blot
After a 72-h transfection, proteins were harvested
from cells using RIPA lysis buffer (Beyotime Institute of
Biotechnology, Haimen, China). The protein concentration was
quantified using a bicinchoninic acid protein assay kit (Pierce;
Thermo Fisher Scientific, Inc.). Equal amount of proteins (20 µg)
were subjected to 10% SDS-PAGE and electrotransferred to
polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA,
USA). The membranes were blocked with 5% non-fat milk in
Tris-buffered saline (TBS) at room temperature for 2 h. Then, the
membranes were incubated with primary antibodies, including a mouse
anti-human monoclonal ZEB1 antibody (1:1,000 dilution; cat. no.
sc-81428; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and an
anti-human monoclonal GADPH antibody (1:1,000 dilution; cat. no.
sc-59540; Santa Cruz Biotechnology, Inc.), overnight at 4°C.
Subsequent to washing with TBS/Tween-20 three times, the membranes
were incubated with corresponding horseradish peroxidase-conjugated
secondary antibodies (1:3,000 dilution; cat. no. A0192; Beyotime
Institute of Biotechnology) at room temperature for 1 h. The
protein blots were visualized with enhanced chemiluminescence
(Pierce; Thermo Fisher Scientific, Inc.). GADPH was used as a
loading control.
Dual-Luciferase reporter assay
PGL3-ZEB1-3′UTR wild type (Wt) and PGL3-ZEB1-3′UTR
mutant (Mut) was obtained from GenePharma Co., Ltd. HEK293T cells
were seeded into 12-well plates and transfected with miR-150 mimics
or NC, and PGL3-ZEB1-3′UTR Wt or PGL3-ZEB1-3′UTR Mut using
Lipofectamine® 2000. After 48 h of transfection, firefly and
Renilla luciferase activities were measured using a
Dual-Luciferase Reporter Assay system (Promega Corporation,
Madison, WI, USA), according to the manufacturer's protocol.
Firefly luciferase activities were normalized to Renilla
luciferase activities for each well.
Statistical analysis
The data are presented as the mean ± standard
deviation, and were compared with Student's t-tests or one-way
analysis of variance and multiple comparisons using the SPSS
version 16.0 statistical software package (SPSS, Inc., Chicago, IL,
USA). SNK was utilized to compare the two groups in multiple groups
studies. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-150 expression is decreased in OS
tissues and cell lines
RT-qPCR was performed in order to evaluate miR-150
expression in OS tissues, NATs, OS cell lines and the human hFOB
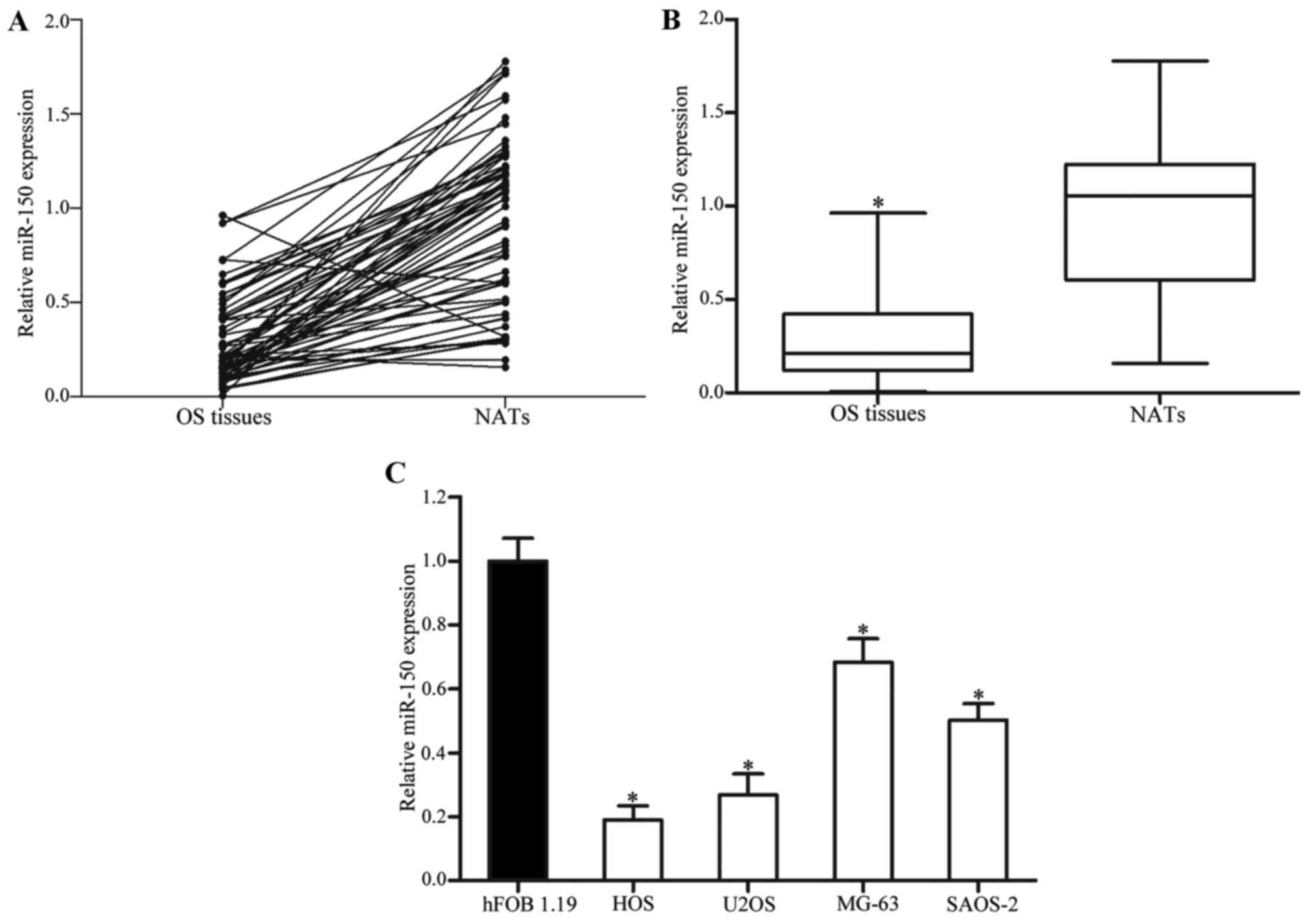
1.19 normal osteoblastic cell line. As presented in Fig. 1A and B, miR-150 expression levels in
OS tissues were significantly lower compared with in NATs
(P<0.05). miR-150 was also downregulated in HOS, U2OS, MG-63 and
SAOS-2 cell lines, as compared with in hFOB 1.19 cells (P<0.05;
Fig. 1C). These results suggested
that miR-150 may serve an important role in OS.
Correlation between miR-150 expression
and clinicopathological features in patients with OS
In the present study, an investigation was performed
into whether the expression levels of miR-150 were associated with
clinicopathological features in patients with OS. As presented in
Table I, statistical analysis
revealed that miR-150 expression was significantly associated with
the clinical stage (P=0.016) and distant metastasis (P=0.027) in
patients with OS. However, no correlation was observed between
miR-150 expression and other clinicopathological factors, including
sex, age, anatomical location and tumor size.
 | Table I.Correlation between expression of
miR-150 and clinicopathological features in patients with
osteosarcoma. |
Table I.
Correlation between expression of
miR-150 and clinicopathological features in patients with
osteosarcoma.
|
|
| miR-150
expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
features | Patient no. | Low (n=38) | High (n=29) | P-value |
|---|
| Sex |
|
|
| 0.803 |
|
Male | 39 | 23 | 16 |
|
|
Female | 28 | 15 | 13 |
|
| Age |
|
|
| 0.204 |
| <50
years | 40 | 26 | 14 |
|
| ≥50
years | 27 | 12 | 15 |
|
| Anatomical
location |
|
|
| 1.000 |
|
Tibia/femur | 39 | 22 | 17 |
|
|
Elsewhere | 28 | 16 | 12 |
|
| Tumor size
(cm) |
|
|
| 0.624 |
| <8
cm | 33 | 20 | 13 |
|
| ≥8
cm | 34 | 18 | 16 |
|
| Clinical stage |
|
|
| 0.016a |
|
I–II | 35 | 15 | 20 |
|
|
III | 32 | 23 | 9 |
|
| Distant
metastasis |
|
|
| 0.027a |
|
Present | 34 | 24 | 10 |
|
|
Absent | 33 | 14 | 19 |
|
miR-150 inhibits the proliferation of
OS cells
To investigate the functional roles of miR-150 in
OS, the present study transfected miR-150 mimics into human OS
cells. miR-150 expression levels in HOS and U2OS cells were low, as
compared with in the other cell lines investigated. Thus, HOS and
U2OS cells were selected for transfection with miR-150 mimics or
the NC. Subsequent to a transfection of 48 h, miR-150 expression
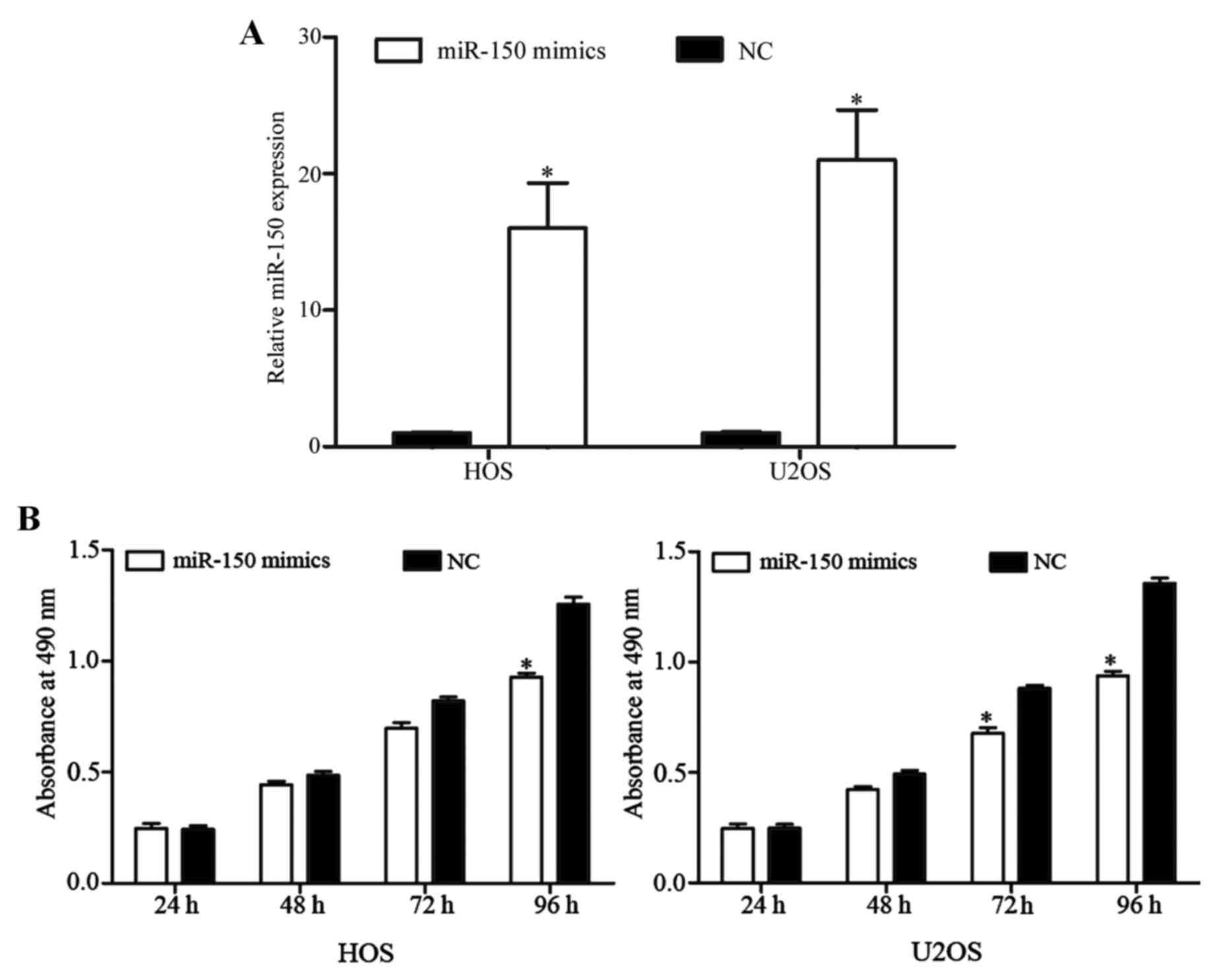
was quantified by RT-qPCR. As presented in Fig. 2A, miR-150 expression level was
markedly elevated by miR-150 mimics in HOS and U2OS cells
(P<0.05).
MTT assays were used to measure OS cell
proliferation subsequent to transfection with miR-150 mimics or NC.
As depicted in Fig. 2B, miR-150
inhibited the growth of HOS and U2OS cells. After 96 h of
transfection, the rate at which miR-150 suppresses cell
proliferation reached 26.05±4.24% in HOS cells and 30.87±5.57% in
U2OS cells. These results indicated that miR-150 may function as a
novel tumor suppressor in OS.
miR-150 inhibits the migration and
invasion abilities of OS cells
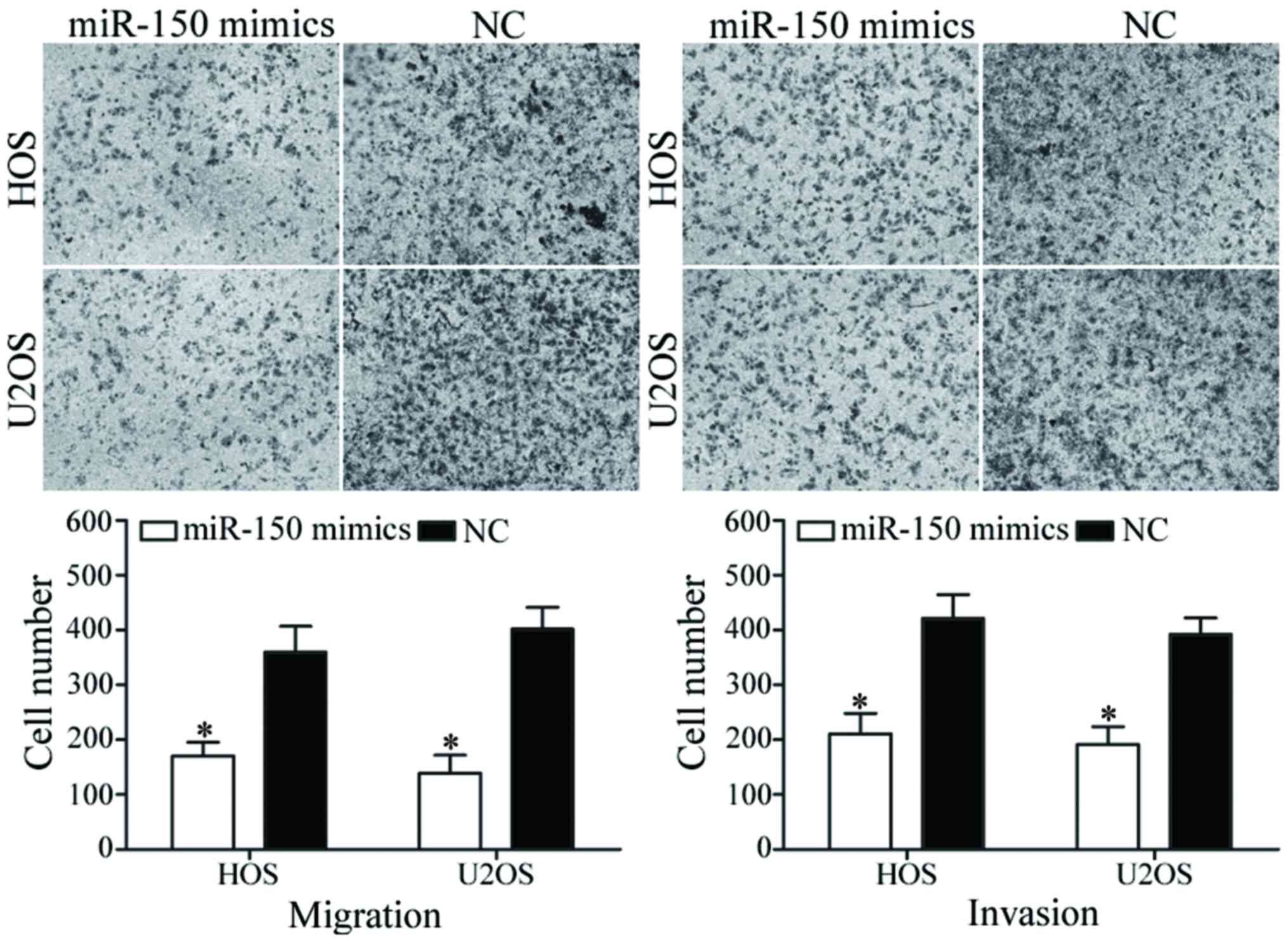
To evaluate the functions of miR-150 in OS
metastasis, migration and invasion assays were performed using
Transwell chambers. As presented in Fig.
3, miR-150 inhibited HOS and U2OS cell migratory and invasive
abilities (P<0.05). These findings suggest that miR-150 may
serve a critical role in OS metastasis.
ZEB1 is a direct target of miR-150 in
vitro
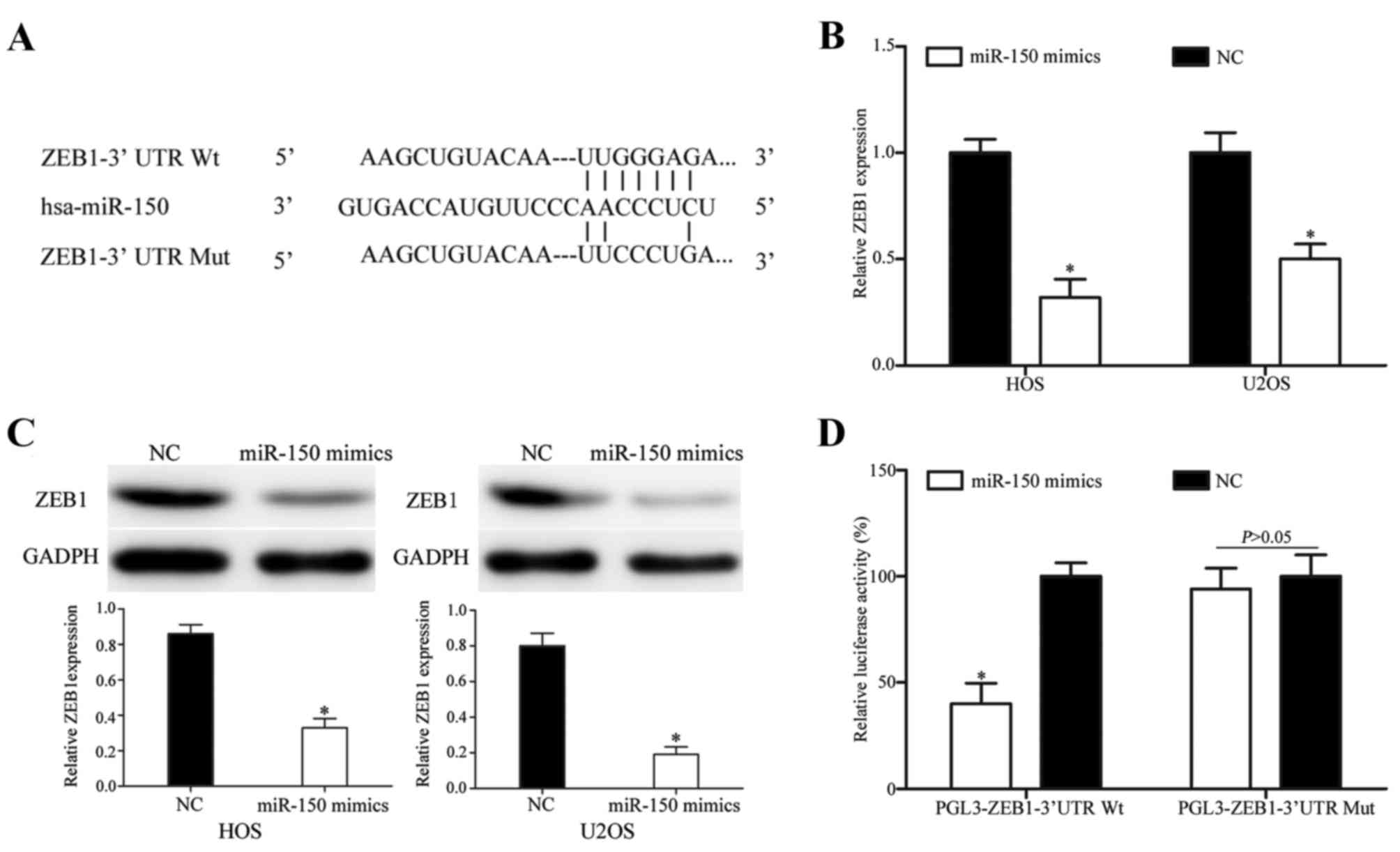
TargetScan, PicTar and miRanda were used to explore
the molecular mechanism of miR-150 in OS. ZEB1 was identified as a
target of miR-150 in all three prediction programs (Fig. 4A). RT-qPCR and western blotting were
then performed to measure ZEB1 expression at the mRNA and protein
levels subsequent to transfection with miR-150 mimics. As indicated
in Fig. 4B, ZEB1 was significantly
downregulated at the mRNA level in HOS and U2OS cells subsequent to
transfection with miR-150 mimics (P<0.05). Similarly, western
blotting revealed that ZEB1 protein expression was downregulated in
miR-150 mimic-transfected HOS and U2OS cells (P<0.05; Fig. 4C).
Finally, Dual-Luciferase reporter assays were
performed to explore whether miR-150 directly targets the 3′UTR of
ZEB1. As presented in Fig. 4D,
miR-150 significantly inhibited PGL3-ZEB1-3′UTR Wt luciferase
activity, but not the PGL3-ZEB1-3′UTR Mut luciferase activity, in
HEK293T cells (P<0.05). These results demonstrate that ZEB1 is a
direct target gene of miR-150 in vitro.
ZEB1 is involved in miR-150-mediated
tumor suppression functions in OS cells
To determine whether ZEB1 serves as a critical
mediator of the suppressive functions of miR-150 on OS cell
proliferation, migration and invasion, the present study
transfected ZEB1 siRNA or NC siRNA into HOS and U2OS cells. After
72 h of transfection, western blot analysis was performed to
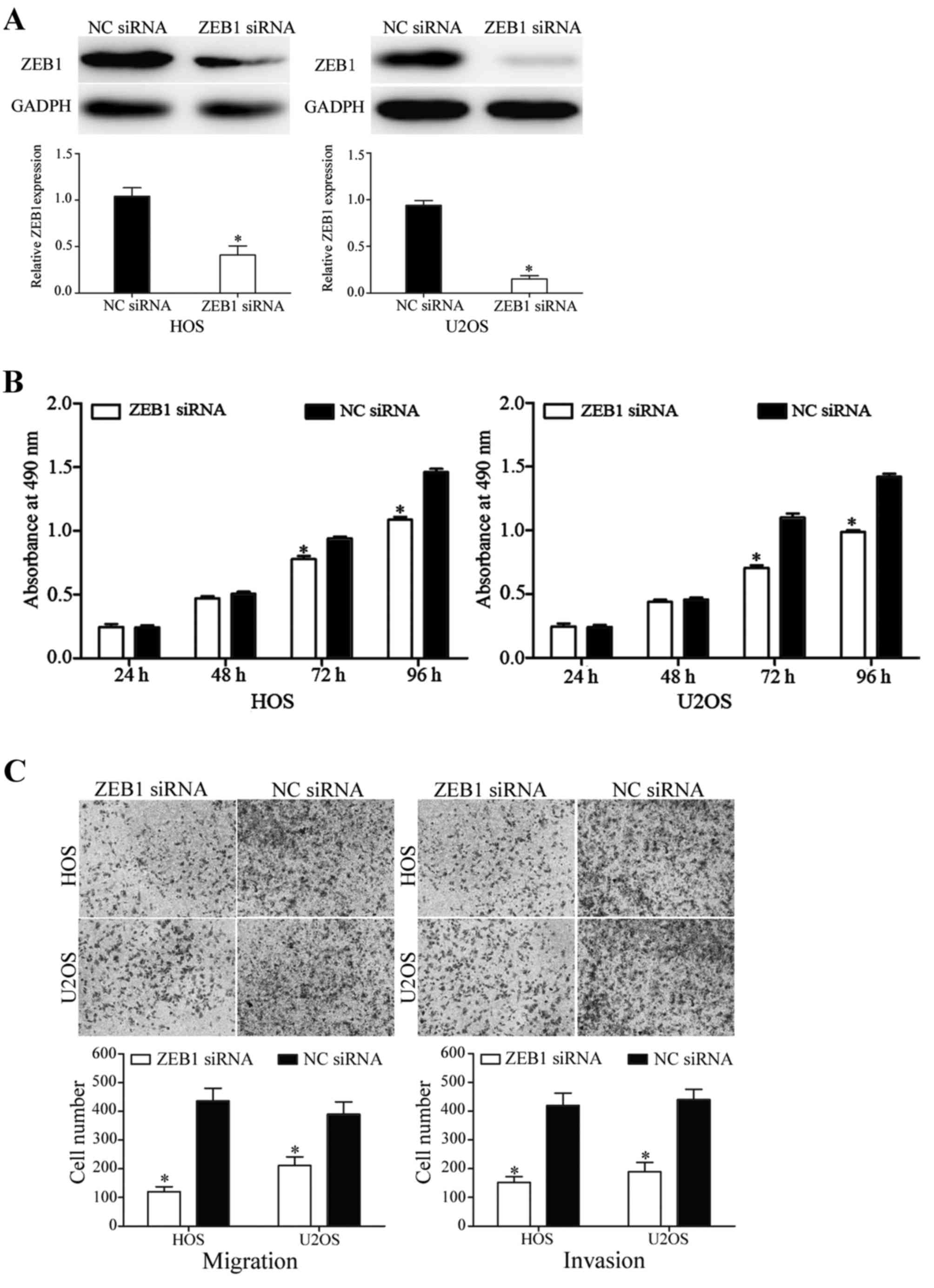
determine ZEB1 protein expression. As indicated in Fig. 5A, ZEB1 was significantly downregulated
in miR-150 mimic-transfected HOS and U2OS cells (P<0.05).
In the MTT assay, the knockdown of ZEB1 decreased
HOS and U2OS cell proliferation (P<0.05; Fig. 5B). In addition, in migration and
invasion assays, silencing of ZEB1 inhibited HOS and U2OS cell
migratory and invasive abilities (P<0.05; Fig. 5C). These results demonstrated that the
functions of ZEB1 siRNA were similar to those induced by miR-150 in
HOS and U2OS cells, suggesting ZEB1 may be a functional target of
miR-150 in OS.
Discussion
Since their discovery, miRNAs have received
considerable attention (19). Several
studies have indicated that miRNAs contribute to various
physiological and pathological processes, and participate in the
initiation and progression of cancer (20,21).
Numerous studies have demonstrated that miR-150 is downregulated in
certain types of human cancer, including pancreatic cancer
(22), esophageal squamous cell
carcinoma (23), colorectal cancer
(24), hepatocellular carcinoma
(25), ovarian cancer (26) and malignant lymphoma (27). However, miR-150 was also reported to
be upregulated in prostate (28),
non-small cell lung (29), breast
(30) and gastric cancer (31). These conflicting studies suggest that
miR-150 expression levels in cancer exhibit tissue specificity.
In the present study, miR-195 was revealed to be
significantly downregulated in OS tissues and cell lines. In
addition, a low expression level of miR-150 was significantly
associated with clinical stage and distant metastasis. These
results suggest that miR-150 may exhibit tumor-suppressive roles in
OS carcinogenesis and development.
The collective results from numerous previous
functional studies demonstrated that miR-150 may be a tumor
suppressor. For example, in pancreatic cancer, patients whose
tumors were associated with low miR-150 expression exhibited higher
mortality rates, compared with patients whose tumors exhibited high
miR-150 expression. In addition, the upregulation of miR-150
decreased pancreatic cancer cell proliferation, migration,
invasion, clonogenicity and cell cycle progression, and promoted
apoptosis via the blockade of c-Myb and mucin 4, cell surface
associated (32). In colorectal
cancer, a low miR-150 expression group exhibited shorter survival
rate and worse response to adjuvant chemotherapy compared with a
high miR-150 expression group (33).
miR-150 inhibited colorectal cancer cell growth and induced cell
apoptosis by directly targeting c-Myb (34). Yokobori et al (23) revealed that low expression levels of
miR-150 in esophageal squamous cell carcinoma were significantly
associated with tumor depth, lymph node metastasis, lymphatic
invasion, venous invasion, clinical staging and poor prognosis. In
the aforementioned study, the upregulation of miR-150 inhibited
esophageal squamous cell carcinoma cell proliferation and
tumorigenicity in vivo. Therefore, upregulating miR-150 or
providing analogous pharmaceutical compounds exogenously, may be an
effective therapy for tumors resulting from the activation or
overexpression of these oncogenes.
The functions of miRNAs are tissue-type dependent.
miR-150 has been verified as an oncogene in a number of different
types of cancer (28,29,35). For
example, in prostate cancer, miR-150 was markedly upregulated, and
the high expression of miR-150 was positively associated with tumor
recurrence and metastasis in prostate cancer (28). In addition, patients with prostate
cancer and high miR-150 expression exhibited significantly poorer
overall survival and disease-free survival compared with those
patients with low miR-150 expression (28). The 5-year overall survival rate was
55.93% in patients with prostate cancer with low miR-150
expression, whereas it was 35.19% in patients with high miR-150
expression (28). In non-small cell
lung cancer, a high expression level of miR-150 was correlated with
lymph node metastasis, distant metastasis and clinical tumor node
metastasis stage. The 5-year overall survival rate was 69.2% in the
low miR-150 expression group; however, in the high miR-150
expression group, it was 40.8% (36).
In addition, the downregulation of miR-150 enhanced non-small cell
lung cancer proliferation and migration, and inhibited cell
apoptosis through targeting B-cell lymphoma 2 antagonist/killer 1,
SRC kinase signaling inhibitor 1 and tumor protein 53 (29,36,37). Huang
et al (30) revealed that the
ectopic expression of miR-150 induced breast cancer cell
proliferation and clonogenicity, and suppressed cell apoptosis by
directly targeting PX27. These findings also suggested that miR-150
may have important functions in these types of cancer, and may be
investigated as a potential therapeutic gene for the treatment of
these cancer types.
In the present study, miR-150 was revealed to
inhibit OS cell proliferation, migration and invasion in
vitro. Identification of miR-150 target mRNAs is important for
understanding the functions of miR-150 in OS carcinogenesis and
progression, and to investigate novel targeted therapies for OS.
The present study identified ZEB1 as a direct target gene of
miR-150 in vitro. ZEB1 is a member of the zinc finger
family, which is located on the short arm of human chromosome 10
(38). Wang et al (39) verified that ZEB1 is involved in cancer
progression, and that it is considered an important transcriptional
regulator of E-cadherin. In OS, ZEB1 was revealed to be upregulated
in patients with lung metastases compared with patients without
lung metastases. In addition, the expression of ZEB1 in OS tissues
was increased with increasing Enneking stage (38). These results indicated that ZEB1 may
contribute to OS metastasis. Therefore, additional studies are
required with respect to ZEB1 as a potential target for the
inhibition of OS metastasis.
In conclusion, the present study demonstrated that
miR-150 was significantly downregulated in OS tissues and cell
lines. Low expression of miR-150 was associated with clinical stage
and distant metastasis. In addition, miR-150 inhibited OS cell
growth, migration and invasion, and ZEB1 was identified as a direct
target of miR-150 in vitro. These findings suggest that
miR-150 targets ZEB1 to inhibit OS growth and metastasis, a
mechanism that may be investigated as a therapeutic regimen to
prevent rapid growth and early metastasis in OS.
References
|
1
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the surveillance, epidemiology, and end results program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jin J, Cai L, Liu ZM and Zhou XS:
miRNA-218 inhibits osteosarcoma cell migration and invasion by
down-regulating of TIAM1, MMP2 and MMP9. Asian Pac J Cancer Prev.
14:3681–3684. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Han K, Chen X, Bian N, Ma B, Yang T, Cai
C, Fan Q, Zhou Y and Zhao TB: MicroRNA profiling identifies MiR-195
suppresses osteosarcoma cell metastasis by targeting CCND1.
Oncotarget. 6:8875–8889. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chou AJ, Geller DS and Gorlick R: Therapy
for osteosarcoma: where do we go from here? Paediatr Drugs.
10:315–327. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meyers PA, Heller G, Healey J, Huvos A,
Lane J, Marcove R, Applewhite A, Vlamis V and Rosen G: Chemotherapy
for nonmetastatic osteogenic sarcoma: The memorial sloan-kettering
experience. J Clin Oncol. 10:5–15. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lv H, Guo J, Li S and Jiang D: inhibitor
reduces the proliferation and migration in osteosarcoma MG-63
cells. Exp Ther Med. 8:1575–1580. 2014.PubMed/NCBI
|
|
7
|
PosthumaDeBoer J, Witlox MA, Kaspers GJ
and van Royen BJ: Molecular alterations as target for therapy in
metastatic osteosarcoma: A review of literature. Clin Exp
Metastasis. 28:493–503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Diao CY, Guo HB, Ouyang YR, Zhang HC, Liu
LH, Bu J, Wang ZH and Xiao T: Screening for metastatic osteosarcoma
biomarkers with a DNA microarray. Asian Pac J Cancer Prev.
15:1817–1822. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bentwich I, Avniel A, Karov Y, Aharonov R,
Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al:
Identification of hundreds of conserved and nonconserved human
microRNAs. Nat Genet. 37:766–770. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Winter J, Jung S, Keller S, Gregory RI and
Diederichs S: Many roads to maturity: MicroRNA biogenesis pathways
and their regulation. Nat Cell Biol. 11:228–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu W, Zhao ZY, Shi L and Yuan WD: Tissue
microRNA-126 expression level predicts outcome in human
osteosarcoma. Diagn Pathol. 10:1162015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: microRNAs in cancer management. Lancet Oncol.
13:e249–e258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tahara H, Kay MA, Yasui W and Tahara E:
MicroRNAs in Cancer: The 22nd Hiroshima Cancer Seminar/the 4th
Japanese Association for RNA Interference Joint International
Symposium, 30 August 2012, Grand Prince hotel Hiroshima. Jpn J Clin
Oncol. 43:579–582. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: MicroRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang F, Ren X and Zhang X: Role of
microRNA-150 in solid tumors. Oncol Lett. 10:11–16. 2015.PubMed/NCBI
|
|
20
|
Weiland M, Gao XH, Zhou L and Mi QS: Small
RNAs have a large impact: Circulating microRNAs as biomarkers for
human diseases. RNA Biol. 9:850–859. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ebert MS and Sharp PA: Roles for microRNAs
in conferring robustness to biological processes. Cell.
149:515–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Srivastava SK, Bhardwaj A, Singh S, Arora
S, Wang B, Grizzle WE and Singh AP: MicroRNA-150 directly targets
MUC4 and suppresses growth and malignant behavior of pancreatic
cancer cells. Carcinogenesis. 32:1832–1839. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yokobori T, Suzuki S, Tanaka N, Inose T,
Sohda M, Sano A, Sakai M, Nakajima M, Miyazaki T, Kato H and Kuwano
H: MiR-150 is associated with poor prognosis in esophageal squamous
cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci.
104:48–54. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pizzini S, Bisognin A, Mandruzzato S,
Biasiolo M, Facciolli A, Perilli L, Rossi E, Esposito G, Rugge M,
Pilati P, et al: Impact of microRNAs on regulatory networks and
pathways in human colorectal carcinogenesis and development of
metastasis. BMC Genomics. 14:5892013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu F, Lu Z, Chen B, Dong P and Zheng J:
microRNA-150: A promising novel biomarker for hepatitis B
virus-related hepatocellular carcinoma. Diagn Pathol. 10:1292015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jin M, Yang Z, Ye W, Xu H and Hua X:
MicroRNA-150 predicts a favorable prognosis in patients with
epithelial ovarian cancer, and inhibits cell invasion and
metastasis by suppressing transcriptional repressor ZEB1. PLoS One.
9:e1039652014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Watanabe A, Tagawa H, Yamashita J, Teshima
K, Nara M, Iwamoto K, Kume M, Kameoka Y, Takahashi N, Nakagawa T,
et al: The role of microRNA-150 as a tumor suppressor in malignant
lymphoma. Leukemia. 25:1324–1334. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dezhong L, Xiaoyi Z, Xianlian L, Hongyan
Z, Guohua Z, Bo S, Shenglei Z and Lian Z: miR-150 is a factor of
survival in prostate cancer patients. J BUON. 20:173–179.
2015.PubMed/NCBI
|
|
29
|
Gu XY, Wang J, Luo YZ, Du Q, Li RR, Shi H
and Yu TP: Down-regulation of miR-150 induces cell proliferation
inhibition and apoptosis in non-small-cell lung cancer by targeting
BAK1 in vitro. Tumour Biol. 35:5287–5293. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang S, Chen Y, Wu W, Ouyang N, Chen J,
Li H, Liu X, Su F, Lin L and Yao Y: miR-150 promotes human breast
cancer growth and malignant behavior by targeting the pro-apoptotic
purinergic P2X7 receptor. PLoS One. 8:e807072013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu Q, Jin H, Yang Z, Luo G, Lu Y, Li K,
Ren G, Su T, Pan Y, Feng B, et al: MiR-150 promotes gastric cancer
proliferation by negatively regulating the pro-apoptotic gene EGR2.
Biochem Biophys Res Commun. 392:340–345. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang K, He M, Cai Z, Ni C, Deng J, Ta N,
Xu J and Zheng J: A decrease in miR-150 regulates the malignancy of
pancreatic cancer by targeting c-Myb and MUC4. Pancreas.
44:370–379. 2015.PubMed/NCBI
|
|
33
|
Ma Y, Zhang P, Wang F, Zhang H, Yang J,
Peng J, Liu W and Qin H: miR-150 as a potential biomarker
associated with prognosis and therapeutic outcome in colorectal
cancer. Gut. 61:1447–1453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Feng J, Yang Y, Zhang P, Wang F, Ma Y, Qin
H and Wang Y: miR-150 functions as a tumour suppressor in human
colorectal cancer by targeting c-Myb. J Cell Mol Med. 18:2125–2134.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang N, Wei X and Xu L: miR-150 promotes
the proliferation of lung cancer cells by targeting P53. FEBS Lett.
587:2346–2351. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yin QW, Sun XF, Yang GT, Li XB, Wu MS and
Zhao J: Increased expression of microRNA-150 is associated with
poor prognosis in non-small cell lung cancer. Int J Clin Exp
Pathol. 8:842–846. 2015.PubMed/NCBI
|
|
37
|
Cao M, Hou D, Liang H, Gong F, Wang Y, Yan
X, Jiang X, Wang C, Zhang J, Zen K, et al: miR-150 promotes the
proliferation and migration of lung cancer cells by targeting SRC
kinase signalling inhibitor 1. Eur J Cancer. 50:1013–1024. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shen A, Zhang Y, Yang H, Xu R and Huang G:
Overexpression of ZEB1 relates to metastasis and invasion in
osteosarcoma. J Surg Oncol. 105:830–834. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Y, Yan S, Liu X, Zhang W, Li Y, Dong
R, Zhang Q, Yang Q, Yuan C, Shen K and Kong B: miR-1236-3p
represses the cell migration and invasion abilities by targeting
ZEB1 in high-grade serous ovarian carcinoma. Oncol Rep.
31:1905–1910. 2014.PubMed/NCBI
|