Introduction
Glioma is the most common subgroup type of brain
tumor, with an incidence of 5–6/100,000 cases/year in the United
States (1). Among all types of
glioma, glioblastoma multiforme (GBM) accounts for ~50% of cases
and has the most malignant phenotype (2). Despite the extensive developments that
have been invested in surgical techniques and therapeutic agents,
88% of patients with GBM succumb to this disease within 3 years
(3). GBM remains one of the most
challenging malignancies worldwide.
Embryonic stem cells (ESCs) are known for their
potent pluripotency and are able to differentiate into >220 cell
types in the adult body. The human homologue of the drosophila
spalt-like transcription factor 4 (SALL4) is a zinc-finger
transcription factor, which is responsible for maintaining
pluripotency and longevity of ESCs (4–6).
Previously, cancer stem cells (CSCs) have been identified in
various types of malignancies (7,8) and
demonstrate high self-renewal capabilities able to sustain tumor
growth (9). Thus far, ESCs and CSCs
have been revealed to share numerous biological similarities with
the SALL4 expression pattern being one of them. In various types of
tumors, high SALL4 expression level has been associated with
increased malignancy, including increased metastasis, enhanced
proliferation (10–12) and poor differentiation (13,14).
In the authors' previous study, it was demonstrated
that SALL4 was highly expressed in glioma and significantly
associated with poor survival (15).
The present study further investigated the biological role of SALL4
in the tumorigenesis of glioma and explored the underlying
mechanism of action. It was revealed that SALL4 regulated the cell
cycle, apoptosis, tumor cell invasion and temozolomide (TMZ)
treatment response in the U251 GBM cell line. Furthermore,
decreased SALL4 expression level was associated with a decreased
expression level of core transcription factors, including POU class
5 homeobox 1 (OCT4), SRY-box 2 (SOX-2) and Nanog homeobox (NANOG).
Additionally, knockdown of SALL4 inhibited O-6-methylguanine-DNA
methyltransferase (MGMT) and adenosine triphosphate-binding
cassette subfamily G member 2 (ABCG2) expression levels, which may
serve essential roles in GBM chemoresistance. These results suggest
that SALL4 serves an important role in tumorigenesis and may be a
useful therapeutic target for GBM.
Materials and methods
Cell culture, construction and
transfection
The human malignant glioma cell line U251 was
purchased from the Type Culture Collection of the Chinese Academy
of Science (Shanghai, China). Cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) containing 8% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.) and antibiotics (100
U/ml penicillin G and 100 µg/ml streptomycin). A total of four
different short interfering RNA (siRNA) sequences against SALL4
(GeneBank accession no. NM-020436) and one scrambled siRNA were
purchased from Shanghai GenePharma Co. Ltd. (Shanghai, China). The
siRNA sequences were as follows: siRNA-1,
5′-GCTAGACACATCCAAGAAAGG-3′; siRNA-2, 5′-GCCGAAAGCATCAAGTCAAAG-3′;
siRNA-3, 5′-GCCGACCTATGTCAAGGTTGA-3′; siRNA-4,
5′-GGAAGTTGGCCATCGAGAACA-3′; and scrambled siRNA,
5′-TTCTCCGAACGTGTCACGT-3′. Cells were transfected with siRNAs using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Following knockdown efficacy evaluation, the 3rd and 4th siRNA
variants at 72 h were selected for subsequent experiments. The
groups were named as blank control (BC; untransfected), negative
control (NC; scrambled siRNA), SALL4/siRNA-1 and SALL4/siRNA-2.
Details of cell culture, construction and transfection siRNA, and
evaluation of the siRNAs intervention efficacy were performed as
previously described (15).
Protein extraction and western
blotting
U251 cells were washed with 1X PBS and pelleted at
12,000 × g for 10 min at 4°C. Total protein concentration was
measured using a Bicinchoninic Assay Protein Assay kit (Beyotime
Institute of Biotechnology, Haimen, China). Proteins extracted from
cells (40 µg per lane) were separated using 10% SDS-PAGE gel
(Beyotime Institute of Biotechnology) and transferred to a
polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA,
USA) at 100 V for 1 h. Following blocking with 5% non-fat milk in
PBS with 0.1% Tween (PBST) for 1 h at room temperature, the
membranes were incubated at 4°C overnight with primary antibodies
and washed three times with 1X TBST. Next, cells were incubated at
room temperature for 2 h with the secondary antibody. The primary
antibodies used were as follows: Mouse polyclonal anti-SALL4
(Abgent, Inc., San Diego, CA, USA; cat no. AP1488b, dilution,
1:500); mouse polyclonal anti-OCT4 (cat no. 11263-1-AP), mouse
polyclonal anti-NANOG (cat no. 14295-1-AP), mouse polyclonal
anti-SOX2 (cat no. 20118-1-AP), mouse polyclonal anti-ABCG2 (cat
no. 10051-1-AP) and mouse polyclonal anti-MGMT (cat no. 17195-1-AP)
(all from ProteinTech Group, Inc., Chicago, IL, USA; dilution,
1:500); and polyclonal rabbit anti-β-actin (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA; cat no. sc-130656, 1:1,000)
used as a gel loading control. The secondary antibody used was
horseradish peroxidise-conjugated Goat Anti-Rabbit IgG (H&L) AP
(BioVision, Inc., San Francisco, CA, USA; cat no. 93-6923-100,
dilution, 1:2,000). Immunoblotting bands were visualized visualized
using ECL Substrates (Tanon Science and Technology Co., Ltd,
Shanghai, China) and quantified using ImageJ software version 1.48
(National Institutes of Health, Bethesda, MA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from U251 glioma cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). Reverse transcription was performed using
the Thermoscript RT-PCR system (Invitrogen; Thermo Fisher
Scientific, Inc.) with random hexamer primers and the Superscript
II Reverse Transcriptase kit (Invitrogen; Thermo Fisher Scientific,
Inc.), for 30 min at 25°C, 30 min at 42°C and 10 min at 85°C. qPCR
was performed using the quantitative Real Time PCR 7500 sequence
detection system (Applied Biosystems, Foster city, CA, USA).
RT-qPCR was performed as follows: 40 cycles of denaturation at 95°C
(12 sec) and annealing/extension at 60°C (40 sec). Primers were
designed using Primer 5.0 software (Applied Biosystems). The primer
sequences were as follows: SALL4 forward (F),
5′-ATAGTCAAGCCGAAAGCATCAAGTC-3′ and reverse (R),
5′-CTCCGACCTTCCATCTCAGTGC-3′; OCT4 F, 5′-ACCTATTCAGCCAAACGACCAT-3′
and R, 5′-CTGCTTCCTCCACCCACTTCT-3′; SOX2 F,
5′-GCTCGCAGACCTACATGAAC-3′ and R, 5′-GGGAGGAAGAGGTAACCACA-3′; NAN
OGF, 5′-ATACCTCAGCCTCCAGCAGATG-3′ and R,
5′-TTCTGCCACCTCTTAGATTTCATTC-3′; MGM T F,
5′-TCTTCACCATCCCGTTTTCCAG-3′ and R,
5′-CTTCTCCGAATTTCACAACCTTCAG-3′; ABCG2 F,
5′-GAAACCTGGTCTCAACGCCATC-3′ and R,
5′-ACTTGGATCTTTCCTTGCAGCTAAG-3′; and GAP DH F,
5′-CATGAGAAGTATGACAACAGCCT-3′ and R, 5′-AGTCCTTCCACGATACCAAAGT-3′.
SALL4 mRNA expression was normalized to GAPDH and analyzed using
the 2−ΔΔCq method (16).
Cell cycle analysis
Cell cycle analysis was performed by flow cytometry
using transfected and control U251 cells in log-phase growth. Cells
were washed with PBS, fixed with 90% ethanol overnight at 4°C and
incubated with RNase at 37°C for 30 min. Cell nuclei were stained
with propidium iodide (PI) at 4°C for an additional 30 min. The
stained nuclei were then analyzed using a FACSCalibur flow
cytometer (BD Biosciences, Franklin Lakes, NJ, USA). The
populations of cells distributed in the
G0/G1, S and G2/M cell cycle
phases were evaluated using WinMDI version 2.9 software (The
Scripps Research Institute, San Diego, CA, USA).
Apoptosis assay
Apoptosis was evaluated using an annexin V
fluorescein isothiocyanate (FITC)/PI apoptosis detection kit
(Thermo Fisher Scientific, Inc.), according to the manufacture's
protocol. Briefly, following incubation (16 h) at 37°C in a 5%
CO2 humidified atmosphere, U251 cells cultured in 6-cm
dishes were digested with trypsin without EDTA, washed twice with
PBS and suspended in 100 µl binding buffer, followed by staining
with 5 µl Annexin V-FITC and 5 µl PI for 15 min in the dark at room
temperature and then analyzed by flow cytometry as
aforementioned.
Invasion assay
Equal numbers (1×105) of transfected and
control U251 cells were seeded in separate 24-well cell culture
inserts coated with Matrigel with 8 µm pores. 500 µl DMEM
supplemented with 10% FBS was added into the lower chamber as a
chemoattractant. Following a 24-h incubation at 37°C with 5%
CO2, cells that were adhered to the upper surface of the
filter were removed using a cotton applicator. The cells on the
lower surface of the membrane (the migrated cells) were fixed with
3.7% formaldehyde for 10 min at −20°C, stained with 0.1% crystal
violet (Beyotime Institute of Biotechnology) for 30 min at 37°C.
The cell number was determined in at least five randomly selected
fields under a light microscope (magnification, ×200; SZ61;
Olympus, Tokyo, Japan). The invasion rate was determined for three
independent experiments as follows: No. of migrated cells/total no.
of cells ×100.
Cytotoxicity assay
The cytotoxicity of TMZ (Merck & Co., Inc.,
Whitehouse Station, NJ, USA) on glioma cells was detected using a
Total Superoxide Dismutase Assay kit and WST-8 assay according to
manufacturers' protocol. Briefly, U251 cells were plated at a
density of 5×103 cells/well in 96-well plates and
allowed to attach overnight at 37°C. Various concentrations of TMZ
(2, 4, 8, 16, 32, 64, 128 and 256 µg/ml) were subsequently added,
and the cells were cultured for 72 h at 37°C. A total of 4-h prior
to harvest, 10 µl/well of the Cell Counting kit-8 reagent (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan) was added and the
cells were incubated for 2 h at 37°C. The optical density (OD) at
450 nm was recorded using a microplate reader (Bio-Rad). The cell
survival rate was determined by comparing the OD values of the
treated samples with those of the untreated controls within each
group. The concentration of TMZ required to inhibit cell growth by
50% (the toxic concentration, TC50) was evaluated using
survival curves.
Statistical analysis
Data are expressed as the mean ± standard deviation
of three independent experiments. Statistical analysis was
performed using the Student's t-test between two groups, whereas
the comparison between ≥ three groups was performed using one-way
analysis of variance. Post hoc tests were used for comparisons
between groups. Student-Neuman-Keuls method was used when equal
variances assumed. If equal variances not assumed, Dunnett's T3
method was used. All analyses were performed using SPSS version
13.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
SALL4 knockdown induces cell cycle
arrest, enhances apoptosis and invasion inhibition
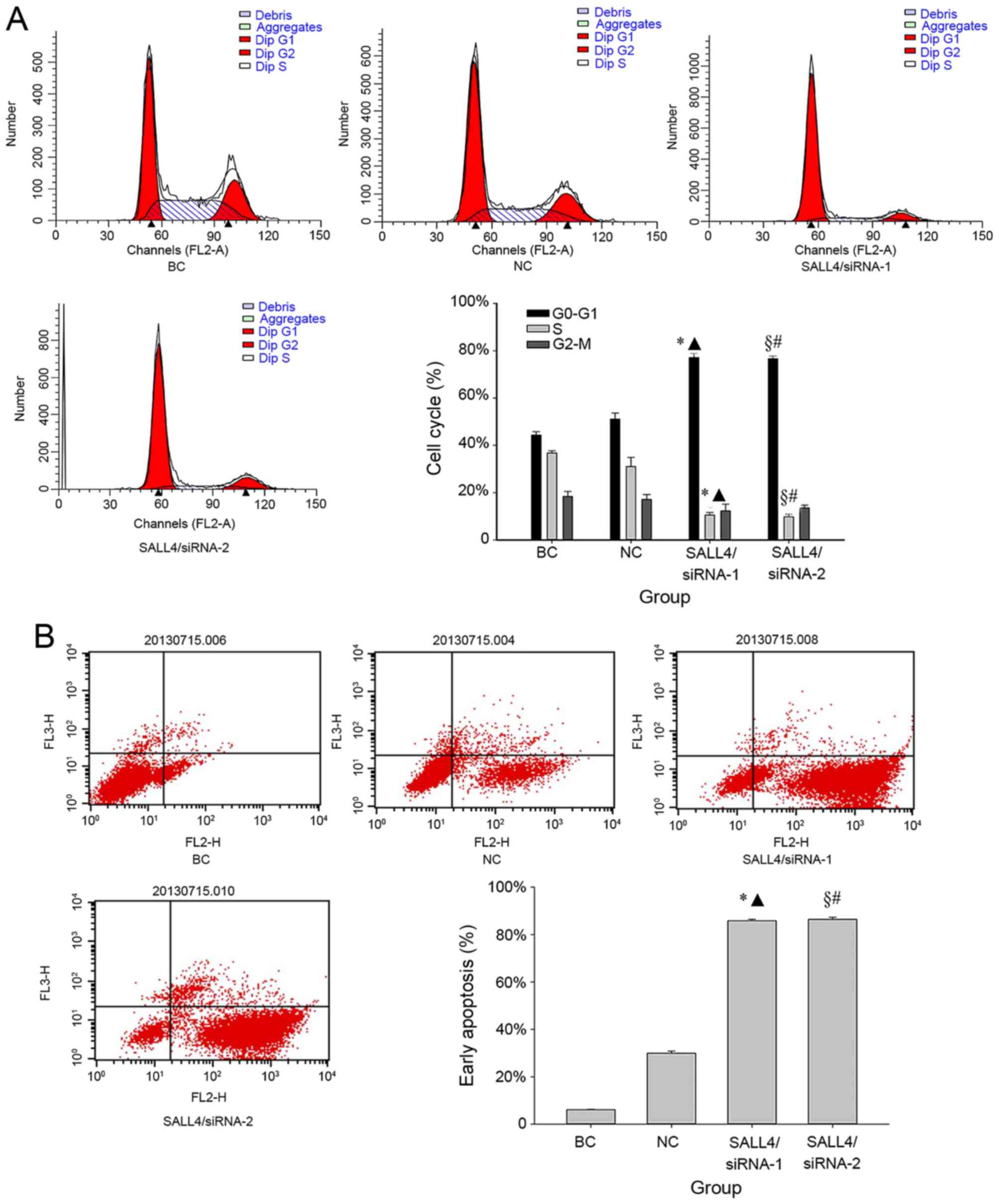
The cell cycle distribution in various transfection
groups of U251 glioma cells were analyzed by flow cytometry 72 h
after transfection. No significant differences in the fraction of
G1 phase cells were identified between the NC group
(52.11±1.92%) and BC group (42.30±0.42%) (P=0.14; Fig. 1A). However, following treatment with
siRNAs, the G1 phase cell fraction in the SALL4/siRNA-1
(77.17±1.77%) and SALL4/siRNA-2 (76.67±1.12%) groups were
significantly higher compared with that of the BC (both P<0.001)
and NC groups (siRNA-1, P=0.002; siRNA-2, P=0.005) (Fig. 1A). Correspondingly, the fraction of
cells in the S phase were 36.75±0.95, 31.03±3.88, 10.54±1.08 and
9.76±1.16% in the BC, NC, SALL4/siRNA-1, and SALL4/siRNA-2 groups,
respectively. Additionally, the reduction in the fraction of cells
in the G2-M phase were 18.41±2.10, 17.17±2.01,
12.28±2.84 and 13.58±1.10% in the BC, NC, SALL4/siRNA-1, and
SALL4/siRNA-2 groups, respectively. SALL4/siRNA-1 and SALL4/siRNA-2
demonstrated significantly lower S phase cell proportions compared
with those in the BC (both P<0.001) and NC groups (siRNA-1,
P=0.047; siRNA-2, P=0.041). However, no significant differences in
the G2-M cell cycle phase were identified in the
SALL4/siRNA-1 and SALL4/siRNA-2 groups, when compared with the BC
(siRNA-1, P=0.24; siRNA-2, P=0.21), and NC (siRNA-1, P=0.39;
siRNA-2, P=0.36) groups.
Subsequently, the present study compared the
apoptosis rate prior to and following SALL4 knockdown (Fig. 1B). It was revealed that SALL4/siRNA-1
(85.88±0.54%) and SALL4/siRNA-2 (86.41±0.87%) induced significantly
increased early apoptosis rates compared with that in the BC, and
NC groups (all P<0.001).
The Transwell migration was performed to further
examine the effect of SALL4 on glioma cell invasion (Fig. 1C). The mean invasion cell number was
94.33±3.51 and 91.33±1.53 in the BC, and NC groups, respectively,
compared with 45.00±1.00 and 47.67±3.06 in the SALL4/siRNA-1, and
SALL4/siRNA-2 groups, respectively. The siRNA treatment groups
significantly reduced the migratory ability of cells compared with
that in BC (siRNA-1, P=0.005; siRNA-2, P<0.001) and NC (siRNA-1,
P=0.001; siRNA-2, P<0.001) groups.
The Janus kinase (Jak)-signal
transducer and activator of transcription 3 (Stat3) signaling
pathway is utilized by SALL4 to fulfill its function
In order to investigate the reduction of malignancy
of glioma detected in the present study, the expression levels of
OCT4, SOX2 and NANOG were analyzed, which are essential factors of
the Jak-Stat3 signaling pathway. It was demonstrated that
SALL4/siRNA-1 and SALL4/siRNA-2 suppressed the expression levels of
OCT4, SOX2 and NANOG, which were all statistically lower compared
with the BC and NC groups (Fig. 2A).
Compared with the BC group, the inhibition rates of SALL4/siRNA-1
on OCT4, SOX2 and NANOG mRNA expression levels were 59% (P=0.004),
38% (P=0.001) and 39% (P=0.002), respectively, whereas the
inhibition rates of SALL4/siRNA-2 were 65% (P<0.001), 26%
(P=0.038) and 29% (P=0.036), respectively. Compared with the NC
group, the inhibition rates of SALL4/siRNA-1 were 65.54%
(P<0.001), 37.37% (P=0.038) and 40.20% (P=0.013), respectively,
whereas the inhibition rates of SALL4/siRNA-2 were 70.59%
(P<0.001), 25.25% (P=0.037) and 30.40% (P=0.032) (Table I; Fig.
2A). Western blot analysis revealed similar results (Fig. 2B).
 | Figure 2.SALL4 knockdown altered the expression
levels of OCT4, SOX2, NANOG, MGMT and ABCG2. (A) The mRNA
expression levels of OCT4, SOX2, NANOG, MGMT and ABCG2 in siRNA
groups was markedly lower compared with in BC and NC groups at 72 h
post-transfection. (B) Western blotting revealed that changes in
expression levels at the protein level were qualitatively similar
to the changes in mRNA expression levels. BC group; NC group;
SALL4/siRNA-1 group; SALL4/siRNA-2 group. *P<0.05 siRNA-1 vs.
BC, ▲P<0.05 siRNA-1 vs. NC, §P<0.05
siRNA-2 vs. BC, #P<0.05 siRNA-2 vs. NC. SALL4,
spalt-like transcription factor 4; OCT4, POU class 5 homeobox 1;
SOX2, SRY-box 2; NANOG, Nanog homeobox; MGMT, O-6-methylguanine-DNA
methyltransferase; ABCG2, adenosine triphosphate-binding cassette
subfamily G member 2; siRNA, short interfering RNA; BC, blank
control; NC, negative control. |
 | Table I.mRNA expression levels relative to
GAPDH. |
Table I.
mRNA expression levels relative to
GAPDH.
|
| Gene |
|---|
|
|
|
|---|
| Group | SALL4 | OCT4 | SOX2 | NANOG | MGMT | ABCG2 |
|---|
| BC | 1 | 1 | 1 | 1 | 1 | 1 |
| NC | 0.81±0.05 | 1.19±0.11 | 0.99±0.02 | 1.02±0.08 | 1.02±0.08 | 1.35±0.16 |
| SALL4/siRNA-1 | 0.39±0.03 | 0.41±0.08 | 0.62±0.02 | 0.61±0.01 | 0.61±0.01 | 0.72±0.09 |
| SALL4/siRNA-2 | 0.36±0.05 | 0.35±0.02 | 0.74±0.04 | 0.71±0.05 | 0.71±0.05 | 0.61±0.07 |
SALL4 knockdown increases
chemosensitivity to TMZ
Subsequently, the present study aimed to investigate
the effect of SALL4 on chemotherapeutic TMZ during glioma
treatment. It was revealed that the TC50 of TMZ in
SALL4/siRNA-1 and SALL4/siRNA-2 groups were 68.34±3.52, and
67.44±4.71 µg/ml, respectively. Compared with that in the BC
(113.66±23.07 µg/ml) and NC groups (114.93±20.91 µg/ml), the
TC50 value of SALL4/siRNA-1 (BC, P=0.047; NC, P=0.030)
and SALL4/siRNA-2 were significantly decreased (BC, P=0.043; NC,
P=0.026) (data not shown).
In order to describe this phenomenon, the present
study hypothesized that the knockdown of SALL4 suppressed glioma
tumor resistance via MGMT and ABCG2 downregulation. The present
study revealed that SALL4/siRNA-1 decreased MGMT and ABCG2 mRNA
expression levels by 39, and 28%, respectively, which were
significantly lower compared with that of the BC (MGMT, P=0.021;
ABCG2, P=0.032) and NC (MGMT, P<0.001; ABCG2, P=0.024) groups
(Table I; Fig. 2A). Similarly, when compared with the
BC and NC groups, SALL4/siRNA-1 decreased MGMT protein expression
level significantly (BC, P=0.009; NC, P=0.010), and SALL4/siRNA-2
significantly decreased expression levels of MGMT (BC, P=0.007; BC,
P=0.009) and ABCG2 (BC, P=0.009; BC, P=0.006) (Fig. 2B).
Discussion
Since SALL4 was first detected as an oncogene in
leukemia (17), the function of SALL4
had been studied in various types of cancer. For example, in lung,
breast, gastric and colorectal cancer cells (10–12), SALL4
has been demonstrated to regulate cell viability, apoptosis, and
tumorigenicity (12). In the authors'
previous study, it was reported that SALL4 served an essential role
in glioma and its expression was negatively associated with
prognosis (15); however, the
detailed mechanisms remain unclear. The present study aimed to
investigate the detailed mechanisms underlying SALL4 in glioma
tumorigenesis. Following knockdown of SALL4, cell cycle arrest,
significantly increased levels of early apoptosis and invasion
inhibition were observed in GBM cells. Furthermore, decreased SALL4
expression level significantly decreased mRNA and protein
expression levels of OCT4, SOX2, and NANOG. In addition, inhibition
of SALL4 decreased the TC50 of chemotherapeutic agent
TMZ. These results partially elucidated the specific mechanisms
utilized by SALL4, indicating that SALL4 may serve an important
role in GBM tumorigenesis.
The CSC theory hypothesizes that CSCs and ESCs share
numerous malfunctioned proliferation signal pathways (18), and the Jak-Stat3 signaling pathway is
considered as one of the most important (19). At the top hierarchy of the Jak-Stat3
signaling pathway, OCT4, SOX2 and NANOG, are considered to serve
the most important function in maintaining ESC properties (20). Previously, increased glioma
progression was identified to be associated with upregulated OCT4,
SOX2 and NANOG (21,22). Additionally, combinatorial expression
levels of OCT4, NANOG and SOX2 were positively associated with
increasing glioma malignancy (22).
The present study demonstrated that knockdown of SALL4
significantly downregulated the mRNA and protein expression levels
of OCT4, SOX2, and NANOG, which supports the results regarding
SALL4 involvement in cell cycle arrest, enhanced apoptosis and
invasion inhibition. These data suggest that SALL4 inhibits glioma
tumorigenesis by participating in the Jak-Stat3 signaling pathway.
However, it remains unclear how and which factors SALL4 interacts
with in the Jak-Stat3 signaling pathway; therefore, further
research is required for elucidation.
Notably in the present study, the knockdown of SALL4
significantly reduced the TMZ TC50. As previously
demonstrated, cancer utilizes various mechanisms enabling
resistance to anticancer drugs, including DNA repairing and
reducing toxin uptake via pumping-out chemotherapeutics (23). TMZ is a classical chemotherapy drug
for glioma, which is normally used by patients with GBM followed by
surgical resection and radiotherapy (24). However, the majority of patients with
glioma develop resistance to TMZ during treatment (25). Thus, it was hypothesized that glioma
may use or develop ‘DNA repairing’ and ‘toxin pumping-out’
strategies to remain resistant. MGMT is a DNA repair protein, which
removes alkylating adducts from the O6 position of guanine and
protects cells from cytotoxic, and mutagenic effects, resulting in
a resistance of tumor cells to alkylating agent-based chemotherapy
(26). Following the knockdown of
SALL4, a significant decrease in the TC50 of TMZ with
simultaneous inhibition of MGMT expression was observed. A previous
study demonstrated that the expression level of SALL4 was
positively associated with histone deacetylase activity in
EpCAM-positive hepatocellular carcinomacell Hep3B and HuH7 cell
lines (27). As the present study
knocked down SALL4 in glioma cells, there would be less potent
histone deacetylase activity, which may inhibit various protein
expression levels. Although it remained unclear whether MGMT
promoter would be deacetylated, the present study observed a
lowered TC50 for TMZ as well as decreased MGMT protein
expression levels simultaneously. Thus, it was reasonable to
hypothesize that the knockdown of SALL4 decreased histone
deacetylase activity, which inhibited MGMT expression and the
subsequent TMZ TC50 in glioma cells. This hypothesis is
supported by the results of a previous study that also used glioma
cells (28) and the results of the
present study. Additionally, pumping out chemotherapeutics is
another popular mechanism used by various types of cancer. ABC drug
transporters have been accepted to reduce the efficacy of
chemotherapeutics by pumping out toxic drugs and thus contribute to
aggressive tumor behaviors, and poor prognosis (29). As previously demonstrated to be
enriched in GBM (30), ABCG2, which
is a member of the ABC family, could also be involved in tumor
resistance. The present study demonstrated that following knockdown
of SALL4, ABCG2 exhibited significantly reduced expression levels,
which may explain the reduction of TMZ TC50. Thus, the
results of the present study suggest that MGMT and ABCG2 serve
important roles in GBM resistance, and SALL4 is responsible for
maintaining high expression and activity levels of MGMT and ABCG2.
Further studies should focus on where and how SALL4 interacts with
MGMT and ABCG2.
In conclusion, the present study revealed that the
knockdown of SALL4 significantly decreased the malignancy and
increased the sensitivity of TMZ to U251 cells. These results
indicate that SALL4 may serve an essential role in the tumorigenic
properties of glioma cells. Thus, SALL4 may be a potential
therapeutic target for glioma, particularly for GBM.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 30930094) and the
Natural Science Foundation of Shanghai (grant no. 13ZR1414200).
References
|
1
|
Weller M: Novel diagnostic and therapeutic
approaches to malignant glioma. Swiss Med Wkly.
141:w132102011.PubMed/NCBI
|
|
2
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wen P and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kohlhase J, Schuh R, Dowe G, Kühnlein RP,
Jäckle H, Schroeder B, Schulz-Schaeffer W, Kretzschmar HA, Köhler
A, Müller U, et al: Isolation, characterization, and organ-specific
expression of two novel human zinc finger genes related to the
Drosophila gene spalt. Genomics. 38:291–298. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY,
Soh BS, Lou Y, Yang J, Ma Y, Chai L, et al: Sall4 modulates
embryonic stem cell pluripotency and early embryonic development by
the transcriptional regulation of Pou5f1. Nat Cell Biol.
8:1114–1123. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang J, Gao C, Chai L and Ma Y: A novel
SALL4/OCT4 transcriptional feedback network for pluripotency of
embryonic stem cells. PLoS One. 5:e107662010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Singh SK, Clarke ID, Terasaki M, Bonn VE,
Hawkins C, Squire J and Dirks PB: Identification of a cancer stem
cell in human brain tumors. Cancer Res. 63:5821–5828.
2003.PubMed/NCBI
|
|
8
|
Galli R, Binda E, Orfanelli U, Cipelletti
B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F and Vescovi
A: Isolation and characterization of tumorigenic, stem-like neural
precursors from human glioblastoma. Cancer Res. 64:7011–7021. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kobayashi D, Kuribayashi K, Tanaka M and
Watanabe N: Overexpression of SALL4 in lung cancer and its
importance in cell proliferation. Oncol Rep. 26:965–970.
2011.PubMed/NCBI
|
|
11
|
Kobayashi D, Kuribayshi K, Tanaka M and
Watanabe N: SALL4 is essential for cancer cell proliferation and is
overexpressed at early clinical stages in breast cancer. Int J
Oncol. 38:933–939. 2011.PubMed/NCBI
|
|
12
|
Yong KJ, Gao C, Lim JS, Yan B, Yang H,
Dimitrov T, Kawasaki A, Ong CW, Wong KF, Lee S, et al: Oncofetal
gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med.
368:2266–2276. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Aguila JR, Liao W, Yang J, Avila C, Hagag
N, Senzel L and Ma Y: SALL4 is a robust stimulator for the
expansion of hematopoietic stem cells. Blood. 118:576–585. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang J, Aguila JR, Alipio Z, Lai R, Fink
LM and Ma Y: Enhanced self-renewal of hematopoietic stem/progenitor
cells mediated by the stem cell gene Sall4. J Hematol Oncol.
4:382011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang L, Yan Y, Jiang Y, Cui Y, Zou Y,
Qian J, Luo C, Lu Y and Wu X: The expression of SALL4 in patients
with gliomas: High level of SALL4 expression is correlated with
poor outcome. J Neurooncol. 121:261–268. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma Y, Cui W, Yang J, Qu J, Di C, Amin HM,
Lai R, Ritz J, Krause DS and Chai L: SALL4, a novel oncogene, is
constitutively expressed in human acute myeloid leukemia (AML) and
induces AML in transgenic mice. Blood. 108:2726–2735. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cheng L, Alexander R, Zhang S, Pan CX,
MacLennan GT, Lopez-Beltran A and Montironi R: The clinical and
therapeutic implications of cancer stem cell biology. Expert Rev
Anticancer Ther. 11:1131–1143. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu H, Lee H, Herrmann A, Buettner R and
Jove R: Revisiting STAT3 signalling in cancer: New and unexpected
biological functions. Nat Rev Cancer. 14:736–746. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Boyer LA, Lee TI, Cole MF, Johnstone SE,
Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG,
et al: Core transcriptional regulatory circuitry in human embryonic
stem cells. Cell. 122:947–956. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo Y, Liu S, Wang P, Zhao S, Wang F, Bing
L, Zhang Y, Ling EA, Gao J and Hao A: Expression of embryonic stem
cell-associated genes Oct4, Sox2 and Nanog in human gliomas.
Histopathology. 59:763–775. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Holmberg J, He X, Peredo I, Orrego A,
Hesselager G, Ericsson C, Hovatta O, Oba-Shinjo SM, Marie SK,
Nistér M and Muhr J: Activation of neural and pluripotent stem cell
signatures correlates with increased malignancy in human glioma.
PLoS One. 6:e184542011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shen DW, Pouliot LM, Hall MD and Gottesman
MM: Cisplatin resistance: A cellular self-defense mechanism
resulting from multiple epigenetic and genetic changes. Pharmacol
Rev. 64:706–721. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hiddingh L, Raktoe RS, Jeuken J, Hulleman
E, Noske DP, Kaspers GJ, Vandertop WP, Wesseling P and Wurdinger T:
Identification of temozolomide resistance factors in glioblastoma
via integrative miRNA/mRNA regulatory network analysis. Sci Rep.
4:52602014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kaina B, Christmann M, Naumann S and Roos
WP: MGMT: Key node in the battle against genotoxicity,
carcinogenicity and apoptosis induced by alkylating agents. DNA
Repair (Amst). 6:1079–1099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zeng SS, Yamashita T, Kondo M, Nio K,
Hayashi T, Hara Y, Nomura Y, Yoshida M, Hayashi T, Oishi N, et al:
The transcription factor SALL4 regulates stemness of EpCAM-positive
hepatocellular carcinoma. J Hepatol. 60:127–134. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kitange GJ, Mladek AC, Carlson BL,
Schroeder MA, Pokorny JL, Cen L, Decker PA, Wu W, Lomberk GA, Gupta
SK, et al: Inhibition of histone deacetylation potentiates the
evolution of acquired temozolomide resistance linked to MGMT
upregulation in glioblastoma xenografts. Clin Cancer Res.
18:4070–4079. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim HS, Kim NC, Chae KH, Kim G, Park WS,
Park YK and Kim YW: Expression of multidrug resistance-associated
protein 2 in human gallbladder carcinoma. Biomed Res Int.
2013:5275342013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chamberlain MC, Bota DA, Linskey ME and
Schwartz PH: Neural stem/progenitors and glioma stem-like cells
have differential sensitivity to chemotherapy. Neurology.
77:e135–e136. 2011. View Article : Google Scholar : PubMed/NCBI
|