Introduction
Tissue trauma is a typical pathological disease in
which normal tissue undergoes structural destruction resulting in
functional impairment (1). Therefore,
rapid wound healing is an important topic for further
investigation. An established hypothesis is to use stem cells to
repair the destroyed tissue (2).
Mesenchymal stem cells (MSCs) obtained from Wharton's jelly are
adult stem cells that have multiple differentiation pathways and
self-renewal potential. Compared with other tissue-derived stem
cells, the umbilical cord is readily available and has limited
immunogenicity and bioethical considerations (3). In addition, previous studies have
identified that microRNA (miRNA/miR) may serve an important role in
wound healing (4,5). However, the role of miRNAs in human
umbilical cord mesenchymal stem cells (HUMSCs) for wound healing
remains to be elucidated.
miRNAs are a type of small non-coding RNA of between
19 and 25 nucleotides in length, which regulate gene expression
through targeting the 3′-untranslated region (3′-UTR) of mRNA
resulting in translation repression or degradation (6). miR-590 has been identified as a tumor
promoter by repressing proliferation-associated genes and enhancing
cell growth in renal carcinoma cancer (7), hepatocellular carcinoma (8), acute myeloid leukemia (9), cervical cancer (10) and lung cancer (11). According to previous studies,
eukaryotic translation elongation factor 1ε1 (12), transforming growth factor (TGF) β
receptor II (8), S100 calcium-binding
protein A10 (9), polybromo 1
(7) and cell adhesion molecule with
homology with L1 cell adhesion molecule (10) are targets of miR-590. The
downregulated expression of miR-590 induces fibrosis in
nicotine-induced atrial fibrosis (13); however, the potential effects of
miR-590 on cell proliferation and extracellular matrix (ECM)
synthesis are poorly understood in stem cells. Smad7 has been
established to serve important roles in the TGFβ signaling pathway
(14). Previous studies have
demonstrated that Smad7 is a vital factor in adult homoeostasis and
embryonic development, and abnormal expression of Smad7 is
associated with human diseases via ECM synthesis and the
development of tumors (15,16). However, the role of miRNA/Smad7
regulation in traumatic disease is poorly understood. Jia et
al (17) demonstrated that
miR-17-5p regulated osteoblastic differentiation and proliferation
by targeting Smad7, and Liu et al (18) identified that miR-21 affected the ECM
deposit in lung by targeting Smad7. In addition, miR-195 (19), miR-16 (20), miR-132 (21), miR-15 (22) and miR-92a (23) regulate Smad7 expression levels. The
results of the present study demonstrate that miR-590 promotes the
proliferation of HUMSCs and induces ECM expression in HUMSCs by
targeting Smad7. miR-590 may be a key regulator in stem cell growth
and tissue repair.
Materials and methods
Cell culture
A HUMSC human stem cell line, was purchased from the
American Type Culture Collection (cat. no. CSC-7700W; Manassas, VA,
USA) and cultured in MSC growth medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with low-glucose
Dulbecco's modified Eagle's medium (HyClone; GE Healthcare Life
Sciences, Logan, UT, USA) plus 10% heat-inactivated fetal bovine
serum (HyClone; GE Healthcare Life Sciences), 100 U/ml penicillin
and 100 U/ml streptomycin (Beyotime Institute of Biotechnology,
Haimen, China). Cells were cultured in a humidified incubator at
37°C in at atmosphere containing 5% CO2.
Transfection
The miR-590 mimic and small interfering RNAs
(siRNAs) were synthesized by Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). The oligonucleotide sequence of Smad7 siRNA was
as follows: 5′-AAGCUCAAUUCGGACAACAAG-3′. miRNA mimic and siRNA
transfections were performed using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. A total of 50 or 100 nM miR-590 mimic or
negative control, and 50 nM siRNA, were used for transfection in
Opti-MEM serum-free medium (Gibco; Thermo Fisher Scientific, Inc.).
A total of 5×106 HUMSCs were plated in each well of
24-well plates (Invitrogen; Thermo Fisher Scientific, Inc.). Total
RNA and protein were prepared at 36 or 48 h respectively following
transfection for further experiments.
Quantitative analysis of miRNAs and
mRNAs
Total RNA was extracted from cultured HUMSCs using
TRizol (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. The reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) was used to analyze the
expression of miR-590 with primers from Guangzhou RiboBio Co.,
Ltd., and U6 was selected as internal reference. For quantitative
analysis of Smad7 expression, 500 ng total RNA was used for the
synthesis of random-primed single-stranded cDNA using a
First-Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.)
and qPCR was performed using SYBR-Green S Mixture (CWBIO, Beijing,
China). The relative quantity of Smad7 was normalized to 18S as per
a previously described method (24).
The thermocycling conditions were; pre-degeneration at 95°C for 5
min, degeneration at 94°C for 20 sec, annealing at 56°C for 30 sec,
extension at 72°C for 10 sec (25 cycles), extension at 72°C for 2
min and a final round at 16°C for 5 min. The sequence of PCR
primers were as follows: 18S forward, 5′-GTAACCCGTTGAACCCCATT-3′;
18S reverse, 5′-CCATCCAATCGGTAGTAGCG-3′; Smad7 forward,
5′-CCTCCTTACTCCAGATACC-3′; Smad7 reverse,
5′-GTCTTCTCCTCCCAGTATG-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′, U6
reverse, 5′-AACGCTTCACGAATTTGCGT-3′. Three independent experiments
were performed.
Protein extraction and western blot
analysis
At 48 h after transfection, HUMSCs were lysed using
radioimmunoprecipitation lysis buffer containing 2 mM
phenylmethylsulfonyl fluoride (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). Each sample protein concentration was
determined using a Bicinchoninic Protein Assay kit (Beyotime
Institute of Biotechnology) and equal amounts of total protein (80
µg) were separated by SDS-PAGE (12% gels) and transferred onto
polyvinylidene membranes (Merck KGaA). Membranes were blocked with
5% non-fat milk in Tris-buffered saline containing 0.05% Tween-20
for 1 h at room temperature, incubated overnight with primary
antibody at 4°C, washed three times with 1X TBST buffer for 5 min
each time at room temperature, incubated with secondary antibody
for 1 h at room temperature and finally visualized by
Chemiluminescence Horseradish Peroxidase (HRP) Substrate reagent
(Merck KGaA). The antibodies used in the present study were as
follows: Mouse anti-β-tubulin (HC101; 1:5,000; Beijing Transgen
Biotech Co., Ltd., Beijing, China), rabbit anti-Smad7 (25840-1-AP;
1:4,000; ProteinTech Group, Inc., Chicago, IL, USA), rabbit
anti-collagen I (ab209539; 1:4,000; Abcam, Cambridge, UK), rabbit
anti-fibronectin (FN) (ab6584; 1:4,000; Abcam), HRP-labeled goat
anti-mouse Immunoglobulin G (IgG; 10004302-1; 1:5,000; Beijing
Zhongshan Jinqiao Biotechnology Co., Ltd., Beijing, China) and
HRP-labeled goat anti-rabbit IgG (A21020; 1:5,000; Beijing
Zhongshan Jinqiao Biotechnology Co., Ltd.). Western blotting was
quantified using the greyscale scanning method (Image J v1.48;
National Institutes of Health, Bethesda, MD, USA).
Luciferase assay
Predictions for the potential binding sites of
miR-590 in the 3′-UTR of Smad7 mRNA was performed using 3 different
online programs: TargetScan (http://www.targetscan.org), microRNA.org (http://www.microrna.org) and the miRBase (http://www.mirbase.org). Fragments of the 3′-UTR of
Smad7 mRNA, including the wild-type (wt) or the mutated (mt) site,
was amplified and cloned downstream of the firefly luciferase in
the pCDNA3.1-luciferase vector (Invitrogen; Thermo Fisher
Scientific, Inc.). The mutation of the predicted binding site was
produced using PCR-mediated site-specific mutation. For construct
the vectors, the following primer sequences were used: wt 3′-UTR of
Smad7 forward 5-CGCGGATCCCCGCGTGCGGAGGGGACAGA-3 and reverse
5-CCGCTCGAGGGAGTCCTTTCTCTCTCAAAGC-3. In the 3′UTR mutants, Smad7
3′UTR mutants from the 122–1,129 nucleotide sequence complementary
to nucleotides 2–5 of miR-590 was mutated to the same sequence as
that in miR-590 (from ATAAGCTA to TTATGCAA). For transfection,
HUMSCs (5×106 cells/well) were seeded on 24-well plates
were transfected with 250 ng pCDNA3.1-luciferase-Smad7 (wt and mt),
pRL-SV40-Renilla and pCDNA3.1-luciferase, and 50 nM miR-590
mimic or negative control. After 24 h, cells were lysed using 200
µl radioimmunoprecipitation assay lysis buffer (Beyotime Institute
of Biotechnology, Shanghai, China) at room temperature for 15 min,
centrifuged at 8,500 × g at room temperature for 5 min to collect
the supernatant and assayed by Dual-Luciferase Reporter assay kit
(Promega Corporation, Madison, WI, USA), according to the
manufacturer's protocol. Three independent experiments were
performed.
5-Ethynyl-20-deoxyuridine (EdU)
assay
For determination of cell proliferation, HUMSCs were
suspended in MSC growth medium consisting of low-glucose Dulbecco's
modified Eagle's medium containing 10% fetal bovine serum and
cultured in 24-well (5×104 cells/100 μl) plates 1 day
before transfection. After 12 h, HUMSCs were transfected with
miR-590 mimic (10 nM), miRNA control (10 nM), siRNA-Smad7 (10 nM)
or siRNA-control (10 nM). Following transfection, cell
proliferative ability was evaluated at 36 h using a EdU DNA
Proliferation and Detection kit to label cellular DNA (Cell-Light
5-ethynyl-20-deoxyuridine Apollo 567 kit; Guangzhou RiboBio Co.,
Ltd.), according to the manufacturer's protocol. The percentage of
HUMSCs proliferation was determined by counting the EdU
proliferation cells in 1,000 cells. Subsequently, the fluorescence
was used to detect DNA synthesis for cell proliferation at an
ultraviolet light wavelength of 480 nm. All experiments were
performed in triplicate and independently.
Immunofluorescence microscopy
Following transfection with si-Smad7 or si-control
(5×106 cells per well), cells cultured on glass
coverslips were fixed with pre-chilled acetone for 10 min at −20°C.
Cells were permeabilized and blocked with 1% Triton X-100 (in PBS)
and 5% bovine serum albumin in PBS, respectively, each for 1 h at
room temperature. The rabbit polyclonal antibody against collagen I
(dilution, 1:500; ab209539; Abcam) was used to probe collagen I
overnight at 4°C and goat anti-rabbit IgG-CruzFluor™ 594 (dilution,
1:1,000; SC362282; Santa Cruz Biotechnology, Inc., Dallas, TX, USA)
was used to determine rabbit IgG for 1 h at room temperature.
Subsequently, the glass covers were washed with 1X PBS at room
temperature for 3×5 min.
Finally, nuclei were stained with DAPI (dilution,
1:4,000; Santa Cruz Biotechnology, Inc.) at room temperature for 10
min. The coverslips were immobilized on the glass slides using 50%
glycerol in PBS and viewed by fluorescence microscopy (ECLIPSE
Ti-s; Nikon Corporation, Tokyo, Japan; 8–10 fields of view;
magnification, ×200) and images were captured with a SPOT
Diagnostic charge-coupled device camera (Spot RT Color Diagnostic
Instruments, Sterling Heights, MI, USA; ~500 cells per glass
coverslip were counted under fluorescence microscopy). The
intensity of fluorescence was evaluated using Image J v1.48
software following image capture (National Institutes of
Health).
Statistical analysis
All experimental data were from triplicate
experiments independently and data were expressed as a mean ±
standard deviation. GraphPad Prism (version 5; GraphPad Software,
Inc., La Jolla, CA, USA) was used for data analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-590 induces the proliferation of
HUMSCs and ECM synthesis
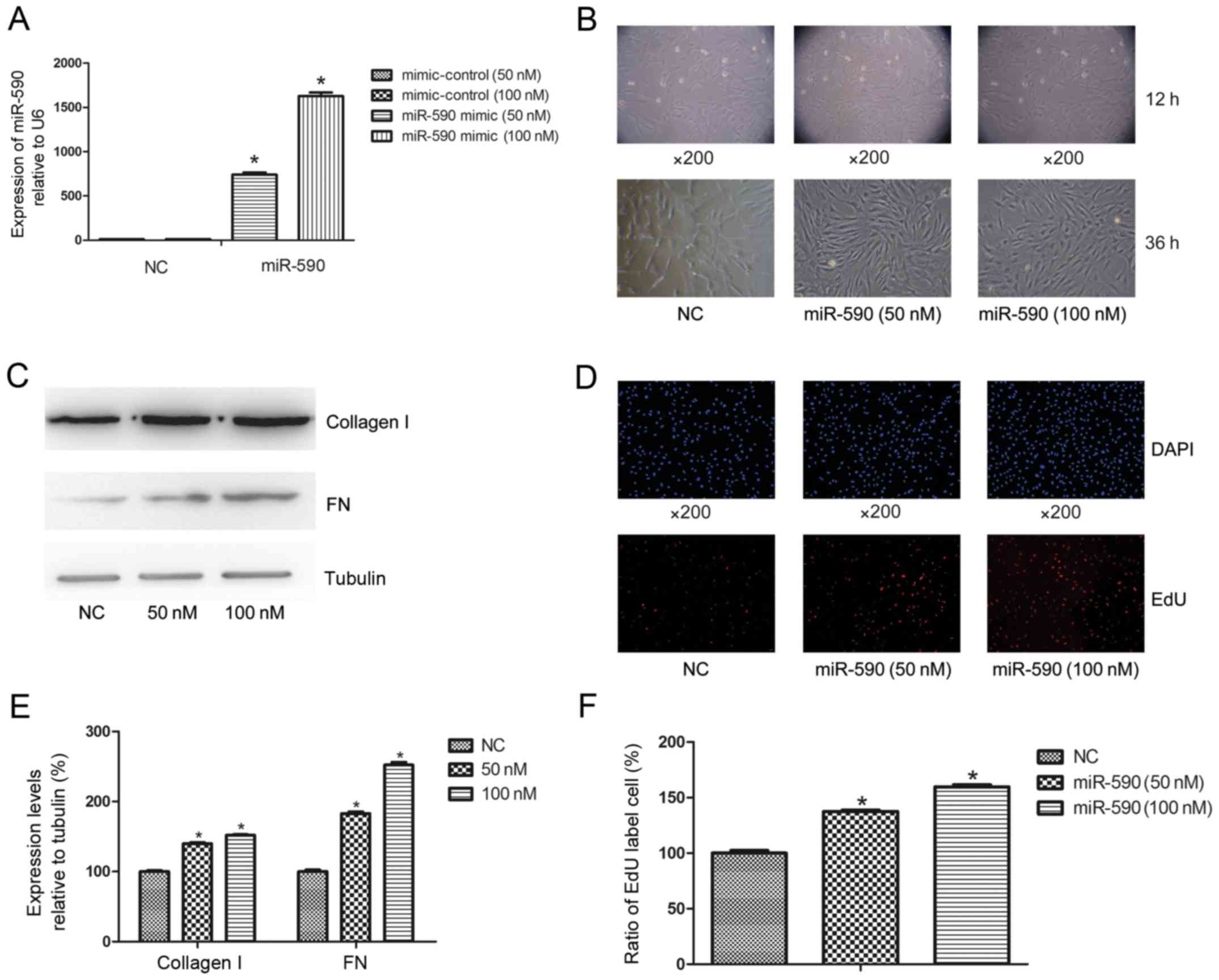
The effects of miR-590 overexpression on HUMSCs,
including cell morphological changes, proliferation and ECM
synthesis, were evaluated. Following transfection with miR-590
mimic, the expression level of miR-590 in HUMSCs significantly
increased at 50 nM (66.00±4.20-fold; P<0.0003) and 100 nM
(138.00±11.40-fold; P<0.0001) compared with the negative control
at 36 h (Fig. 1A), whereas cell
morphology altered from the typical mesenchymal cell to
fibroblast-like cell morphology (Fig.
1B). Western blot analysis indicated that there was increased
expression levels of miR-590 leading to increases in the levels of
collagen I and FN of 39.86±2.18 and 82.99±3.16% for 50 nM, and
52.21±2.46 and 152.03±4.58% for 100 nM, respectively, compared with
the control group (Fig. 1C and E).
Cell EdU analysis demonstrated that the proliferation of HUMSCs
were increased to 38.15±2.14 and 60.14±4.52% at 50 nM and 100 nM,
respectively, compared with the controls (Fig. 1D and F).
Predicting miR-590-binding sites in
the 3′-UTR of Smad7 mRNA
To identify the downstream targets of miR-590, 3
independent online databases (TargetScan, microRNA.org and the miRBase) were selected to predict
potential targets. In total, 110 putative mRNAs were predicted by
the 3 databases (Fig. 2A) and among
these Smad7 was of particular interest as previous studies have
demonstrated its role in the cell proliferation and its effect on
ECM (25). Following analysis of the
sequence of Smad7 3′-UTR [1,520 base pairs (bp)], the only
predicted binding site of miR-590 was identified at 1,122–1,129 bp
(Fig. 2B).
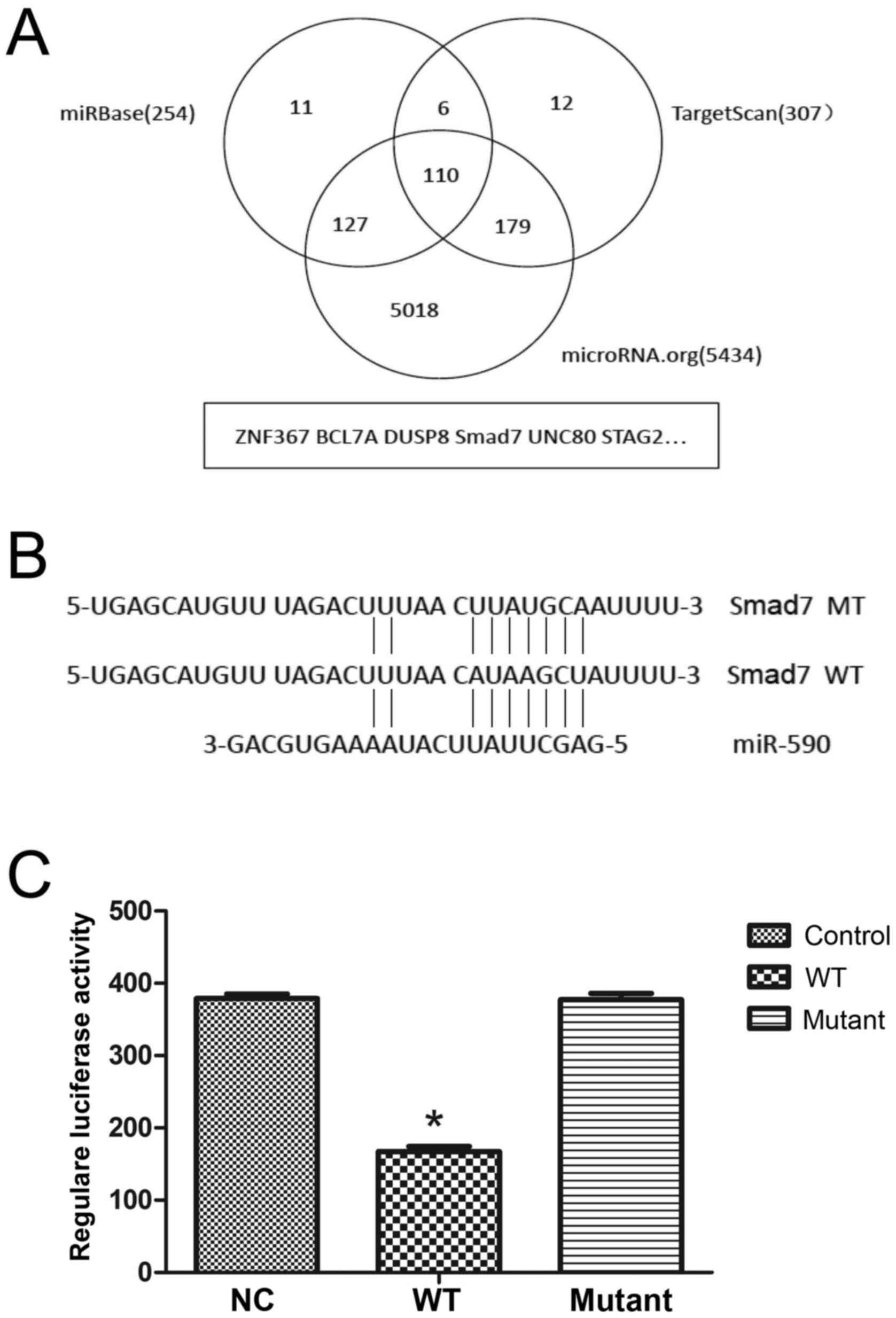 | Figure 2.miR-590 directly binds to the 3′-UTR
of Smad7 mRNA. (A) Bioinformatic analyses predicted 110 potential
target genes for miR-590 using 3 independent databases. (B) The
predicted binding site of miR-590 in the 3′-UTR of Smad7 mRNA
according to the miR-590 sequence and the fragment containing the
mutated binding site was amplified. (C) In the pCDNA3.1-luciferase
vector, the Smad7 mRNA 3′-UTR fragment containing the wild-type or
mutated site was inserted downstream into the reporter gene. The
plasmids were cotransfected with miR-590 mimic or mimic control and
the relative luciferase activity, normalized by Renilla, was
significantly decreased using the wild-type sequence compared with
that of the mutated site (46.70±5.81%). *P<0.05 vs. NC. NC,
negative control; WT, wild-type; MT, mutated site; Smad7, Mothers
Against Decapentaplegic Homolog 7; UTR, untranslated region; miR,
microRNA; ZNF367, zinc finger 367; BCL7A, B-cell lymphoma 7A;
DUSP8, dual-specificity phosphatase 8; UNC80, uncoordinated 80;
STAG2, stromal antigen 2. |
Luciferase assay of miR-590 and Smad7
3′-UTR in HUMSCs
To determine whether Smad7 is a target of miR-590, a
luciferase reporter vector was inserted with wt or mt Smad7 mRNA
3′-UTR, and this was cotransfected with miR-590 mimic.
Overexpression of miR-590 in HUMSCs for 36 h significantly
decreased (P<0.05) the luciferase activity of the luciferase
reporter plasmid fused to wt Smad7 mRNA 3′-UTR (Fig. 2C), whereas the luciferase activity
with mt Smad7 mRNA 3′-UTR was not affected (Fig. 2C).
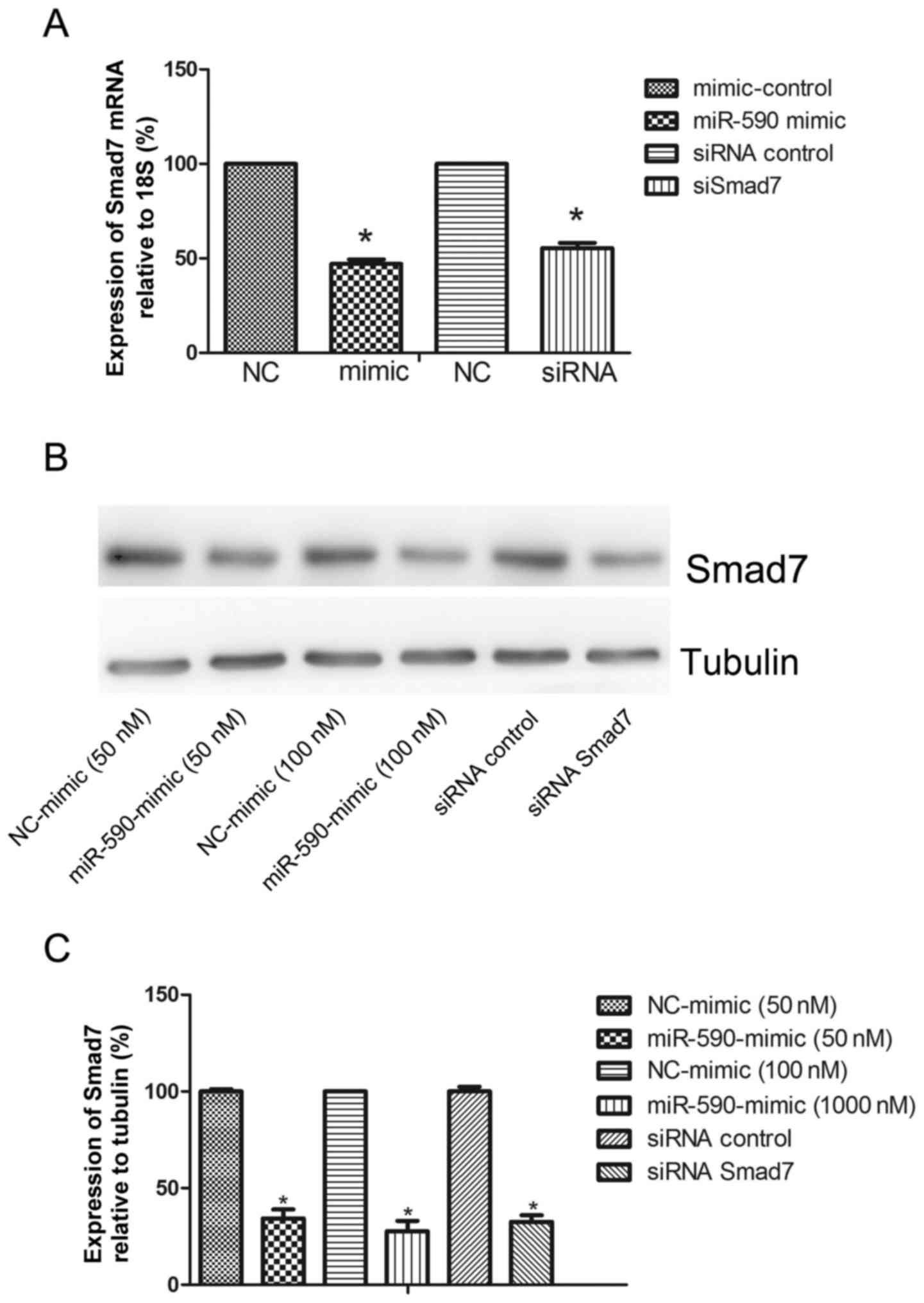
Overexpression of miR-590 reduces
Smad7 expression
To verify whether Smad7 is a functional upstream
gene of miR-590, the mRNA and protein expression levels of Smad7
were analyzed. The miR-590 mimic and the negative control were
transfected into HUMSCs. The expression of Smad7 at 36 and 48 h
after transfection was detected using qPCR and western blot
analysis for mRNA and protein, respectively. The overexpression of
miR-590 downregulated the expression levels of Smad7 mRNA at 36 h
(51.55±3.83%; Fig. 3A). A similar
effect was also identified on the protein level of Smad7
(65.73±8.09% at 50 nM and 72.36±9.38% at 100 nM; Fig. 3B and C).
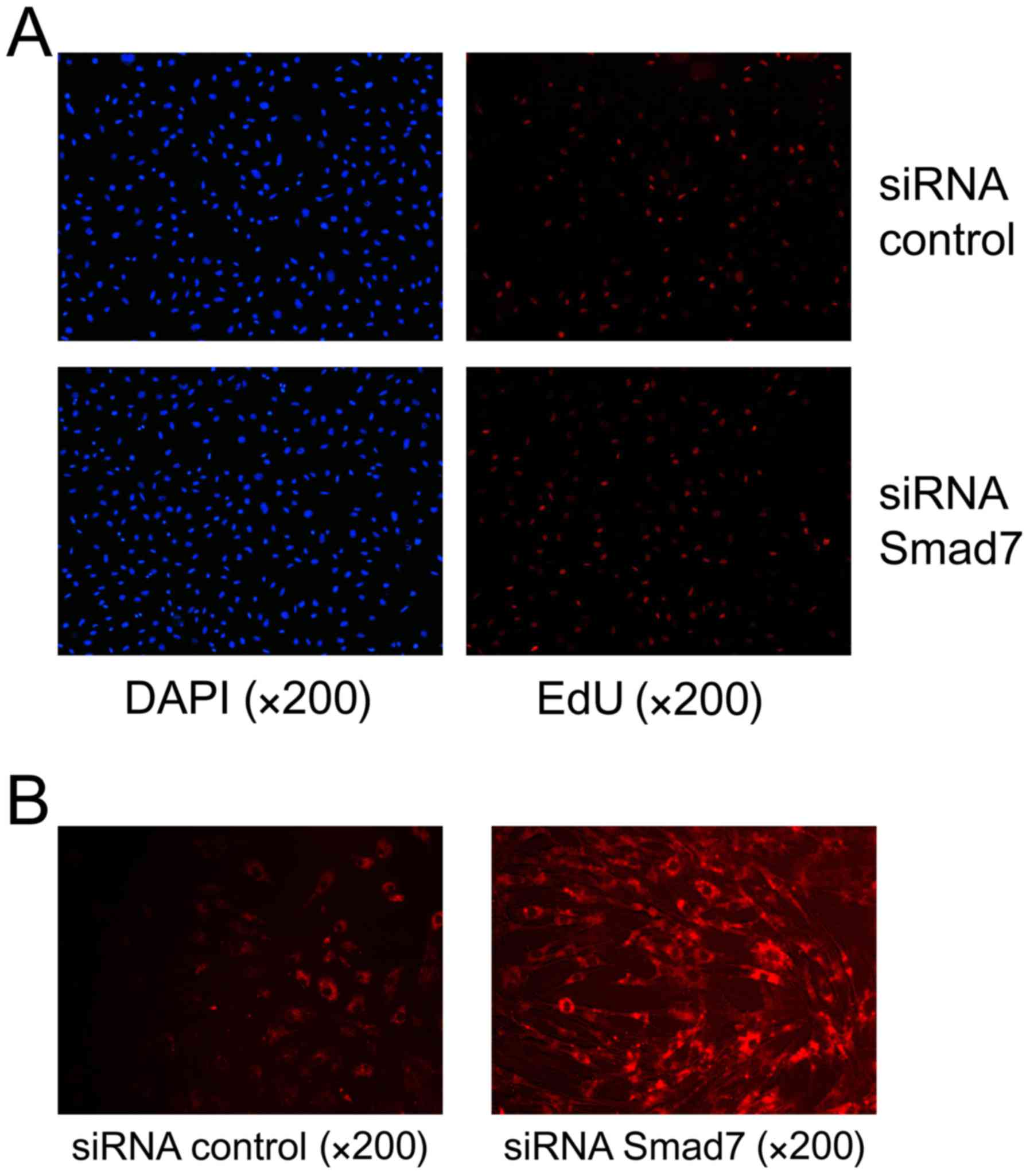
Smad7 siRNA facilitates the
proliferation of HUMSCs and ECM enhancement
The importance of Smad7 as a functional target for
miR-590 was evaluated. HUMSCs were transfected with Smad7 siRNA or
scramble control for 36 or 48 h, and the mRNA (43.72±6.48%;
Fig. 3A) and protein (67.49±6.01%;
Fig. 3C) levels of Smad7 were
significantly decreased using Smad7 siRNA. EdU assay and
immunofluorescence microscopy were used to evaluate the
proliferative ability and ECM expression in the HUMSCs,
respectively. The proliferative ability of HUMSCs was increased to
30.17±2.31% compared with the control at 36 h (Fig. 4A). Immunofluorescence analysis
identified decreased expression of Smad7 promoted collagen I
upregulation and the proportion of collagen I increased to
67.86±8.18% compared with the control (Fig. 4B).
Discussion
Trauma is a predominant surgical outcome and
therefore the problem of effective wound healing is a focus of
investigation for researchers and clinicians. A series of
pathological alterations, which are overlapping and interrelated,
are involved in wound healing. For example, a number of cells and
ECM components participate in the reconstruction (26). In tissue engineering, stem cell
therapy is typically investigated. The theory of stem cell therapy,
including cell-replacement (27) and
ECM enhancement (28), may explain
the underlying mechanisms involved in effective wound healing.
miRNAs are a type of small non-coding RNA that regulate gene
expression by directly binding to the 3′-UTR of mRNA (29). An increasing number of studies have
indicated that miRNAs have vital roles in numerous biological
processes and in pathological disease, and previous studies have
suggested that miRNAs participate in cell proliferation, apoptosis,
differentiation and fibrosis (29,30).
However, the distinct role and underlying molecular mechanism of
the involvement of miRNAs in stem cell therapy for the repair of
trauma are yet to be elucidated.
In the past decades (2005 to 2015), miR-590 has been
identified in numerous types of disease, including atherosclerosis
(31), myocarditis (32), hepatocellular carcinoma (8) and renal cancer (7). In the present study, the effect of
miR-590 on HUMSCs, including proliferation and ECM accumulation,
were evaluated. Wound healing is a complex process involving the
proliferation, migration, differentiation and ECM accumulation of
local unwounded cells. In diabetic ulcers, a classic model of wound
healing, miR-198 was significantly increased and inhibited
migratory ability by targeting plasminogen activator urokinase,
diaphanous related formin 1 and laminin subunit γ2 (33). Considering rapid wound healing is
critical and stem cell therapy is considered promising, the
investigation of miR-590 effect on wound healing was required. In
the present study, miR-590 overexpression promoted cell
proliferation in HUMSCs and ECM enhancement was demonstrated using
western blotting and immunofluorescence microscopy. To explain the
effects of miR-590, the potential targets of miR-590 were
identified using bioinformatic analyses. Notably, Smad7 was
identified as one of the potential targets and selected for further
investigation. Further experiments identified that the expression
level of Smad7 is repressed at the mRNA and protein level by
miR-590 overexpression. According to the luciferase assay, one
predicted binding site of miR-590 on 3′-UTR of Smad7 mRNA was
considered to be a functional site. On the basis of the
identification of Smad7 as a novel target of miR-590, the function
of Smad7 mRNA in cell proliferation and ECM synthesis were
assessed. The transfection of Smad7 siRNA resulted in a similar
stimulation of cell proliferation and ECM promotion in HUMSCs
compared with miR-590 overexpression.
In conclusion, the results of the present study
suggested that miR-590 induces stem cell proliferation by targeting
Smad7. Therefore, future studies may be able to determine whether
miR-590 regulates other genes for the promotion of stem cell
differentiation or migration.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81302540).
References
|
1
|
Lazarus GS, Cooper DM, Knighton DR,
Percoraro RE, Rodeheaver G and Robson MC: Definitions and
guidelines for assessment of wounds and evaluation of healing.
Wound Repair Regen. 2:165–170. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Boateng JS, Matthews KH, Stevens HN and
Eccleston GM: Wound healing dressings and drug delivery systems: A
review. J Pharm Sci. 97:2892–2923. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Seshareddy K, Troyer D and Weiss ML:
Method to isolate mesenchymal-like cells from Wharton's Jelly of
umbilical cord. Methods Cell Biol. 86:101–119. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Banerjee J, Chan YC and Sen CK: MicroRNAs
in skin and wound healing. Physiol Genomics. 43:543–556. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sand M, Gambichler T, Sand D, Skrygan M,
Altmeyer P and Bechara FG: MicroRNAs and the skin: Tiny players in
the body's largest organ. J Dermatol Sci. 53:169–175. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stefani G and Slack FJ: Small non-coding
RNAs in animal development. Nat Rev Mol Cell Biol. 9:219–230. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xiao X, Tang C, Xiao S, Fu C and Yu P:
Enhancement of proliferation and invasion by microRNA-590-5p via
targeting PBRM1 in clear cell renal carcinoma cells. Oncol Res.
20:537–544. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jiang X, Xiang G, Wang Y, Zhang L, Yang X,
Cao L, Peng H, Xue P and Chen D: MicroRNA-590-5p regulates
proliferation and invasion in human hepatocellular carcinoma cells
by targeting TGF-b RII. Mol Cells. 33:545–551. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shan X, Miao Y, Fan R, Qian H, Chen P, Liu
H, Yan X, Li J and Zhou F: MiR-590-5P inhibits growth of HepG2
Cells via decrease of S100A10 expression and inhibition of the Wnt
pathway. Int J Mol Sci. 14:8556–8569. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chu Y, Ouyang Y, Wang F, Zheng A, Bai L,
Han L, Chen Y and Wang H: MicroRNA-590 promotes cervical cancer
cell growth and invasion by targeting CHL1. J Cell Biochem.
115:847–853. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li K, Li Z, Zhao N and Xu Y, Liu Y, Zhou
Y, Shang D, Qiu F, Zhang R, Chang Z and Xu Y: Functional analysis
of microRNA and transcription factor synergistic regulatory network
based on identifying regulatory motifs in non-small cell lung
cancer. BMC Syst Biol. 7:1222013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee S, Yu KR, Ryu YS, Oh YS, Hong IS, Kim
HS, Lee JY, Kim S, Seo KW and Kang KS: miR-543 and miR-590-3p
regulate human mesenchymal stem cell aging via direct targeting of
AIMP3/p18. Age (Dordr). 36:97242014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shan H, Zhang Y, Lu Y, Zhang Y, Pan Z, Cai
B, Wang N, Li X, Feng T, Hong Y and Yang B: Downregulation of
miR-133 and miR-590 contributes to nicotine-induced atrial
remodelling in canines. Cardiovasc Res. 83:465–472. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kavsak P, Rasmussen RK, Causing CG, Bonni
S, Zhu H, Thomsen GH and Wrana JL: Smad7 binds to Smurf2 to form an
E3 ubiquitin ligase that targets the TGF beta receptor for
degradation. Mol Cell. 6:1365–1375. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fukasawa H, Yamamoto T, Togawa A, Ohashi
N, Fujigaki Y, Oda T, Uchida C, Kitagawa K, Hattori T, Suzuki S, et
al: Down-regulation of Smad7 expression by ubiquitin-dependent
degradation contributes to renal fibrosis in obstructive
nephropathy in mice. Proc Natl Acad Sci USA. 101:8687–8692. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Afrakhte M, Morén A, Jossan S, Itoh S,
Sampath K, Westermark B, Heldin CH, Heldin NE and ten Dijke P:
Induction of inhibitory Smad6 and Smad7 mRNA by TGF-beta family
members. Biochem Biophys Res Commun. 249:505–511. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jia J, Feng X, Xu W, Yang S, Zhang Q, Liu
X, Feng Y and Dai Z: MiR-17-5p modulates osteoblastic
differentiation and cell proliferation by targeting SMAD7 in
non-traumatic osteonecrosis. Exp Mol Med. 46:e1072014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu G, Friggeri A, Yang Y, Milosevic J,
Ding Q, Thannickal VJ, Kaminski N and Abraham E: miR-21 mediates
fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J
Exp Med. 207:1589–1597. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen G, Cao S, Liu F and Liu Y: miR-195
plays a role in steroid resistance of ulcerative colitis by
targeting Smad7. Biochem J. 471:357–367. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu B, Wei XX, Wang TB, Zhou YC, Liu AM
and Zhang GW: Increased miR-16 expression induced by hepatitis C
virus infection promotes liver fibrosis through downregulation of
hepatocyte growth factor and Smad7. Arch Virol. 160:2043–2050.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang ZH, Zhang QS, Duan YL, Zhang JL, Li
GF and Zheng DL: TGF-β induced miR-132 enhances the activation of
TGF-β signaling through inhibiting SMAD7 expression in glioma
cells. Biochem Biophys Res Commu. 463:187–192. 2015. View Article : Google Scholar
|
|
22
|
Liu N, Jiao T, Huang Y, Liu W, Li Z and Ye
X: Hepatitis B virus regulates apoptosis and tumorigenesis through
the microRNA-15a-Smad7-transforming growth factor β pathway. J
Virol. 89:2739–2749. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu J, Yao W, Yao Y, Du X, Zhou J, Ma B,
Liu H, Li Q and Pan Z: miR-92a inhibits porcine ovarian granulosa
cell apoptosis by targeting Smad7 gene. FEBS Lett. 588:4497–4503.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Trang NV, Choisy M, Nakagomi T, Chinh NT,
Doan YH, Yamashiro T, Bryant JE, Nakagomi O and Anh DD:
Determination of cut-off cycle threshold values in routine RT-PCR
assays to assist differential diagnosis of norovirusin children
hospitalized for acute gastroenteritis. Epidemiol Infect.
143:3292–3299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Su Y, Yang CY, Li Z, Xu F, Zhang L, Wang F
and Zhao S: Smad7 siRNA inhibit expression of extracellular matrix
intrabecular meshwork cells treated with TGF-β2. Mol Vis.
18:1881–1884. 2012.PubMed/NCBI
|
|
26
|
Shakespeare P: Burn wound healing and skin
substitutes. Burns. 27:517–522. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Newman RE, Yoo D, LeRoux MA and
Danilkovitch-Miagkova A: Treatment of inflammatory diseases with
mesenchymal stem cells. Inflamm Allergy Drug Targets. 8:110–123.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Arno AI, Amini-Nik S, Blit PH, Al-Shehab
M, Belo C, Herer E, Tien CH and Jeschke MG: Human Wharton's
jelly-mesenchymal stem cells promote skin wound healing through
paracrine signaling. Stem Cell Res Ther. 5:282014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bushati N and Cohen SM: microRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He PP, OuYang XP, Li Y, Lv YC, Wang ZB,
Yao F, Xie W, Tan YL, Li L, Zhang M, et al: MicroRNA-590 inhibits
lipoprotein lipase expression and prevents atherosclerosis in apoE
knockout mice. PLoS One. 10:e01387882015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao S, Yang G, Liu PN, Deng YY, Zhao Z,
Sun T, Zhuo XZ, Liu JH, Tian Y, Zhou J, et al: miR-590-3p is a
novel MicroRNA in myocarditis by targeting nuclear factor Kappa-B
in vivo. Cardiology. 132:182–188. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sundaram GM, Common JE, Gopal FE, Srikanta
S, Lakshman K, Lunny DP, Lim TC, Tanavde V, Lane EB and Sampath P:
'See-saw' expression of microRNA-198 and FSTL1 from a single
transcript in wound healing. Nature. 495:103–106. 2013. View Article : Google Scholar : PubMed/NCBI
|