Introduction
Hepatocellular carcinoma (HCC) is a common malignant
tumor; its incidence ranks fifth and it has the third highest death
rate among cancers (1). The
pathogenesis of HCC is very complex; excessive alcohol drinking,
viral hepatitis and nonalcoholic fatty liver have all been shown to
induce carcinogenesis in normal liver cells (2–4). Studies
on molecular physiology have found a dynamic balance between
hepatocyte proliferation and apoptosis. When this balance is
broken, apoptosis does not function well and tumor cell
proliferation and even metastasis occur (5,6).
The inhibitor of apoptosis (IAP) family includes
protein factors that inhibit apoptosis in vivo (7). Among the many factors of the IAP family,
the activity of the X-linked IAP protein (XIAP) is the strongest.
XIAP is mainly composed of baculovirus IAP repeat (BIR) and
zinc finger domains. BIR domains can inhibit the functions of
caspase-3 and −7, while the zinc finger domains inhibit the
functions of caspase-9; together these domains have the capacity
for inhibiting all the caspases that induce apoptosis (8). Additionally, studies have also shown
that XIAP plays a key role in other biological processes such as
cell signal transduction and protein ubiquitination (9).
The p53 genes were some of the first factors shown
to control cell proliferation and apoptosis, thus inhibiting
tumors. However, mutations in the wild-type p53 gene can lead to
inhibition of tumor cell apoptosis (10). Studies have confirmed that mutation of
p53 gene happens in 50% of tumor tissues in humans, so p53 gene is
used as tumor biomarker (11).
The present study was designed to measure expression
of XIAP and p53 in HCC patients and to identify their relationships
to clinicopathological parameters and prognosis of patients.
Quantitative polymerase chain reaction (qPCR) and
immunohistochemistry (IHC) were used to detect the expression
levels of XIAP and p53 mRNA and proteins in HCC tumors and
tumor-adjacent normal samples. Clinicopathological variables from
the clinical history of 5 years from the day of surgery were used
in the statistical analysis to calculate prognosis of patients.
Materials and methods
Specimens
Frozen and paraffin specimens of tumor tissues from
70 patients with HCC and the accompanying tumor adjacent normal
tissues in 30 of those patients were used for the measurements.
Patients admitted to Weifang People's Hospital (Weifang, China)
from January 2009 to December 2011 were enrolled in the study. All
patients were clinically and pathologically diagnosed with HCC and
they all received surgical treatment for the first time and had no
prior history of radiotherapy or chemotherapy. Patients with
metastatic HCC, chronic hepatitis B or cirrhosis were excluded. In
the end, there were 36 males and 34 females, aged 25–79 years with
an average age of 47. The Clinical Ethics Committee of our hospital
approved all the procedures in the study and all participating
patients or their family members signed the informed consent
forms.
Detection of the expression levels of
XIAP and p53 in specimens by qPCR
Approximately 50 mg of frozen tumor tissues and
tumor-adjacent tissues of patients were used to extract total RNA
according to the method provided in the RNA isolation kit
(Invitrogen Life Technologies, Carlsbad, CA, USA). The UV-Vis
spectrophotometer (Hitachi, Ltd., Tokyo, Japan) was used to detect
the total RNA concentration and purity (when the absorbance value
ratio of A260/A280 was 1.8–2.0, the RNA was deemed of good
quality). cDNAs were obtained via reverse transcription according
to the instructions in the reverse transcription kit (Invitrogen
Life Technologies) and then the expression of XIAP and p53 mRNAs
were detected following the method in the RT-PCR kit (Invitrogen
Life Technologies) using cDNAs as templates. Glyceraldehyde
3-phosphate dehydrogenase (GAPDH) was used as an internal control.
The primer sequences of XIAP, mutant p53 and GAPDH are shown in
Table I, all primers were synthesized
by Takara Bio (Dalian, China). The PCR conditions included 95°C for
10 min, then a total of 40 cycles of 95, 57 and 72°C for 30 sec and
then a final extension at 72°C for 5 min. The relative expression
levels were calculated using the 2−ΔCq method according
to the following formula: ΔCq (target gene) = Cq (target gene) - Cq
(control gene).
 | Table I.Primer sequences of qPCR. |
Table I.
Primer sequences of qPCR.
| Gene | Primer name | Primer sequence |
|---|
| XIAP | Forward |
5′-AACCTTGTGATCGTGCCT-3′ |
|
| Reverse |
5′-ACCCTGGATACCATTTAGC-3′ |
| p53 | Forward |
5′-TGCGTGTGGAGTATTTGGATG-3′ |
|
| Reverse |
5′-TGGTACAGTCAGAGCCAACCTC-3′ |
| GAPDH | Forward |
5′-ATGGCACCGTCAAGGCTGAG-3′ |
|
| Reverse |
5′-GCAGTGATGGCATGGACTGT-3′ |
Detection of the expression levels of XIAP and p53
proteins in patient specimens by IHC. The procedure was carried out
according to the instructions of the IHC SP-9001 kit (Beijing
Zhongshan Golden Bridge Biological Technology, Beijing, China).
Briefly, after dewaxing of the paraffin sections, the endogenous
peroxidase was inactivated with 3% H2O2. The
citrate buffer was used for thermal remediation and proteins were
blocked with 10% goat serum. Primary rabbit polyclonal XIAP
antibody (dilution, 1:500, cat. no. ab21278) and rabbit polyclonal
p53 antibody (dilution, 1:500; cat. no. ab1431) were added and
incubated at 4°C overnight. Then proteins were washed with
phosphate-buffered saline (PBS) and secondary goat anti-rabbit
(HRP) IgG antibody (dilution, 1:2,000; cat. no. ab6721) were added.
All the antibodies were all purchased from Abcam (Cambridge, MA,
USA). Under this condition, samples were incubated for 15 min.
After that, proteins were washed with PBS and diaminobenzidine
solution was used for development in the dark. Then hematoxylin was
used for restaining the samples. Finally, pictures were taken under
the light microscope (TE2000-U; Nikon, Tokyo, Japan).
Three representative regions from each sample were
selected and the results of immunohistochemical staining were
evaluated by rating and grading the expression according to the
staining intensity and the percentage of positive cells. The
staining intensity was classified as stainless, brownish brown, tan
and sepia (recorded as 0, 1, 2 and 3 points, respectively). In
addition, the number of positive cells (magnification, ×400) was
classified as <5, 5–25, 26–50 and >50% (recorded as 0, 1, 2
and 3 points). In the end, the scores of the two indexes were
added. Total points >3 were considered as positive for
expression and ≤2 as negative for expression. The final scores were
used in the statistical analyses.
Relationship between the expression
levels of XIAP and p53 in HCC tissues and the prognosis in
patients
The 70 patients in the study were divided into a
XIAP positive or a XIAP negative group and also into a p53-positive
or -negative group, according to the expression levels of XIAP and
p53 in HCC tissues. There were no differences in statistical
significance in terms of age, sex, patient condition and other
factors between the two groups (P>0.05). The relationship of
XIAP and p53 with clinicopathological parameters was analyzed using
the available recorded clinical data of patients. All the 70
patients were followed up for 5 years and the follow-up rate was
100%. The survival duration was recorded from the first day after
surgery to the date of death of patients or the deadline of the
follow-up. The statistical analysis was performed on a monthly
basis.
Statistical analysis
SPSS 17.0 (IBM SPSS, Armonk, NY, USA) was used for
statistical analyses in this study. Measurement data were expressed
as mean ± standard deviation and the t-test was used for
comparisons between groups. The enumeration data were compared
between the two groups using the χ2 test. Clinical
prognostic data were calculated using the Kaplan-Meier survival
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
Detection of the expression levels of
XIAP and p53 mRNAs in tissue specimens by qPCR
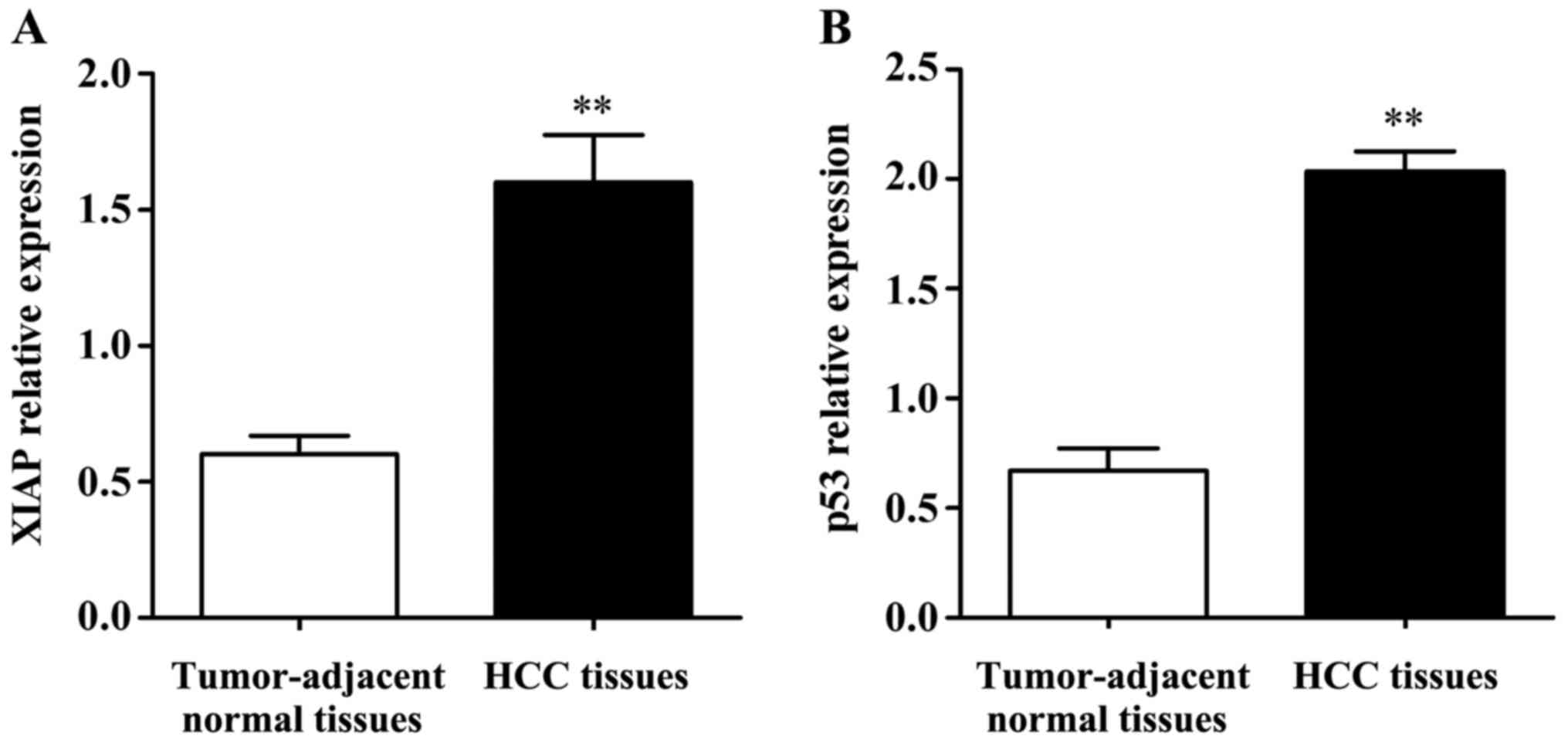
qPCR results (Fig. 1)
showed that the expression levels of XIAP and p53 mRNAs in HCC
tissues were significantly higher than those in tumor-adjacent
normal tissues and the differences were statistically significant
(P<0.01).
Detection of the expression levels of
XIAP and p53 proteins in clinicopathological tissues by IHC
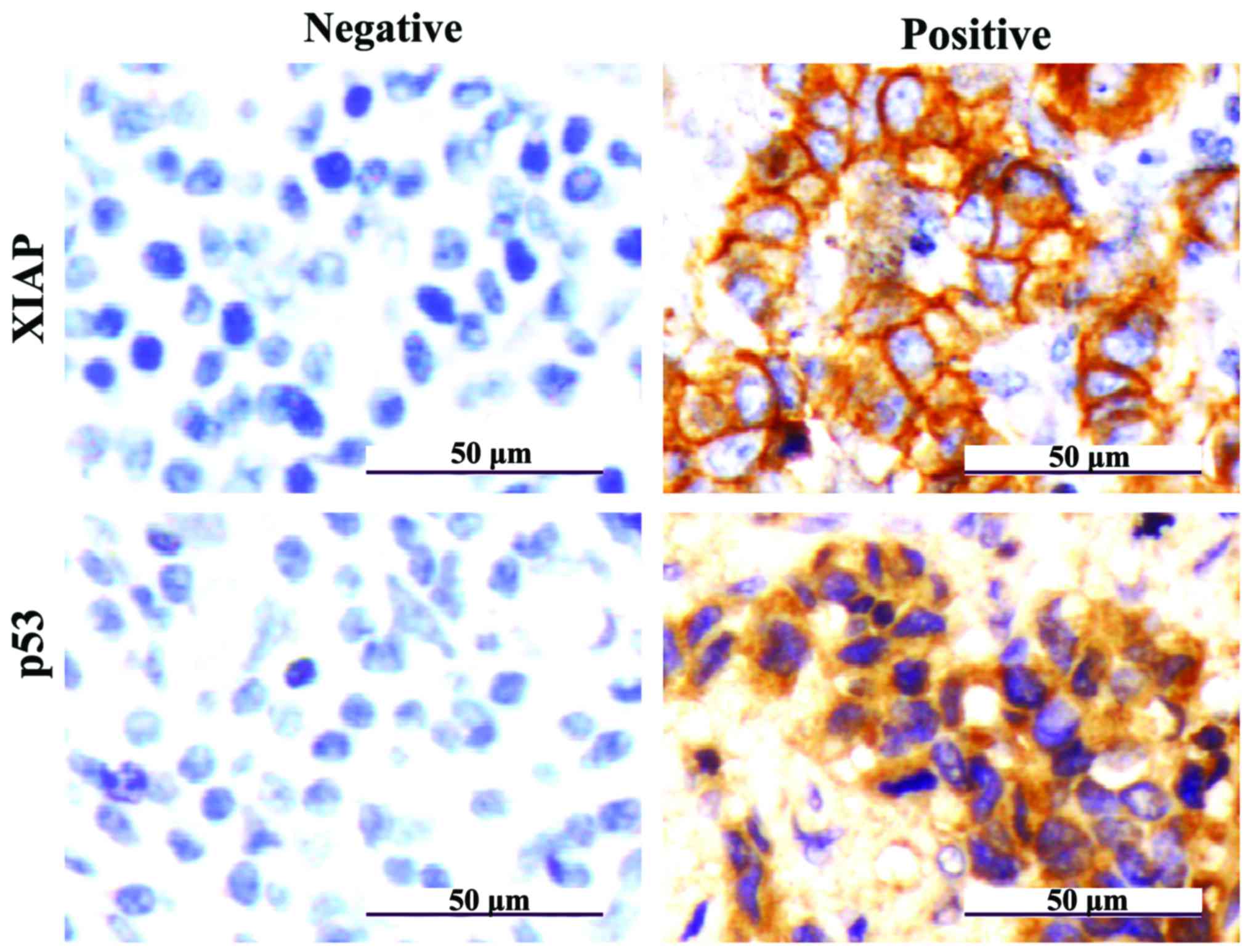
In IHC results the color of stained XIAP and p53 by
IHC was tan, with XIAP proteins mainly being present in the
cytoplasm and partly in cell membranes and p53 proteins mainly
present in the nucleus (Fig. 2).
The IHC scores were counted, the positive expression
rates of XIAP in HCC and tumor-adjacent normal tissues were 81.4%
(57/70) and 13.3% (4/30), respectively; and the difference was
statistically significant (P<0.01). The positive expression
rates of p53 in HCC and tumor-adjacent normal tissues were 72.9%
(51/70) and 10.0% (3/30), respectively; and the difference was
statistically significant (P<0.01) (Table II).
 | Table II.Positivity of expression of XIAP and
p53 proteins in HCC and tumor-adjacent normal tissues. |
Table II.
Positivity of expression of XIAP and
p53 proteins in HCC and tumor-adjacent normal tissues.
|
|
| XIAP | p53 |
|---|
|
|
|
|
|
|---|
| Group | Case | Positive
expression | Positive expression
rate (%) | P-value | Positive
expression | Positive expression
rate (%) | P-value |
|---|
| HCC tissues | 70 | 57 | 81.4 | <0.01 | 51 | 72.9 | <0.01 |
| Tumor-adjacent normal
tissues | 30 | 4 | 13.3 |
| 3 | 10.0 |
|
Relationship of clinicopathological
parameters of HCC and the expression levels of XIAP and p53
The statistical analysis results showing the
relationship between clinicopathological parameters of HCC patients
and the expression levels of XIAP and p53 are shown in Table III and Fig. 2. The χ2 test showed that
the positive expression of XIAP and p53 correlated with the
occurrence of metastasis and higher tumor stage (P<0.01), but
were not correlated with sex, age or tumor size (P>0.05).
 | Table III.Relationship between high expression
levels of XIAP and p53 with clinicopathological parameters of HCC
patients. |
Table III.
Relationship between high expression
levels of XIAP and p53 with clinicopathological parameters of HCC
patients.
|
|
| XIAP | p53 |
|---|
|
|
|
|
|
|---|
| Clinical data | Case | Positive
expression | Positive expression
rate (%) | P-value | Positive
expression | Positive expression
rate (%) | P-value |
|---|
| Sex |
|
|
|
|
|
| >0.05 |
| Male | 36 | 30 | 83.3 | >0.05 | 26 | 72.2 |
|
|
Female | 34 | 27 | 79.4 |
| 25 | 73.5 |
|
| Age, years |
|
|
|
|
|
| >0.05 |
| ≥50 | 42 | 34 | 81.0 | >0.05 | 32 | 76.2 |
|
|
<50 | 28 | 23 | 82.1 |
| 19 | 67.9 |
|
| Tumor size |
|
|
|
|
|
| >0.05 |
| ≥5
cm | 39 | 31 | 79.5 | >0.05 | 28 | 71.8 |
|
| <5
cm | 31 | 26 | 83.9 |
| 23 | 74.2 |
|
| Invasion and
metastasis |
|
|
|
|
|
| <0.01 |
| Yes | 46 | 43 | 93.5 | <0.01 | 41 | 89.1 |
|
| No | 24 | 14 | 58.3 |
| 10 | 41.7 |
|
| Tumor staging |
|
|
|
|
|
| <0.01 |
| I–II | 26 | 15 | 57.7 | <0.01 | 11 | 42.3 |
|
|
III–IV | 44 | 42 | 95.5 |
| 40 | 90.9 |
|
Single factor analysis of the survival
time and prognosis of patients
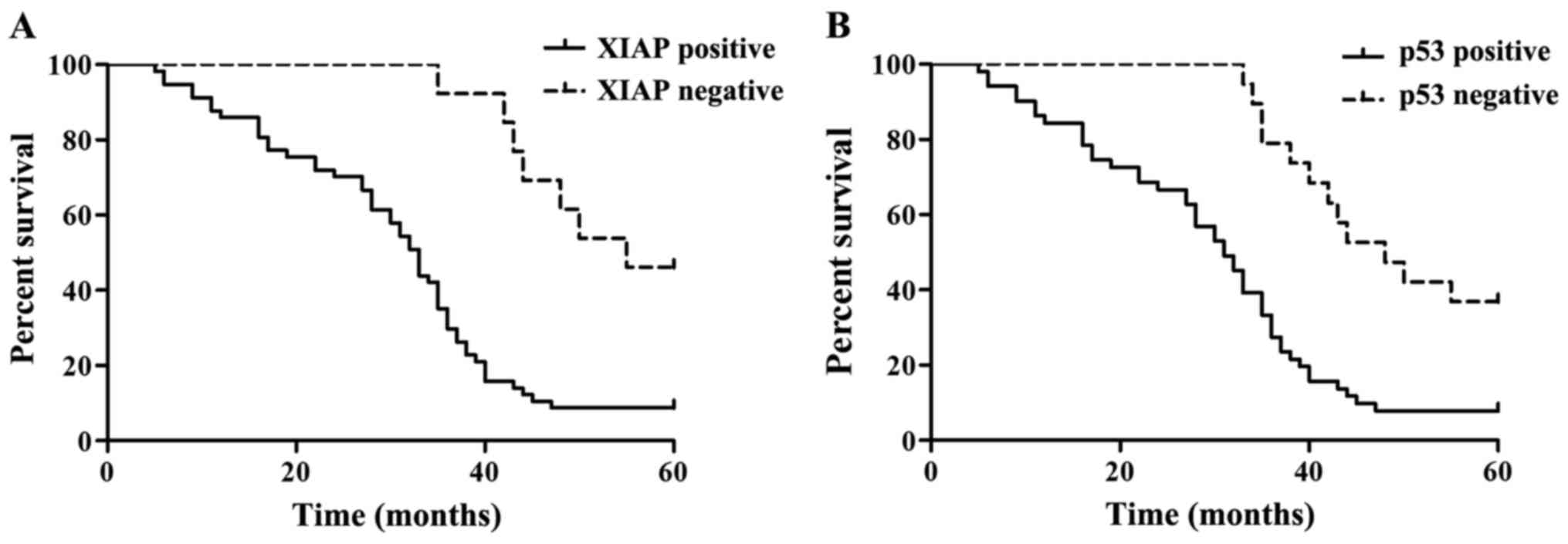
A total of 70 patients with HCC were followed up for
a maximum of 5 years. The number of 5-year survivors was 11 and the
number of deaths was 59, as shown in Table IV. Fig.
3 shows the Kaplan-Meier survival curves of 70 patients with
different expression of XIAP and p53. The results showed that HCC
patients with negatively expressed XIAP and p53 had a relatively
better survival prognosis. Differences in the overall survival
curve were analyzed using the log-rank test, as shown in Table III. The results of single factor
survival analysis showed that the effects of XIAP and p53 on the
overall survival rate of patients with HCC were statistically
significant (P<0.01).
 | Table IV.Single factor analysis of the
relationship of the expression levels of XIAP and p53 with the
overall survival rate of HCC patients. |
Table IV.
Single factor analysis of the
relationship of the expression levels of XIAP and p53 with the
overall survival rate of HCC patients.
| Group | Case | 5-year survival
case | 5-year survival
rate (%) | Wald
(log-rank) | P-value |
|---|
| XIAP |
|
|
|
| <0.01 |
|
Positive | 57 | 5 | 8.8 | 15.14 |
|
|
Negative | 13 | 6 | 46.2 |
|
|
| p53 |
|
|
|
| <0.01 |
|
Positive | 51 | 4 | 7.8 | 15.68 |
|
|
Negative | 19 | 7 | 36.8 |
|
|
Discussion
HCC is one of the most common malignant tumors in
China (12). Presently, more than 100
oncogenes have been found in studies, but the number of tumor
suppressor genes is very small. Mutations, deletions and
inactivations of tumor suppressor genes are events closely related
to the occurrence and development of tumors (13).
XIAP, as an important member of the IAP family, can
specifically inhibit the activity of caspase-3, −7 and −9, and
inhibit the apoptosis of tumor cells (14). Clinical studies have shown that XIAP
expression levels are significantly higher in a variety of
malignant tumor tissues than in normal tissues (15).
The p53 gene is located on human chromosome 17p13
and is a kind of tumor suppressor and pro-apoptosis gene. Wild-type
p53 plays a key role in cell gene transcription, cell cycle
regulation, apoptosis, proliferation and differentiation (16,17).
Studies have found that p53 gene mutations are closely related to
the occurrence and development of HCC (18,19). The
amount of wild-type p53 proteins is low in normal cells and their
half-life periods are short, so IHC and other methods cannot detect
them. However, the half-life periods of mutant p53 proteins are
relatively longer and the amounts are relatively higher, allowing
for their detection (20,21). Studies have found that mutant p53
proteins are highly expressed in tumor tissues of patients with
breast cancer and the degree of malignancy of the cancer in these
patients is high resulting in poor prognosis after surgery
(22).
In order to find if tumor tissue levels of XIAP and
p53 in HCC patients are also useful for prognoses, the expressions
of XIAP and p53 mRNAs in tumor tissues of HCC patients were
detected by qPCR. Our results showed the expression levels of XIAP
and p53 mRNAs in HCC tissues were significantly higher than those
in tumor-adjacent normal tissues. Furthermore, the IHC results
showed that the positive expression rates of XIAP and p53 proteins
in HCC tissues were 81.4% (57/70) and 72.9% (51/70), respectively,
which were significantly higher than those in tumor-adjacent normal
tissues. A further analysis, in combination with the
clinicopathological features of patients, revealed that the
expression of XIAP and p53 was related to the occurrence of
metastasis and tumor staging; but not related to sex, age or the
tumor size. Single factor Kaplan-Meier survival analysis was used
to analyze the effects of different mRNA expression levels of XIAP
and p53 on the overall survival rate curve of patients. The results
showed the overall survival period of HCC patients with high
expression levels of XIAP and p53 was relatively short.
A large number of experiments have shown that XIAP
and p53 are closely related to the occurrence and development of a
variety of tumors. A study found that XIAP protein levels in kidney
clear cell carcinoma tissue specimens were high and the expression
levels in the advanced stages were significantly higher than those
in the early stages (23). Another
study found the expression levels of p53 in 62 cases of primary HCC
to be higher in cancer tissues than those in tumor-adjacent normal
tissues (24). Our findings
corroborate these studies and further confirmed that XIAP and p53
are highly expressed in HCC, and the expression of XIAP and p53
were related to the metastasis, staging and prognosis of HCC
patients.
In conclusion, the positive expression of XIAP and
p53 are closely related to the occurrence and development of HCC,
especially to tumor metastasis and tumor staging. XIAP and p53 can
be used as reference markers to guide the treatment of HCC and
estimate its prognosis.
References
|
1
|
Yang JD and Roberts LR: Epidemiology and
management of hepatocellular carcinoma. Infect Dis Clin North Am.
24:899–919. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Duan XY, Zhang L, Fan JG and Qiao L: NAFLD
leads to liver cancer: Do we have sufficient evidence? Cancer Lett.
345:230–234. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Karagozian R, Derdák Z and Baffy G:
Obesity-associated mechanisms of hepatocarcinogenesis. Metabolism.
63:607–617. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nakagawa H, Umemura A, Taniguchi K,
Font-Burgada J, Dhar D, Ogata H, Zhong Z, Valasek MA, Seki E,
Hidalgo J, et al: ER stress cooperates with hypernutrition to
trigger TNF-dependent spontaneous HCC development. Cancer Cell.
26:331–343. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han XJ, Dong BW, Liang P, Yu XL and Yu DJ:
Local cellular immune response induced by ultrasound-guided tumor
bed superantigen injection after percutaneous microwave coagulation
therapy for liver cancer. Zhonghua Zhong Liu Za Zhi. 31:602–606.
2009.(In Chinese). PubMed/NCBI
|
|
7
|
Deveraux QL and Reed JC: IAP family
proteins - suppressors of apoptosis. Genes Dev. 13:239–252. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Holcik M, Gibson H and Korneluk RG: XIAP:
Apoptotic brake and promising therapeutic target. Apoptosis.
6:253–261. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liston P, Fong WG and Korneluk RG: The
inhibitors of apoptosis: There is more to life than Bcl2. Oncogene.
22:8568–8580. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maniwa Y, Yoshimura M, Obayashi C, Inaba
M, Kiyooka K, Kanki M and Okita Y: Association of p53 gene mutation
and telomerase activity in resectable non-small cell lung cancer.
Chest. 120:589–594. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Graziano SL, Tatum A, Herndon JE II, Box
J, Memoli V, Green MR and Kern JA: Use of neuroendocrine markers,
p53 and HER2 to predict response to chemotherapy in patients with
stage III non-small cell lung cancer: A cancer and leukemia group B
study. Lung Cancer. 33:115–123. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Green DR and Kroemer G: Cytoplasmic
functions of the tumour suppressor p53. Nature. 458:1127–1130.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song LP, Li YP, Wang N, Li WW, Ren J, Qiu
SD, Wang QY and Yang GX: NT4(Si)-p53(N15)-antennapedia induces cell
death in a human hepatocellular carcinoma cell line. World J
Gastroenterol. 15:5813–5820. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu W, Wang F, Tang J, Liu X, Yuan Z, Nie C
and Wei Y: Proapoptotic protein Smac mediates apoptosis in
cisplatin-resistant ovarian cancer cells when treated with the
antitumor agent AT101. J Biol Chem. 287:68–80. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hussain AR, Uddin S, Ahmed M, Bu R, Ahmed
SO, Abubaker J, Sultana M, Ajarim D, Al-Dayel F, Bavi PP, et al:
Prognostic significance of XIAP expression in DLBCL and effect of
its inhibition on AKT signalling. J Pathol. 222:180–190. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shangary S and Wang S: Targeting the
MDM2-p53 interaction for cancer therapy. Clin Cancer Res.
14:5318–5324. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shangary S, Qin D, McEachern D, Liu M,
Miller RS, Qiu S, Nikolovska-Coleska Z, Ding K, Wang G, Chen J, et
al: Temporal activation of p53 by a specific MDM2 inhibitor is
selectively toxic to tumors and leads to complete tumor growth
inhibition. Proc Natl Acad Sci USA. 105:3933–3938. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Scoumanne A and Chen X: Protein
methylation: A new mechanism of p53 tumor suppressor regulation.
Histol Histopathol. 23:1143–1149. 2008.PubMed/NCBI
|
|
19
|
Lu C and El-Deiry WS: Targeting p53 for
enhanced radio- and chemo-sensitivity. Apoptosis. 14:597–606. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hall PA, Ray A, Lemoine NR, Midgley CA,
Krausz T and Lane DP: p53 immunostaining as a marker of malignant
disease in diagnostic cytopathology. Lancet. 338:5131991.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martinazzi M, Crivelli F, Zampatti C and
Martinazzi S: Relationship between p53 expression and other
prognostic factors in human breast carcinoma. An
immunohistochemical study. Am J Clin Pathol. 100:213–217. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen W, Li J, Liu C, Chen X, Zhu Y, Yang
Y, Gong Y, Wang T, Miao X and Nie X: A functional p53 responsive
polymorphism in KITLG, rs4590952, does not affect the risk of
breast cancer. Sci Rep. 4:63712014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ramp U, Krieg T, Caliskan E, Mahotka C,
Ebert T, Willers R, Gabbert HE and Gerharz CD: XIAP expression is
an independent prognostic marker in clear-cell renal carcinomas.
Hum Pathol. 35:1022–1028. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hong Y, Zhong R and Hang Z: The
association study and its clinical significance of the expression
of P53, AR and ER in the primary hepatocelluar carcinoma (liver
cancer) and surrounding tissue. Int Med Health Guid News. 12:12–14.
2006.
|