Introduction
Lung cancer remains the primary cause of
cancer-associated mortality worldwide with a 5-year survival rate
of <15% (1), despite advances in
diagnostic and therapeutic approaches. Significant advances have
been made in understanding cancer biology, including lung cancer,
with the identification of oncogenic drivers of the disease
(2).
In the present study, the cytotoxicity of
L-securinine in human lung cancer A549 cells was evaluated.
Securinines, including the two optical isomers L-securinine and
D-securinine, are a class of natural products isolated from the
plants of the Euphorbiaceae family. Securinine was extracted and
isolated from Securinega suffruticosa (3) and structurally characterized in 1963
(4). Securinine has been applied as a
treatment for blood (5,6) and nervous system diseases (7–9). Previous
publications have reported that L-securinine can induce cell
apoptosis in human breast cancer MCF-7 cells (10) and human promyelocytic leukemia HL-60
cells (11).
The Wnt signaling pathway is involved in a wide
range of developmental and physiological processes, including cell
fate specification, tissue morphogenesis and homeostasis (12). The loss or gain of function of Wnt
pathway components can result in the development of hepatocellular,
lung or colon cancer (13). Certain
factors of the Wnt signaling pathway have been considered as
potential therapeutic targets for cancer treatment (14). Dickkopf-related proteins (DDKs) are
Wnt signaling pathway inhibitors that are frequently inactivated in
certain types of human cancer (12).
In the present study, the expression of DKK genes in A549 cells
treated by L-securinine was determined by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
Epigenetics is defined as the mechanism by which
cells transfer environmental change-induced phenotypes to their
daughter cells (15). Epigenetic
changes are heritable changes in gene expression that do not alter
the primary DNA sequence (16).
Emerging evidence suggests that epigenetic changes, including
genomic methylation, serve crucial regulatory roles in X-chromosome
inactivation, mammalian development, gene silencing,
retrotransposon silencing and genomic imprinting (17). Aberrant methylation profiles of genes
are associated with cancer, in addition to autoimmune disease,
psychiatric and neurodegenerative disorders, diabetes and heart
disease (18). The addition of a
methyl group to the carbon-5 position of the cytosine present in
the CpG dinucleotides is a common methylation event of DNA in
eukaryotes (19). These CpG
dinucleotides act as either CpG islands or as dispersedly
distributed CpG motifs within gene promoter regions. Multiple
methods, including methylation-specific PCR, methylation-sensitive
restriction enzyme PCR, pyrosequencing and bisulfite sequencing PCR
(BSP), can be used to detect the CpG islands and motifs (20). Nevertheless, BSP is the gold standard
for detecting DNA methylation (21).
BSP was used to detect the DNA methylation profile of DKK1 genes in
the present study.
Material and methods
Extraction, isolation and structure
identification of L-securinine
A total of 700 ml H2SO4
solution (pH 1–2) was added to a leaf of dried Securinega
suffruticosa (obtained from Traditional Chinese Medicine and
Natural Products, Jinan University, Guangzhou, China) in a 1,000 ml
beaker to be moistened sufficiently for 20 min before being
transferred to a percolator and percolated at a rate of 4–5 ml/min.
The acidic extract was collected and passed through a cationic
exchange column, which was packed with 80 g wet resin, at the rate
of 4–5 ml/min. The resin was then washed and dried at room
temperature. The dried resin was alkalified with ammonia water (pH
8–9). Benzin was used to elute the resin for 2.5 h at 60°C. The
resin petroleum extract was parboiled with agitation at 50°C, then
filtered until the crystals were separated out. Spectral
techniques, including infra-red spectroscopy, ultraviolet-visible
spectroscopy, mass spectrometry and nuclear magnetic resonance
spectroscopy were used to identify the structure of the crystal,
and X-ray crystallography, circular dichroism and Mosher chiral
reagents were used to determine the absolute configuration of the
product, as previously described (22,23).
Finally, L-securinine was extracted and isolated, as described in
the previous studies (Fig. 1).
Cell culture
The human lung cancer A549 cell line and human lung
fibroblast HLF-a cell line (Cell Bank of Type Culture Collection of
Chinese Academy of Sciences, Shanghai, China) were cultured in
RPMI-1640 supplemented with 10% heat-inactivated fetal bovine serum
(FBS) (both from Thermo Fisher Scientific, Inc., Waltham, MA, USA),
penicillin (10 U/ml) and streptomycin (10 µg/ml), in a humidified
incubator (Sanyo XD-101; Sanyo Electric Co., Ltd., Osaka, Japan)
with 5% CO2 at 37°C.
Cell cytotoxicity
The exponentially growing human lung cancer cell
suspension (5,000 cells/100 µl) was seeded into each well of
96-well plates. The cells were pre-incubated for 24 h in the
humidified incubator at 37°C with 5% CO2. Various
concentrations of L-securinine from 0 to 160 µg/ml (10 µl) were
added into each well. The cells were then incubated for 24, 36 and
48 h in the incubator. CCK-8 solution (10 µl; cat no. KGA317;
Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) was added to each
well and incubated for 3 h. The absorbance of the wells at 450 nm
was measured using a microplate reader (ELx800; BioTek Instruments,
Inc., Winooski, VT, USA). The inhibition rate (IR) of cell
proliferation was calculated according to the following formula: IR
= [1 - (average A450 values of the experimental group - average
A450 values of the blank group)/(average A450 values of the control
group-average A450 values of the blank group)] ×100%. The
experimental groups and the control groups refer to the cells in
the 96-well plates treated with or without L-securinine,
respectively. The blank groups refer to the lung cancer cell
suspension in the wells of 96-well plates replaced by physiological
saline.
RT-qPCR
The RT-qPCR assays were performed on HLF-a and A549
cells treated with or without L-securinine to determine the
expression of the DKK genes. Total RNA was extracted from the cells
using TRIzol reagent (cat no. 15596-026; Invitrogen; Thermo Fisher
Scientific, Inc.). Complementary DNA (cDNA) was synthesized from
the extracted RNA using a cDNA synthesis kit (cat no. PC0002;
Fermentas; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Each PCR reaction was performed in a final
volume of 20 µl that contained 2 µl cDNA, 10 µl SYBR-Green Master
Mix (cat no. QPK-201; Takara Bio, Inc., Otsu, Japan), 0.8 µl of the
primer mixture (forward and reverse; 10 µmol) and 7.2 µl DEPC
water. PCR reactions were performed using the FTC-3000 real-time
fluorescence quantitative PCR system (Funglyn Biotech, Inc.,
Ontario, Canada). The thermocycling conditions were as follows:
95°C for 180 sec, followed by 40 cycles of 95°C for 5 sec and 60°C
for 30 sec. The experiment was repeated three times and all data
were compared with GAPDH gene using the 2−∆∆Cq method
(24). The sequences of specific
primers for GAPDH and target genes were as follows: GAPDH forward,
5′-TGTTGCCATCAATGACCCCTT-3′ and reverse, 5′-CTCCACGACGTACTCAGCG-3′;
DKK1 forward, 5′-GGGGTGAAGAGTGTTAAAGGTTT-3′ and reverse,
5′-CCCAAAATCCTAACTACAAAAAACA-3′; DKK2 forward,
5′-GCTGTGCTCGTCATTTCTGGA-3′ and reverse,
5′-TTGGAGGAGTAGGTGGCATCTT-3′; DKK3 forward,
5′-GCGAGGTTGAGGAACTGATGG-3′ reverse,
5′-CCTTCGTGTCTGTGTTGGTCTC-3′.
DNA extraction and bisulfite
treatment
The A549 cells were treated with 5.0 µg/ml
L-securinine and 10.0 µg/ml 5-azacytidine (cat no. A2385;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Genomic DNA was
extracted from HLF-a and A549 cells using a DNA extraction kit
(Aidalb Biotechnologies Co., Ltd., Beijing, China) according to the
manufacturer's protocol. Then the genomic DNA was treated with
sodium bisulfite using the EZ DNA Methylation™-GOLD kit (Zymo
Research, Irvine, CA, USA) following the manufacturer's
protocol.
BSP amplifiction
The BSP primer of DKK1 was designed using ABI Methyl
Primer Express 1.0 software (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The primer sequence was as follows: Forward,
5′-GGGGTGAAGAGTGTTAAAGGTTT-3′ and reverse,
5′-CCCAAAATCCTAACTACAAAAAACA-3′. The PCR reaction was performed on
the ABI 7500 Fast Real-Time PCR platform (Applied Biosystems;
Thermo Fisher Scientific, Inc.) in a volume of 50 µl containing 5
µl buffer (10X), 5 µl dNTPs (2 mM), 3 µl MgSO4 (25 mM),
1.5 µl forward primer (10 pmol/µl), 1.5 µl reverse primer (10
pmol/µl), 1 µl KOD-Plus-Neo (1.0 U/µl), 31 µl ddH2O and
2 µl DNA template. The thermocycling conditions were as follows:
94°C for 2 min, 98°C for 10 sec for 25–45 cycles, 68°C for 30 sec,
followed by 94°C for 2 min, 98°C for 10 sec, Tm°C for 30
sec for 25–45 cycles, 68°C for 30 sec. The amplified products were
sent to Sangon Biotech Co., Ltd. (Shanghai, China) for sequence
analysis.
Statistical analysis
The data were processed using Microsoft Excel 2007
(Microsoft Corporation, Redmond, WA, USA) and SPSS 16.0 software
(SPSS, Inc., Chicago, IL, USA) and presented as the mean ± standard
deviation. Statistically significant differences between two groups
were analyzed using the Student's t-test, and pairwise comparison
among the groups was performed using the Student-Newman-Keuls test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
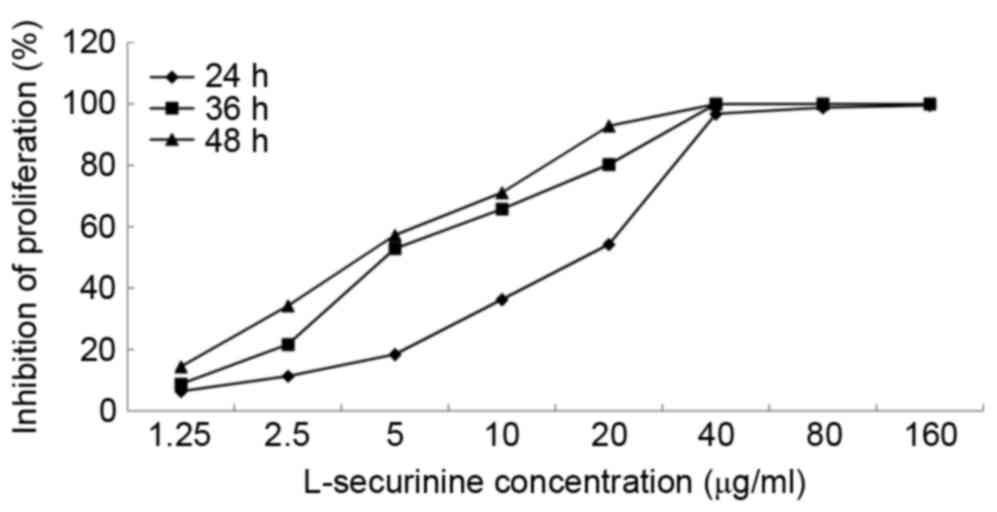
L-securinine inhibited the growth of
A549 cells
The CCK-8 assay was used to determine the effects of
L-securinine on the proliferation of A549 cells. L-securinine
decreased the proliferation of A549 cells in a dose- and
time-dependent manner (Fig. 2), with
half maximal inhibitory concentration (IC50) values of
8.92, 4.73 and 3.81 µg/ml at 24, 36 and 48 h post-treatment,
respectively. An extract is considered cytotoxic when it possesses
an IC50 value ≤20 µg/ml (25); therefore L-securinine is cytotoxic
against A549 cells.
DDK expression in A549 and HLF-a
cells
The A549 and HLF-a cell lines represent human lung
cancer cells and lung fibroblasts, respectively. RT-qPCR assays
were used to measure the expression of DKK1, 2 and 3 in HLF-a and
A549 cell lines. DKK1, DKK2 and DKK3 expression was significantly
increased in A549 lung cancer cells compared with HLF-a lung
fibroblasts (P≤0.001; Table I and
Fig. 3).
 | Table I.Reverse transcription-quantitative
polymerase chain reaction analysis of DKK expression in A549 and
HLF-a cell lines. |
Table I.
Reverse transcription-quantitative
polymerase chain reaction analysis of DKK expression in A549 and
HLF-a cell lines.
| Human lung cell
lines | DKK1/GAPDH | DKK2/GAPDH | DKK3/GAPDH |
|---|
| HLF-a | 1.003±0.02 | 1.004±0.07 | 1.005±0.12 |
| A549 | 2.33±0.17 | 15.71±0.55 | 759.45±141.15 |
| t-value | 13.08 | 45.64 | 9.31 |
| P-value | <0.001 | <0.001 | 0.001 |
DDK expression in A549 cells treated
with L-securinine
A549 cells were treated without or with L-securinine
at 2.5, 5.0 and 10.0 µg/ml for 48 h. DKK1, DKK2 and DKK3 expression
were then measured using RT-qPCR. DKK1 expression was significantly
decreased in a dose-dependent manner following treatment with
L-securinine compared with untreated cells (P<0.05); however, no
significant differences were observed in DKK2 or 3 expression
levels in treated compared with untreated cells (Table II and Fig.
3).
 | Table II.Reverse transcription-quantitative
polymerase chain reaction analysis of DKK expression in A549 cell
lines. |
Table II.
Reverse transcription-quantitative
polymerase chain reaction analysis of DKK expression in A549 cell
lines.
| Concentration of
L-securinine (µg/ml) | DKK1/GAPDH | DKK2/GAPDH | DKK3/GAPDH |
|---|
| 0 | 2.33±0.17 | 15.71±0.55 | 759.45±141.15 |
| 2.5 |
1.53±0.08a | 17.28±5.07 | 838.16±31.67 |
| 5.0 |
1.26±0.02b | 13.26±2.06 | 784.56±227.69 |
| 10.0 |
1.14±0.05c | 14.53±2.74 | 845.29±151.86 |
| F value | 85.33 | 0.94 | 0.217 |
| P-value | <0.001 | 0.467 | 0.882 |
Methylation profile at CpG sites
within the DKK1 promoter region
The DKK1 gene was extracted from HLF-a cells, A549
cells, A549 cells treated with L-securinine and A549 cells treated
with 5-azacytidine. The methylation profiles at 11 CpG sites of the
DKK1 promoter region were detected using BSP (Fig. 4). 5-azacytidine is a demethylating
agent typically used in methylation research for a comparison with
other sources of methylation or demethylation (26). The third, the sixth and eleventh CpG
sites were methylated specifically in the DKK1 gene extracted from
the A549 cells treated with L-securinine compared with untreated
A549 cells. However, only the third and eleventh CpG sites were
methylated specifically in A549 cells treated with L-securinine
compared with cells treated with 5-azacytidine. Combined with the
aforementioned RT-qPCR results, there may be an association between
the methylation changes at CpG sites in the DKK1 promoter region
and changes in the expression level of DKK1 gene following
treatment with L-securinine.
Discussion
Lung cancer remains the leading cause of
cancer-associated mortalities worldwide, despite improved tools for
early detection. Therapeutic stratification would be expected to
increase the survival rate (27,28).
Although certain targeted drugs, including epidermal growth factor
receptor tyrosine-kinase and anaplastic lymphoma
kinase/proto-oncogene tyrosine-protein kinase ROS inhibitors
(29) have been used in the clinic,
their cost (30) and acquired
resistance (31) remain challenges
for their use in the treatment of lung cancer. Therefore further
studies are required to identify alternative treatment strategies.
Securinine has previously been used in the clinic for the treatment
of blood and nervous system diseases (32); therefore, the pharmacokinetic and
pharmacodynamics properties of securinine are well understood.
Identifying alterations in the signal transduction
pathways that promote lung cancer, including epigenetic
modifications, may reveal biomarkers that can be used for the
diagnosis and treatment of lung cancer (33). The proteins encoded by the DKK genes
represent the dickkopf family of proteins, which negatively
regulate the Wnt signaling pathway (12). Data from the present study
demonstrated that DKK1, DKK2 and DKK3 expression is significantly
increased in lung cancer cells compared with lung fibroblasts. A
previous meta-analysis revealed that DKK1 is a potential biomarker
for lung cancer diagnosis (34). It
has previously been reported that DKK1, as a secreted protein with
two cysteine rich regions, negatively regulates the Wnt signaling
pathway (35).
Epigenetic changes are prevalent in all types of
cancer, serve central roles in DNA replication and repair, protein
structure and function, and can be regulated with chemical agents
(36,37). However, the anticancer drugs that can
target gene methylation have not yet been applied to clinical
therapy. The present study revealed that L-securinine regulates DNA
methylation in the DKK1 promoter, therefore L-securinine may be a
potential therapeutic agent that can target changes in gene
methylation in lung cancer cells.
In the present study, the IC50 values and
growth-inhibitory curves of A549 cells treated by L-securinine were
analyzed. According to the IC50 values and
growth-inhibitory curves, the appropriate concentration of
L-securinine was selected to treat A549 cells and analyze of the
expression of DKK genes following treatment. DKK1 expression was
demonstrated to be significantly decreased in A549 cells treated
with L-securinine compared with that in untreated cells. The
methylation profile of the DKK1 promoter was explored to identify
the molecular mechanisms underlying the effect of L-securinine on
DKK1 expression. Compared with 5-azacytidine, L-securinine promoted
the methylation of the DKK1 promoter.
Acknowledgements
The present study was supported the National Natural
Science Foundation of China (grant no. 81241102). The authors would
like to thank the Institute of Traditional Chinese Medicine and
Natural Products, Jinan University (Guangzhou, China) for providing
the sample of Securinega suffruticosa.
References
|
1
|
Wood SL, Pernemalm M, Crosbie PA and
Whetton AD: Molecular histology of lung cancer: From targets to
treatments. Cancer Treat Rev. 41:361–375. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carcereny E, Morán T, Capdevila L, Cros S,
Vilà L, de Los Llanos, Gil M, Remón J and Rosell R: The epidermal
growth factor receptor (EGRF) in lung cancer. Transl Respir Med.
3:12015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li LN, Chen WM and Fang QC: Studies on the
extraction and isolation of securinine from securinega suffruticosa
(PALL.) REHD by adsorption with active carbon. Yao Xue Xue Bao.
9:352–358. 1962.(In Chinese). PubMed/NCBI
|
|
4
|
Saito S, Kotera K, Shigematsu N, Ide A,
Sugimoto N, Horii Z, Hanaoka M, Yamawaki Y and Tamura Y: Structure
of securinine. Tetrahedron. 19:2085–2099. 1963. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gupta K, Chakrabarti A, Rana S, Ramdeo R,
Roth BL, Agarwal ML, Tse W, Agarwal MK and Wald DN: Securinine, a
myeloid differentiation agent with therapeutic potential for AML.
PLoS One. 6:e212032011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Treatment of chronic aplastic anemia with
securinine-analysis of 123 cases. Zhonghua Nei Ke Za Zhi.
22:764–766. 1983.(In Chinese). PubMed/NCBI
|
|
7
|
Neganova ME, Klochkov SG, Afanasieva SV,
Serkova TP, Chudinova ES, Bachurin SO, Reddy VP, Aliev G and
Shevtsova EF: Neuroprotective effects of the securinine-analogues:
Identification of Allomargaritarine as a lead compound. CNS Neurol
Disord Drug Targets. 15:102–107. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Beutler JA, Karbon EW, Brubaker AN, Malik
R, Curtis DR and Enna SJ: Securinine alkaloids: A new class of GABA
receptor antagonist. Brain Res. 330:135–140. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Copperman R, Copperman G and Der
Marderosian A: From Asia securinine-a central nervous stimulant is
used in treatment of amytrophic lateral sclerosis. Pa Med.
76:36–41. 1973.PubMed/NCBI
|
|
10
|
Li M, Han S, Zhang G, Wang Y and Ji Z:
Antiproliferative activity and apoptosis-inducing mechanism of
L-securinine on human breast cancer MCF-7 cells. Pharmazie.
69:217–223. 2014.PubMed/NCBI
|
|
11
|
Han S, Zhang G, Li M, Chen D, Wang Y, Ye W
and Ji Z: L-securinine induces apoptosis in the human promyelocytic
leukemia cell line HL-60 and influences the expression of genes
involved in the PI3K/AKT/mTOR signaling pathway. Oncol Rep.
31:2245–2251. 2014.PubMed/NCBI
|
|
12
|
Choi HJ, Park H, Lee HW and Kwon YG: The
Wnt pathway and the roles for its antagonists, DKKS, in
angiogenesis. IUBMB Life. 64:724–731. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Abdel-Magid AF: Wnt/β-catenin signaling
pathway inhibitors: A promising cancer therapy. ACS Med Chem Lett.
5:956–957. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xue G, Romano E, Massi D and Mandalà M:
Wnt/β-catenin signaling in melanoma: Preclinical rationale and
novel therapeutic insights. Cancer Treat Rev. 49:1–12. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chandra V and Hong KM: Effects of deranged
metabolism on epigenetic changes in cancer. Arch Pharm Res.
38:321–337. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Walter K, Holcomb T, Januario T, Yauch RL,
Du P, Bourgon R, Seshagiri S, Amler LC, Hampton GMS and Shames D:
Discovery and development of DNA methylation-based biomarkers for
lung cancer. Epigenomics. 6:59–72. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ko M, An J, Pastor WA, Koralov SB,
Rajewsky K and Rao A: TET proteins and 5-methylcytosine oxidation
in hematological cancers. Immunol Rev. 263:6–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Carless MA: Investigation of genomic
methylation status using methylation-specific and bisulfite
sequencing polymerase chain reaction. Methods Mol Biol.
1288:193–212. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang M, Zhang Y, Fei J, Chang X, Fan W,
Qian X, Zhang T and Lu D: Rapid quantification of DNA methylation
by measuring relative peak heights in direct bisulfite-PCR
sequencing traces. Lab Invest. 90:282–290. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hughes S and Jones JL: The use of multiple
displacement amplified DNA as a control for methylation specific
PCR, pyrosequencing, bisulfite sequencing and methylation-sensitive
restriction enzyme PCR. BMC Mol Biol. 8:912007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Reed K, Poulin ML, Yan L and Parissenti
AM: Comparison of bisulfite sequencing PCR with pyrosequencing for
measuring differences in DNA methylation. Anal Biochem. 397:96–106.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chirkin E, Atkatlian W, Do Q, Gaslonde T,
Dufat TH, Michel S, Lemoine P, Genta-Jouve G and Porée FH:
Chiroptical study and absolute configuration of securinine
oxidation products. Nat Prod Res. 29:1235–1242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Xiao, Ming Zheng and Hong Yan: The
extraction technology of securinine. Agricultural Food Products
Science and Technology. 5:6–1112. 2011.
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Navanesan S, Wahab Abdul N, Manickam S and
Sim KS: Leptospermum flavescens constituent-LF1 causes cell death
through the induction of cell cycle arrest and apoptosis in human
lung carcinoma cells. PLoS One. 10:e01359952015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sood S and Srinivasan R: Alterations in
gene promoter methylation and transcript expression induced by
cisplatin in comparison to 5-Azacytidine in HeLa and SiHa cervical
cancer cell lines. Mol Cell Biochem. 404:181–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nardi-Agmon I and Peled N: Exhaled breath
analysis for the early detection of lung cancer: Recent
developments and future prospects. Lung Cancer (Auckl). 8:31–38.
2017.PubMed/NCBI
|
|
28
|
Chen Z, Mei J, Liu L, Wang G, Li Z, Hou J,
Zhang Q, You Z and Zhang L: PD-L1 expression is associated with
advanced non-small cell lung cancer. Oncol Lett. 12:921–927.
2016.PubMed/NCBI
|
|
29
|
Ge L and Shi R: Progress of EGFR-TKI and
ALK/ROS1 inhibitors in advanced non-small cell lung cancer. Int J
Clin Exp Med. 8:10330–10339. 2015.PubMed/NCBI
|
|
30
|
Atherly AJ and Camidge DR: The
cost-effectiveness of screening lung cancer patients for targeted
drug sensitivity markers. Br J Cancer. 106:1100–1106. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lantermann AB, Chen D, McCutcheon K,
Hoffman G, Frias E, Ruddy D, Rakiec D, Korn J, McAllister G,
Stegmeier F, et al: Inhibition of casein kinase 1 alpha prevents
acquired drug resistance to erlotinib in EGFR-mutant non-small
celllung cancer. Cancer Res. 75:4937–4948. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang X, Han SW, Liu H, Zhu L, Chen YX and
Ji ZN: Secreted frizzled-related protein 1 (SFRP1) gene methylation
changes in the human lung adenocarcinoma cells treated with
L-securinine. J Asian Nat Prod Res. 1–19. 2017. View Article : Google Scholar
|
|
33
|
Gills JJ, Granville CA and Dennis PA:
Targeting aberrant signal transduction pathways in lung cancer.
Cancer Biol Ther. 3:147–155. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang XT, Ma YY, Guo K, Xia YJ, Wang HJ,
Li L, He XJ, Huang DS and Tao HQ: Assessing the diagnostic value of
serum Dickkopf-related protein 1 levels in cancer detection: A
case-control study and meta-analysis. Asian Pac J Cancer Prev.
15:9077–9083. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tian J, Xu XJ, Shen L, Yang YP, Zhu R,
Shuai B, Zhu XW, Li CG, Ma C and Lv L: Association of serum Dkk-1
levels with β-catenin in patients with postmenopausal osteoporosis.
J Huazhong Univ Sci Technolog Med Sci. 35:212–218. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sonohara F, Inokawa Y, Hayashi M, Kodera Y
and Nomoto S: Epigenetic modulation associated with carcinogenesis
and prognosis of human gastric cancer. Oncol Lett. 13:3363–3368.
2017.PubMed/NCBI
|
|
37
|
Shinjo K and Kondo Y: Targeting cancer
epigenetics: Linking basic biology to clinical medicine. Adv Drug
Deliv Rev. 95:56–64. 2015. View Article : Google Scholar : PubMed/NCBI
|