Introduction
Cervical cancer is the most prevalent type of cancer
in females (1) with >500,000 novel
cases reported annually in developing countries (2), and >200,000 cervical
cancer-associated mortalities each year worldwide (3). Signaling pathway dysregulation is
hypothesized to be involved in cervical cancer tumorigenesis
(4). The highly conserved Wnt
signaling pathways, are involved in cell differentiation, embryonic
development and tissue formation. The canonical Wnt signaling
pathway is also known as the β-catenin pathway and is responsible
for triggering transcription of a subset of downstream genes. The
Wnt signaling pathway serves a crucial function in development and
the dysregulation of this pathway is associated with a number of
disorders, including cancer. The principal reasons for Wnt
signaling dysfunction are mutation or malfunction of proteins
involved in the pathway (5). Abnormal
accumulation of β-catenin is suggested to stimulate the
proliferation and metastasis of cancer cells (6–8).
β-catenin is an 88 kDa multi-function scaffolding
protein encoded at chromosome 3p22-21.3 and is primarily membrane
bound with epithelial (E)-cadherin. β-catenin accounts for the
following two biological functions: i) Acts as a subunit of the
adhering complex when binding with E-cadherin and α-catenin; and
ii) acts as a co-activator of the transcription factor
(TCF)/lymphoid enhancer-binding factor family (LEF) (9,10).
Cytoplasmic and nuclear accumulation of β-catenin are a feature of
colorectal, lung and breast cancer, hepatoma and melanoma (11–13).
Additionally, β-catenin has been identified to be overexpressed in
cervical cancer (8) and the
dysregulation of β-catenin has been suggested to be a biomarker of
cervical squamous cell carcinoma (14). A number of underlying molecular
mechanisms have been suggested to explain how the abnormal
accumulation of β-catenin leads to cancer vulnerability, including
interference with cellular polarity and accelerated cell
proliferation, survival and migration (5,15).
Chibby is a highly conserved protein with a
molecular weight of 14.5 kDa comprising 126 amino acids and encoded
at chromosome 22q12-13. Chibby has been identified to be an
antagonist of β-catenin; it binds to β-catenin in the nucleus, and
exhibits a negative regulating effect on the Wnt signaling pathway
and the transcriptional activities of genes downstream of Wnt
(16). In addition, Chibby has been
hypothesized to be a tumor suppressor that controls β-catenin
through two primary mechanisms, including a direct control,
nuclear-cytoplasmic shuttling of Chibby controls β-catenin
signaling and indirect control, which cooperates with 14-3-3 in
order to regulate β-catenin subcellular distribution and signaling
activity (17,18). Chibby is suggested to be downregulated
in neuroblastoma (19), lung cancer
(20), ependymoma (21) and colorectal cancer (22,23).
Furthermore, mutations of Chibby have been associated with
tumorigenesis (24). However, the
association between Chibby and β-catenin, and the underlying
molecular mechanism by which Chibby may suppress cervical cancer
tumorigenesis, remains unknown. From preliminary data,
overexpression of Chibby may suppress the transcriptional activity
of β-catenin and induce apoptosis. Consequently, cell
proliferation, migration and cancer cell colonization were
inhibited, which indicated a tumor suppressor function of Chibby
(Huang, Y.L., unpublished work). Therefore, we hypothesized that
Chibby may control cancer cell growth by negatively regulating
β-catenin/TCF4 signaling and affecting c-Myc and proliferating cell
nuclear antigen expression. In two cervical cancer cell lines, HeLa
and SiHa, mRNA and protein Chibby expression were downregulated.
These results suggested that Chibby may be a tumor suppressor in
cervical cancer in vitro (Huang, Y.L., unpublished work);
however, the precise in vivo function of Chibby in cervical
cancer, the differences between Chibby expression in carcinoma and
benign tissue, and the association between Chibby and β-catenin
expression all remain unknown. Therefore, in the present study,
paraffin-embedded cervical cancer tissues at distinct cervical
intraepithelial neoplasia (CIN) stages were selected and the mRNA
expression of Chibby and β-catenin was quantified. In addition,
protein expression and localization were determined to explore: i)
The cytosolic and nuclear expression patterns with respect to
Chibby and β-catenin; ii) whether mRNA expression of Chibby and
β-catenin is associated with the progression of cervical cancer;
and iii) whether Chibby and β-catenin expression may be used as a
biomarker of cervical cancer.
Materials and methods
Tissue collection
All paraffin-embedded tissues were retrospectively
collected from the tissue bank of Kaohsiung Armed Forces General
Hospital (KAFGH; Kaohsiung, Taiwan) between June 2004 and October
2012. Patient's age ranged from 19 to 89 years with an average of
49.09 years. A total of 87 cervical tissues were selected and
dissected from cervical conization and total hysterectomy, and no
biopsy samples were used. All participants were females. Tissues
selected were 4 chronic cervicitis, 13 CIN 1, 15 CIN 2, 33 CIN 3
and 22 invasive squamous-cell carcinoma (SCC) samples, based on the
classification system described previously (25). The present study was approved by the
Institutional Review Board (IRB) of KAFGH (Kaohsiung, Taiwan).
Immunohistochemistry of β-catenin and
Chibby
The paraffin-embedded tissue blocks were pre-sliced
using a tissue dissector (thickness, 1–2 µm). Subsequently,
paraffin blocks were cooled in an ice-water mixture for 30 min
prior to sectioning. Tissue sections 4–6 µm thick were cut and
placed on histone-coated (Hate Chemical, Westbury, NY, USA) slides.
Following a brief drying period of approximately 15 min, the tissue
sections were heat-fixed to the slide at 37°C and subsequently
incubated at 70°C for 30 min before deparaffinization. The slides
were then taken directly from the incubator and deparaffinized in
xylene twice for 5 min each, followed by one rinse in absolute
ethanol (ETOH), and then sequentially rinsed in 90, 80 and 70% ETOH
and dH2O for 10 sec each. To eliminate staining due to
endogenous peroxidase, the tissue sections were treated with an
ETOH/H2O2 (45 ml ETOH/3 ml
H2O2) block for 10 min at 42°C (26). Immunohistological staining was
performed using a NovoLink polymer detection kit (Leica
Microsystems, Ltd., Milton Keynes, UK). A mouse monoclonal
immunoglobulin G (IgG) antibody (Ab) was used to detect β-catenin
(monoclonal mouse anti-β-catenin Ab; catalog no. NCL-B-CAT;
dilution, 1:200; Leica Microsystems, Ltd.) and a rabbit polyclonal
IgG Ab (polyclonal rabbit anti-Chibby Ab; catalog no. HRIHFB2025;
dilution, 1:400; ABgene; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) was used to detect Chibby. Slides were incubated for 30
min at 42°C with the primary antibodies. Slides were then rinsed in
PBS and subsequently incubated in the presence of the secondary
antibody at 42°C for 15 min. The secondary antibodies were included
in the NovoLink polymer detection kit. All procedures were
performed according to the manufacture's protocol. The results were
observed under a Nikon Eclipse 50i light microscope (Nikon, Tokyo,
Japan) at magnifications of ×40–400 and were interpreted by senior
pathologists who were blinded to the diagnostic results of each
section. Overall, 90% of the samples were consistent with their
original diagnosis; if any uncertainties were encountered, the
section was re-interpreted by a different pathologist. The protein
expression level was interpreted by two means, staining intensity
and the positive staining area; in addition, the protein location
corresponding to cytosol and nuclear was indicated. For determining
intensity, the following four-level scale was used: No (no protein
positive staining can be observed at ×400 magnification), weak
(protein positive staining can easily be observed at ×400
magnification), intermediate (protein positive staining can easily
be observed at ×200 magnification) and strong staining (protein
positive staining can easily be observed at ×100 magnification).
For defining the area positive staining, the following five-level
scale was used: No staining, <1%, between 1 and 10%, between 11
and 50%, and >50% positivity. For those sections with positive
detecting area <10% was considered as ‘negative’ for protein
expression; in contrast, sections with >10% positive staining
area were viewed as ‘positive’ for target protein expression.
RNA extraction
Total RNA was prepared from tissue sections prepared
from paraffin-embedded tissues using a PureLink FFPE Total RNA
Isolation kit (Invitrogen; Thermo Fisher Scientific, Inc., Waltham,
MA, USA), and de-paraffinization, purification and washing were
conducted according to the manufacturer's protocol. The RNA
concentration was determined by detecting the absorbance at 280 nm
under ultraviolet.
Complementary (c)DNA synthesis
First strand cDNA synthesis was carried out using an
Impron-II Reverse Transcription System (Promega Corporation,
Madison, WI, USA). Total RNA (1 µg) was premixed with oligo(dT) and
random hexamers in a 0.2 ml vial, heated at 70°C for 5 min and
chilled at 4°C for 5 min for pre-denaturation. Subsequently, 4 µl
Impron-II 5× Reaction Buffer, 25 mM MgCl2, 1 µl 10 mM
dNTP Mix, 20 U ribonuclease inhibitor, nuclease-free water and 1 µl
ImProm-II Reverse Transcriptase were added to a final volume of 15
µl. The following temperature protocol was applied: 25°C for 5 min
for primer annealing, 42°C for 60 min for synthesis, followed by
70°C for 15 min to inactivate enzymes. The synthesized cDNA was
stored at −20°C until used.
Quantitative polymerase chain reaction
(qPCR)
A total of 11 CIN 1, 9 CIN 2, 25 CIN 3 and 9 SCC
cancerous tissues underwent target mRNA detection (only 54 out of
83 collected cervical tumor tissues had sufficient quality mRNA for
further tests). Detection of mRNA expression levels with respect to
endogenous Chibby and β-catenin was performed using EZtime
real-time PCR premix (2X SYBR Green premix, Yeastern Biotech Corp.,
Taipei, Taiwan). The thermal cycling protocol was performed using
an IQ5 real-time PCR detection system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The increase in fluorescence emission (Rn) was
measured during PCR amplification, and the difference (ΔRn) between
the fluorescence emission of the product and the baseline was
calculated using IQ5 Optical System Software version 2.1 (Bio-Rad
Laboratories, Inc.) and plotted against the cycle number. Cycle
threshold (CT) values were calculated by determining the number of
thermal cycles at which the emitted fluorescence exceeded the
threshold point, as described previously (27). The reaction mixture contained 10 ng
cDNA diluted in 2.5 µl diethylpyrocarbonate-treated water (Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 12.5
µl (2X) EZtime SYBR Green PCR premix (Yeastern Biotech Corp.) and 2
µl gene-specific primers (final concentration, 50 nM each) in a
final reaction volume of 25 µl. The reaction conditions for Chibby
and β-catenin were as follows: 95°C for 10 min (denaturation of the
template and activation of DNA polymerase), followed by 45 cycles
of 95°C for 20 sec (denaturation of PCR products), 58°C for 1 min
(primer annealing), and 72°C for 15 sec (extension). The reaction
conditions for β-actin were as follows: 95°C for 10 min
(denaturation of the template and activation of DNA polymerase),
followed by 45 cycles of 95°C for 10 sec (denaturation of the PCR
products), 58°C for 1 min (primer annealing), and 55°C for 15 sec
(extension). Each real-time PCR was performed in duplicate to
evaluate data reproducibility. A melting curve analysis of the PCR
products was generated following amplification by heating the
reaction mixtures between 60°C and 95°C with a rate of 0.1°C/sec,
and continuously acquiring fluorescence emission data. The melting
temperatures (Tm) of the target genes and β-actin amplicons were
expected to be ~80.0°C and 85.0°C, respectively, whereas
primer-dimers and/or other non-specific products were characterized
by a decreased Tm (≤75.0°C). Calculations and validation of the
comparative CT (2−ΔΔCq) method (normalizing to an
endogenous reference provides a method for correcting results for
differing amounts of input RNA) were used for target gene mRNA
quantification (28). β-actin was
used as a reference gene to normalize all PCRs for the amount of
loaded RNA. All primer pairs were designed using a web-based
program' GenScript Real-time PCR (TaqMan) Primer Design' provided
by GeneScript.com. The sequences of primer pairs
were as follows: Chibby forward (F), 5′-TCTGGGCTACAGAGTCCTTG-3′;
Chibby reverse (R), 5′-TGTCTTCTTCGGACTGAACG-3′; β-catenin F,
5′-GCAATCCCTGAACTGACAAA-3′; β-catenin R,
5′-TGAGGAGAACGCATGATAGC-3′; β-actin F, 5′-GACATCCGCAAAGACCTGTA-3′;
β-actin R, 5′-GGAGCAATGATCTTGATCTTCA-3′.
Statistical analysis
All statistical procedures were conducted using SPSS
software (version 22.0; IBM Corp, Armonk, NY, USA). The expression
levels of target mRNAs were analyzed using non-parametric
statistical methods, including the Kruskal-Wallis test and median
test, to discriminate whether inter-group differences were
significant among the CIN staging groups (including chronic
cervicitis, CIN 1, CIN 2, CIN 3 and invasive SCC). For
Kruskal-Wallis test, all the mRNA expression levels for the four
CIN groups were entered into one column and rank ordered from
lowest to highest. Once the ranks were assigned, the scores were
split back into the four groups. Subsequently, the mean of the
*ranks* in each group were computed, and Kruskal-Wallis test was
performed to determine whether there was a statistically
significant difference in the mean ranks for each group. The
χ2 test was used to determine whether protein staining
intensity and positive staining area were significantly different
among different CIN groups. The statistical results were presented
separately by the different subcellular locations. In addition,
Chibby/β-catenin ratio was evaluated and compared for any
difference among CIN groups. Finally, logistic regression was
applied to distinguish factors associated with CIN stage and/or
invasive tumor. In addition, the data was categorized as a dummy
variable (29) according to the
criteria 01, by which samples with a Chibby nucleus and cytoplasmic
positive staining area <1% and a β-catenin cytoplasmic intensity
that was greater than or equal to intermediate. Membrane intensity
was identified as strong staining and was indicated as 1; whereas
the counterparts were indicated as 0. The aforementioned dummy
variable of criteria 01 was analyzed using binary logistic
regression to determine whether it was associated with SCC. The
positive prediction value (PPV; the probability that the criteria
01-positive tumors were SCC), the negative prediction value (NPV;
the probability that criteria 01-negative tumors were non-SCC) and
accuracy (refers to the probability that true SCC and non-SCC
samples can be predicted by criteria 01 from overall samples) were
used as performance measures. P<0.05 was considered to indicate
a statistically significant difference.
Results
Chibby mRNA expression is
downregulated in SCC, but not β-catenin
A total of 54/83 paraffin embedded cervical
cancerous tissues were subjected to detection of Chibby and
β-catenin mRNA expression due to a relatively good RNA preparation.
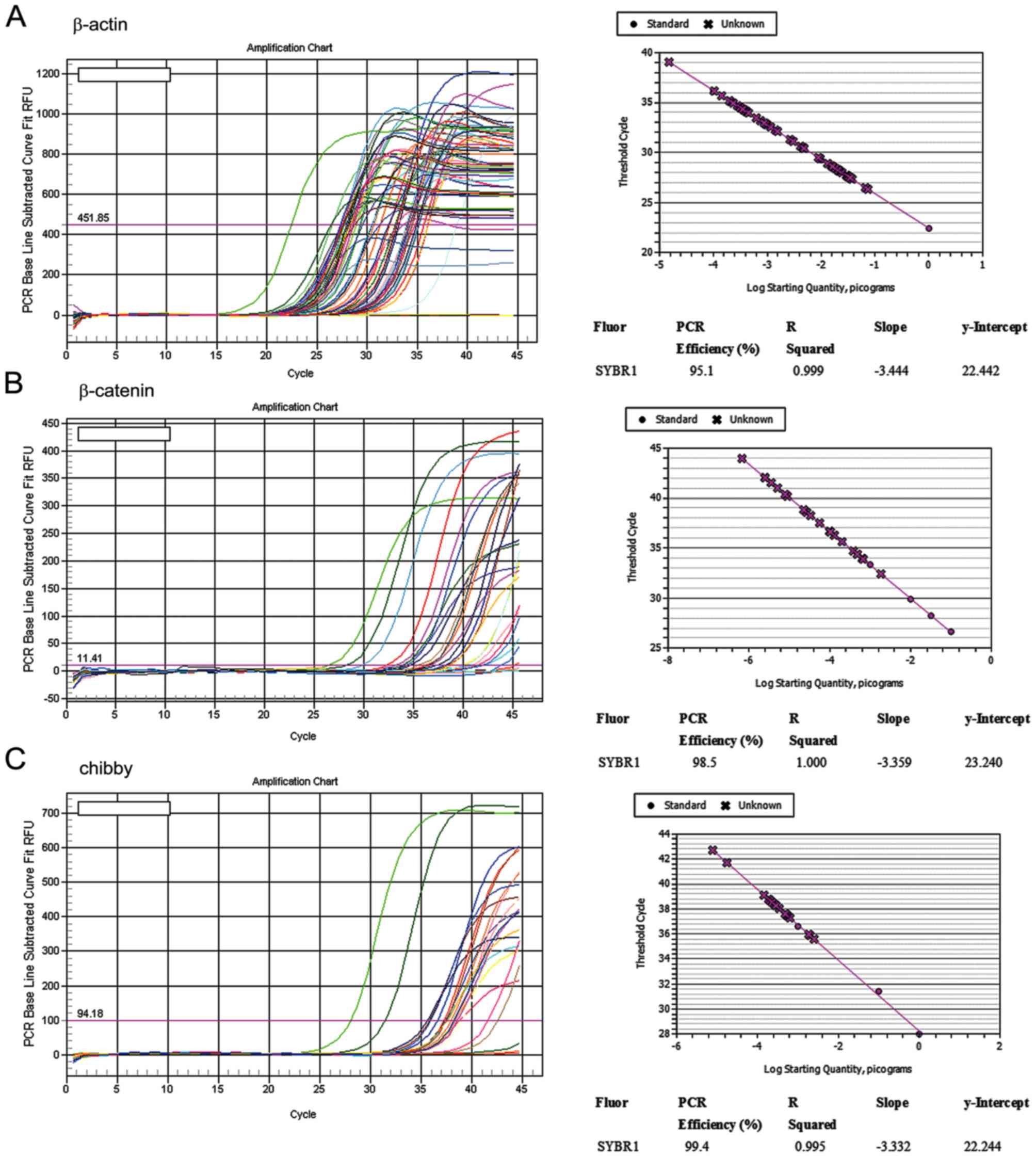
PCR efficiency with respect to β-catenin and Chibby amplification
ranged between 90 and 110%, and the standard curves of the two
genes were obtained with a slope of −3.3 and an R squared >0.99
(the coefficient value of a standard curve), which revealed
successful amplification (Fig. 1). A
non-parametrical approach was adopted for analyzing mRNA
expression, since the expression data was unlikely to be normally
distributed. As presented in the Table
I, using the Kruskal-Wallis test, the raw data of mRNA
expression was converted into ranking values. Chibby mRNA (P=0.001)
and the Chibby/β-catenin ratio (P=0.018), but not β-catenin
(P=0.267), differed among the CIN groups. Progressively decreased
expression levels of Chibby (mean rank, 13.22) and decreased
Chibby/β-catenin ratio (mean rank, 12.44) were identified when
compared with CIN 1 to CIN 3 tumors (>20 mean rank). Using the
median test, Chibby mRNA (Table II,
line 2, 8/9 tumors; 88.89%) and Chibby/β-catenin ratio (Table II, line 6, 9/9 tumors; 100%) revealed
an increased proportion in the mRNA expression level below the
median value in invasive tumors compared with CIN 1, 2 and 3
tumors, which suggested that Chibby mRNA was downregulated
significantly in SCC (Chibby mRNA, P=0.018; Chibby/β-catenin ratio,
P=0.011; Table II).
 | Table I.mRNA expression with respect to
β-catenin and chibby in different CIN stages of cervical
cancer. |
Table I.
mRNA expression with respect to
β-catenin and chibby in different CIN stages of cervical
cancer.
| Variable | CIN | N | Mean rank | χ2 | Df | Asymp.
sig.a,b |
|---|
| Chibby mRNA
expression | CIN 1 | 11 | 39.95 | 15.57 | 3 | 0.001 |
|
| CIN 2 | 9 | 22.22 |
|
|
|
|
| CIN 3 | 25 | 29.06 |
|
|
|
|
| Invasive tumor | 9 | 13.22 |
|
|
|
|
| Total | 54 |
|
|
|
|
| β-catenin mRNA
expression | CIN 1 | 11 | 34.77 | 3.95 | 3 | 0.267 |
|
| CIN 2 | 9 | 22.89 |
|
|
|
|
| CIN 3 | 25 | 25.06 |
|
|
|
|
| Invasive tumor | 9 | 30.00 |
|
|
|
|
| Total | 54 |
|
|
|
|
| Chibby/β-catenin
ratio | CIN 1 | 8 | 30.09 | 10.07 | 3 | 0.018 |
|
| CIN 2 | 9 | 28.78 |
|
|
|
|
| CIN 3 | 25 | 31.32 |
|
|
|
|
| Invasive tumor | 9 | 12.44 |
|
|
|
|
| Total | 51 |
|
|
|
|
 | Table II.mRNA expression level with respect to
β-catenin and chibby in various CIN stages of cervical cancer. |
Table II.
mRNA expression level with respect to
β-catenin and chibby in various CIN stages of cervical cancer.
| Variable |
| CIN 1, n (%) | CIN 2, n (%) | CIN 3, n (%) | Invasive, n (%)
tumor | Median | χ2 | Df | Asymp. sig. |
|---|
| Chibby mRNA
expression | >Median | 9 (81.82) | 4 (44.44) | 13 (52.00) | 1 (11.11) | 486.62 | 10.05 | 3 | 0.018 |
|
| ≤Median | 2 (18.18) | 5 (55.56) | 12 (48.00) | 8
(88.89) |
|
|
|
|
| β-catenin mRNA
expression | >Median | 8 (72.73) | 4(44.44) | 11(44.00) | 4
(44.44) | 663.47 |
2.86 | 3 | 0.415 |
|
| ≤Median | 3 (27.27) | 5 (55.56) | 14 (56.00) | 5
(55.56) |
|
|
|
|
| Chibby/β-catenin
ratio | >Median | 7 (63.64) | 6 (66.67) | 14(56.00) | 0 (0.0) |
1.91 | 11.18 | 3 | 0.011 |
|
| ≤Median | 4 (36.36) | 3 (33.33) | 11(44.00) | 9 (100) |
|
|
|
|
Chibby nucleus and cytoplasmic
staining are associated with CIN stage
Chibby was determined in all chronic cervicitis
samples with a weak to intermediate intensity and <10%
positivity area in the nucleus. In addition, Chibby was identified
with weak intensity and <10% positivity area in the cytoplasm in
half of the chronic cervicitis tissues (Table III; Fig.
2A and B). In the nucleus and cytoplasm, the intensity of
Chibby was identified to be decreased in invasive tumor (45.5% weak
positivity in nucleus; 9.1% weak positivity in the cytoplasm)
compared with the other groups (Table
III, part A, nucleus, P=0.004; part C, cytoplasm, P<0.001).
Notably, the positive staining area of Chibby in the nucleus
(>50% positivity; Table III) and
cytoplasm (11–50% positivity; Table
III) decreased as the CIN stage increased (Table III; P<0.001; Fig. 2C-J).
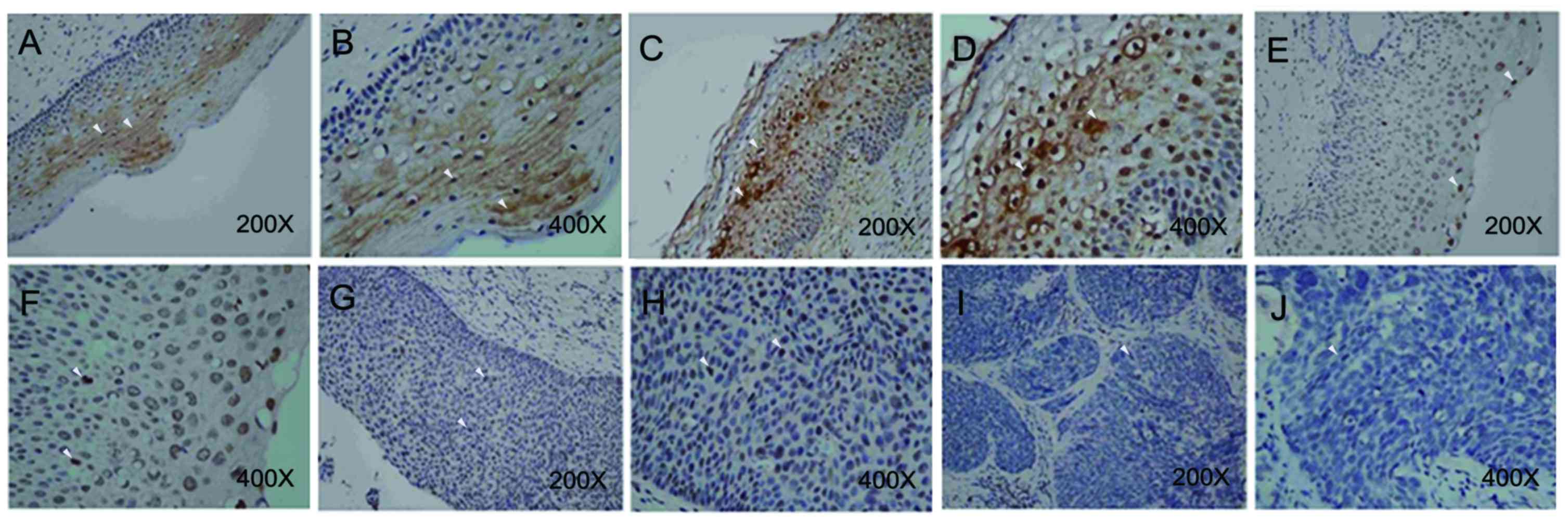 | Figure 2.Expression of Chibby (an antagonist
of β-catenin) in different CIN tumor stages. Immunohistological
staining was performed using a NovoLink polymer detection kit. A
Nikon Eclipse 50i light microscope was used at magnifications of
×40–400. In chronic cervicitis tissues, Chibby was observed either
in nucleus or in cytoplasm at (A) magnification, ×200; and (B)
magnification, ×400. In CIN 1 tumor, Chibby was observed either in
nucleus or in cytoplasm at (C) magnification, ×200; and (D)
magnification, ×400. Chibby was only observed in the nucleus in
partial CIN 2 tumors at (E) magnification, ×200; and (F)
magnification, ×400. G. Nuclear Chibby expression was detected in
CIN 3 tumors at (G) magnification, ×200; and (H) at magnification,
×400. The nucleus and cytoplasm lacked Chibby expression when
observed at (I) magnification, ×200; and (J) at magnification,
×400. CIN, cervical intra-epithelial neoplasia. Left panel of
images (A, C, E, G and I) were captured at ×200 and right panel of
images (B, D, F, H, and J) were captured at ×400. White arrows
indicate positive staining signal. |
 | Table III.Protein expression with respect to
β-catenin and chibby in different CIN stages of cervical
cancer. |
Table III.
Protein expression with respect to
β-catenin and chibby in different CIN stages of cervical
cancer.
| A, Chibby nucleus
intensity |
|---|
|
|---|
|
| Count, n (%) |
|
|
|
|---|
|
|
|
|
|
|
|---|
| Staining | Chronic
cervicitis | CIN 1 | CIN 2 | CIN 3 | Invasive tumor | χ2 | df | P-value |
|---|
| None | 0 (0.0) | 0 (0.0) | 1 (6.7) | 4 (12.1) | 11 (50.0) | 29.258 | 12 | 0.004 |
| Weak | 3 (75.0) | 7 (53.8) | 11 (73.3) | 18 (54.5) | 10 (45.5) |
|
|
|
| Intermediate | 1 (25.0) | 5 (38.5) | 3 (20.0) | 11 (33.3) | 1 (4.5) |
|
|
|
| Strong | 0 (0.0) | 1 (7.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
|
|
|
| Total count, n | 4 | 13 | 15 | 33 | 22 |
|
|
|
|
| B, Chibby
nucleus area |
|
|
| Count, n
(%) |
|
|
|
|
|
|
|
|
|
|
Staining | Chronic
cervicitis | CIN 1 | CIN 2 | CIN 3 | Invasive
tumor |
χ2 | df | P-value |
|
| None | 0 (0.0) | 0 (0.0) | 1 (6.7) | 4 (12.1) | 11 (50.0) | 60.179 | 16 | <0.001 |
| <1 | 3 (75.0) | 1 (7.7) | 0 (0.0) | 2 (6.1) | 1 (4.5) |
|
|
|
| 1–10 | 1 (25.0) | 0 (0.0) | 2 (13.3) | 0 (0.0) | 5 (22.7) |
|
|
|
| 11–50 | 0 (0.0) | 2 (15.4) | 3 (20.0) | 8 (24.2) | 0 (0.0) |
|
|
|
| >50 | 0 (0.0) | 10 (76.9) | 9 (60.0) | 19 (57.6) | 5 (22.7) |
|
|
|
| Total count, n | 4 | 13 | 15 | 33 | 22 |
|
|
|
|
| C, Chibby
cytoplasm intensity |
|
|
| Count, n
(%) |
|
|
|
|
|
|
|
|
|
|
Staining | Chronic
cervicitis | CIN 1 | CIN 2 | CIN 3 | Invasive
tumor |
χ2 | df | P-value |
|
| None | 2 (50.0) | 0 (0.0) | 11 (73.3) | 25 (75.8) | 20 (90.9) | 49.101 | 12 | <0.001 |
| Weak | 2 (50.0) | 4 (30.8) | 1 (6.7) | 6 (18.2) | 2 (9.1) |
|
|
|
| Intermediate | 0 (0.0) | 7 (53.8) | 2 (13.3) | 1 (3.0) | 0 (0.0) |
|
|
|
| Strong | 0 (0.0) | 2 (15.4) | 1 (6.7) | 1 (3.0) | 0 (0.0) |
|
|
|
| Total count, n | 4 | 13 | 15 | 33 | 22 |
|
|
|
|
| D, Chibby
cytoplasm area |
|
|
| Count, n
(%) |
|
|
|
|
|
|
|
|
|
|
Staining | Chronic
cervicitis | CIN 1 | CIN 2 | CIN 3 | Invasive
tumor |
χ2 | df | P-value |
|
| None | 0 (0.0) | 0 (0.0) | 1 (6.7) | 4 (12.1) | 11 (50.0) | 29.258 | 12 | 0.004 |
| None | 2 (50.0) | 0 (0.0) | 11 (73.3) | 25 (75.8) | 20 (90.9) | 60.179 | 16 | <0.001 |
| <1 | 1 (25.0) | 2 (15.4) | 0 (0.0) | 0 (0.0) | 1 (4.5) |
|
|
|
| 1–10 | 1 (25.0) | 2 (15.4) | 0 (0.0) | 0 (0.0) | 1 (4.5) |
|
|
|
| 11–50 | 0 (0.0) | 9 (69.2) | 4 (26.7) | 6 (18.2) | 0 (0.0) |
|
|
|
| >50 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (6.1) | 0 (0.0) |
|
|
|
| Total count, n | 4 | 13 | 15 | 33 | 22 |
|
|
|
|
| E, β-catenin
nucleus intensity |
|
|
| Count, n
(%) |
|
|
|
|
|
|
|
|
|
|
Staining | Chronic
cervicitis | CIN 1 | CIN 2 | CIN 3 | Invasive
tumor |
χ2 | df | P-value |
|
| ND | 0 (0.0) | 0 (0.0) | 2 (13.3) | 0 (0.0) | 3 (13.6) |
|
| ND |
| None | 4 (100.0) | 13 (100.0) | 13 (86.7) | 33 (100.0 | 19 (86.4) |
|
|
|
| Total count, n | 4 | 13 | 15 | 33 | 22 |
|
|
|
|
| F, β-catenin
nucleus area |
|
|
| Count, n
(%) |
|
|
|
|
|
|
|
|
|
|
Staining | Chronic
cervicitis | CIN 1 | CIN 2 | CIN 3 | Invasive
tumor |
χ2 | df | P-value |
|
| ND | 0 (0.0) | 0 (0.0) | 2 (13.3) | 0 (0.0) | 3 (13.6) |
|
| ND |
| None | 4 (100.0) | 13 (100.0) | 13 (86.7) | 33 (100.0 | 19 (86.4) |
|
|
|
| Total count, n | 4 | 13 | 15 | 33 | 22 |
|
|
|
|
| G, β-catenin
cytoplasm intensity |
|
|
| Count, n
(%) |
|
|
|
|
|
|
|
|
|
|
Staining | Chronic
cervicitis | CIN 1 | CIN 2 | CIN 3 | Invasive
tumor |
χ2 | df | P-value |
|
| ND | 0 (0.0) | 0 (0.0) | 2 (13.3) | 0 (0.0) | 3 (13.6) |
|
| ND |
| Weak | 0 (0.0) | 12 (92.3) | 5 (38.5) | 15 (45.5) | 5 (26.3) |
|
|
|
| Intermediate | 0 (0.0) | 1 (7.7) | 4 (30.8) | 8 (24.2) | 7 (36.8) |
|
|
|
| Strong | 0 (0.0) | 0 (0.0) | 4 (30.8) | 10 (30.3) | 7 (36.8) |
|
|
|
| Total count, n | 4 | 13 | 13 | 33 | 19 |
|
|
|
|
| H, β-catenin
cytoplasm area |
|
|
| Count, n
(%) |
|
|
|
|
|
|
|
|
|
|
Staining | Chronic
cervicitis | CIN 1 | CIN 2 | CIN 3 | Invasive
tumor |
χ2 | df | P-value |
|
| None | 4 (100.0) | 0 (0.0) | 2 (13.3) | 0 (0.0) | 3 (13.6) | 71.498 | 16 | <0.001 |
| <1 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (3.0) | 0 (0.0) |
|
|
|
| 1–10 | 0 (0.0) | 1 (7.7) | 1 (6.7) | 0 (0.0) | 0 (0.0) |
|
|
|
| 11–50 | 0 (0.0) | 12 (92.3) | 3 (20.0) | 5 (15.2) | 2 (9.1) |
|
|
|
| >50 | 0 (0.0) | 0 (0.0) | 9 (60.0) | 27 (81.8) | 17 (77.3) |
|
|
|
| Total count, n | 4 | 13 | 15 | 33 | 22 |
|
|
|
|
| I, β-catenin
membrane intensity |
|
|
| Count, n
(%) |
|
|
|
|
|
|
|
|
|
|
Staining | Chronic
cervicitis | CIN 1 | CIN 2 | CIN 3 | Invasive
tumor |
χ2 | df | P-value |
|
| None | 4 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 7.831 | 6 | 0.251 |
| Weak |
| 1 (7.7) | 2 (15.4) | 8 (24.2) | 2 (10.5) |
|
|
|
| Intermediate |
| 6 (46.2) | 4 (30.8) | 12 (36.4) | 3 (15.8) |
|
|
|
| Strong |
| 6 (46.2) | 7 (53.8) | 13 (39.4) | 14 (73.7) |
|
|
|
| Total count, n | 4 | 13 | 13 | 33 | 19 |
|
|
|
|
| J, β-catenin
membrane area |
|
|
| Count, n
(%) |
|
|
|
|
|
|
|
|
|
|
Staining | Chronic
cervicitis | CIN 1 | CIN 2 | CIN 3 | Invasive
tumor |
χ2 | df | P-value |
|
| <1 | 4 (100.0) | 0 (0.0) | 2 (13.3) | 0 (0.0) | 3 (13.6) | 33.206 | 12.0 | 0.001 |
| 1–10 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (3.0) | 0 (0.0) |
|
|
|
| 11–50 | 0 (0.0) | 2 (15.4) | 2 (13.3) | 1 (3.0) | 2 (9.1) |
|
|
|
| >50 | 0 (0.0) | 11 (84.6) | 11 (73.3) | 31 (93.9) | 17 (77.3) |
|
|
|
| Total count, n | 4 | 13 | 15 | 33 | 22 |
|
|
|
β-catenin cytoplasmic and membrane
staining intensity are associated with CIN stage
β-catenin protein expression was detected in the
cytoplasm and membrane, but not in the nucleus (Table III, part E-J). Specifically,
β-catenin was frequently detected at the sites where proximal to
cell membrane (Fig. 3). In chronic
cervicitis tissues, no β-catenin expression was detected in any of
the samples. Cytoplasmic β-catenin expression was increased in CIN
1 (weak intensity, 11–50% positive staining area, 12/13, 92.3%) and
CIN 3 (>50% positive staining area, 27/33, 81.8%) stage tumor
compared with the invasive tumor, with the intensity principally
ranging between intermediate and strong (14/19; 73.7%) and a
positive staining area >50% (17/22; 77.3%) in invasive tumors
(Table III, part G&H;
P<0.001). An identical pattern was observed in membrane
β-catenin, where the intensity was primarily identified as strong
(14/19; 73.7%) and a positive staining area of >50% (17/22;
77.3%) in the invasive tumor group. However, no inter-group
differences were identified in β-catenin membrane intensity between
the different CIN staging groups (Table
III, part I, P=0.251). The results of the present study may
suggest that β-catenin is overexpressed in the cytoplasm and
membrane in advanced CIN tumors, and may be an early event
associated with cervical cancer tumorigenesis. To clarify whether
and which factor was associated with CIN stage and invasive tumors,
multi-nominal logistic regression was performed, and the results
are presented in Table IV. Chibby
nucleus (P=0.001) and cytoplasmic (P<0.001) positive staining
area and β-catenin cytoplasmic (P=0.006) and membrane intensity
(P=0.018) were independently associated with CIN stage, but not the
Chibby cytoplasmic intensity (P=0.062). On the basis of logistical
regression, the data was categorized according to the criteria 01,
by which samples with a Chibby nucleus and cytoplasmic positive
staining area <1%, β-catenin cytoplasmic intensity was greater
than or equal to intermediate, and membrane intensity was
identified as strong staining were indicated as 1; whereas the
counterparts were indicated as 0. The dummy variable of criteria 01
was brought to binary logistic regression to predict SCC. As
Table V presents, criteria 01 was
significantly associated with SCC (P=0.002) and samples that
fulfilled criteria 01 exhibited a 5.364-fold increased risk of
developing SCC. The PPV was 50.0%, the NPV was 84.3% and the
accuracy was 76.1%. The results indicated that determining the
protein expression of Chibby and β-catenin, on the basis of
criteria 01, may serve as biomarker to predict invasive SCC.
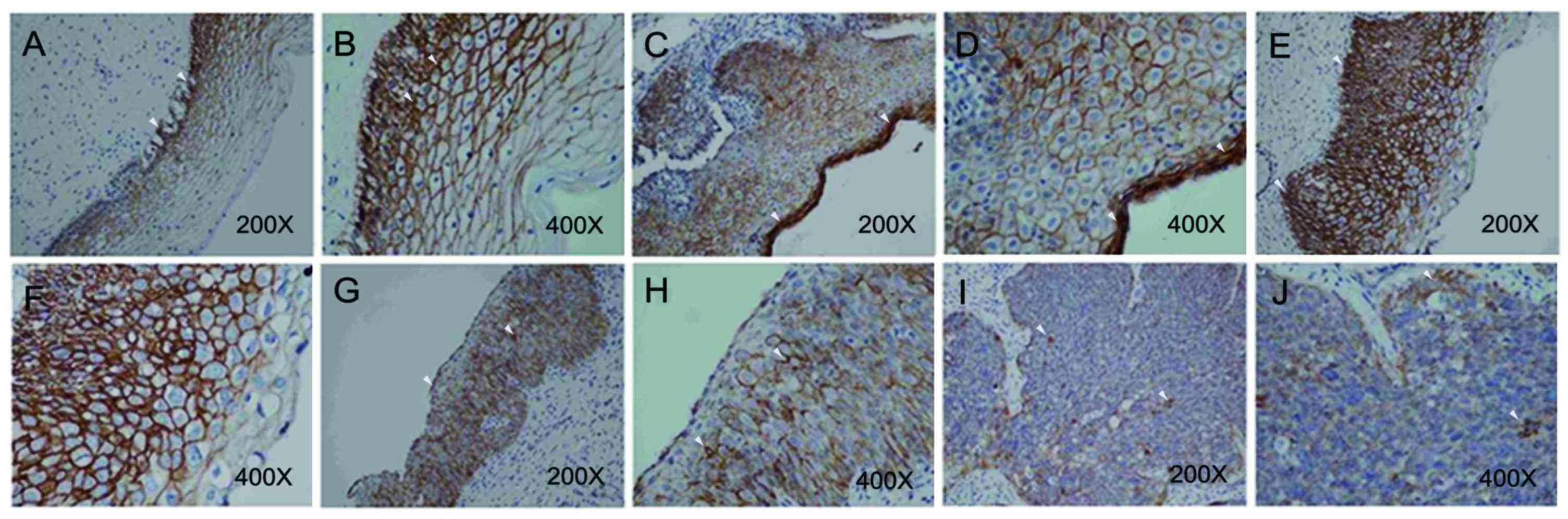 | Figure 3.β-catenin expression in different CIN
stage tumors. Immunohistological staining was performed using a
NovoLink polymer detection kit. A Nikon Eclipse 50i light
microscope was used at magnifications of ×40–400. (A) Nuclear
expression of β-catenin was not observed in normal cervix at
magnification, ×200; however, (B) β-catenin was observed in the
cytoplasm, principally localized in sites adjacent to cell membrane
at magnification, ×400. (C) Nuclear expression of β-catenin was
also not observed in CIN 1 tumors at magnification, ×200; however,
(D) β-catenin was detected in the cytoplasm at magnification, ×400.
β-catenin was observed, primarily in the cell membrane of CIN 2
tumors, at (E) magnification, ×200; and (F) at magnification, ×400.
β-catenin was observed, principally in the cell membrane of CIN 3
tumors, at (G) magnification, ×200; and (H) magnification, ×400.
Cytosolic β-catenin was observed, principally in the cell membrane
of SCC tumors, at (I) magnification, ×200; and (J) magnification,
×400. White arrows indicate positive staining signal. CIN, cervical
intra-epithelial neoplasia; SCC, squamous-cell carcinoma. |
 | Table IV.Multi-nominal regression analysis to
predict factors associated with CIN staging. |
Table IV.
Multi-nominal regression analysis to
predict factors associated with CIN staging.
|
| Model fitting
criteria | Likelihood ratio
tests |
|---|
|
|
|
|
|---|
| Effecta | −2 Log likelihood
of reduced model | χ2 | df | P-value |
|---|
| Intercept | 40.109 |
0.000 | 0 | . |
| Chibby nucleus
area | 72.127 | 32.017 | 12 | 0.001 |
| Chibby cytoplasmic
intensity | 52.130 | 12.021 | 6 | 0.062 |
| Chibby cytoplasmic
area | 73.933 | 33.824 | 9 | <0.001 |
| β-catenin
cytoplasmic intensity | 58.017 | 17.908 | 6 | 0.006 |
| β-catenin membrane
intensity | 55.452 | 15.343 | 6 | 0.018 |
 | Table V.Results of logistical regression that
was aimed at determining whether criteria 01 can predict SCC
development. |
Table V.
Results of logistical regression that
was aimed at determining whether criteria 01 can predict SCC
development.
| b | B | Wald | Sig. | Exp (B) | PPV | NPV | Overall |
|---|
| Criteria 01 | 1.680 | 9.739 | 0.002 | 5.364 | 50.0% | 84.3% | 76.1% |
| Constant | −1.680 | 26.157 | 0.000 | 0.186 |
|
|
|
Discussion
Abnormal activation of the Wnt signaling pathway is
typically observed in a number of types of cancer cell (30). β-catenin stability may determine the
activation of the Wnt signaling pathway. In quiescent cells,
cytosolic β-catenin is decreased due to its degradation by the
ubiquitin-proteasome dependent pathway; however, when β-catenin is
translocated to the nucleus, it binds to LEF/TCF, and consequently
activates basal transcriptional factor and a subset of downstream
genes comprising cyclin D1, c-myc, c-jun, and matrix
metalloproteinases 2 and 7. The activation of these genes may
prompt cancer cell proliferation and have been associated with
cancer cell invasiveness (31–33). In
contrast to β-catenin protein expression and localization,
β-catenin mRNA was less likely to be associated with tumorigenesis
(34). Generally, the nuclear
translocation of β-catenin is thought to be one of major causes of
tumorigenesis (35). Furthermore,
β-catenin is dysregulated and has been identified to accumulate in
cancer cells; therefore, the abnormal accumulation observed in
cervical cancer was not surprising (36–38).
Increased expression of β-catenin may accelerate proliferation and
differentiation in malignant cervical cancer, including CIN 3
tumors. In CIN 3 malignant tumors, increased proliferation and
migration are indicators of the patient outcome. Previous studies
have revealed that β-catenin expression was associated with patient
survival (39–42). Decreased expression of β-catenin is
associated with the long-term survival of patients with brainstem
gliomas (43). In cervical
adenocarcinomas, β-catenin expression has been associated with a
poor 10-year survival, and detecting β-catenin is suggested as a
prognostic marker of cervical adenocarcinoma (44).
In the normal cervix, we suggested that cytosolic
and nuclear expression of β-catenin is hypothesized to be
antagonized by Chibby, as previously a reported phenomenon in a
mammalian cell line (45). Increased
expression of β-catenin is hypothesized to be associated with
highly proliferative and differentiating cancer cells, and expected
to be observed in the majority of types of cancerous tissues
(46). In the present study,
β-catenin was highly expressed in advanced CIN stage tumors, in
particular, in invasive SCC. Additionally, β-catenin gradually
accumulated as the CIN stage increased, between CIN 2 and SCC. This
result is consistent with current hypotheses about β-catenin
expression in cervical cancer (47,48).
Notably, in the present study, β-catenin expression was restricted
to the cytoplasm in all tissues. Furthermore, β-catenin principally
accumulated near the cell membrane and lacked expression within the
nucleus. The nuclear translocation of β-catenin was not detected in
tumors of higher CIN stage, or even in CIN 1 lesions. This finding
is in agreement with a previous study in which β-catenin was
observed to be expressed in the cell membrane of pre-malignant
tumors, but not cancerous tissues (38). In the present study, membrane
β-catenin was identified to be more abundant in CIN 1 lesions
compared with cytosolic β-catenin. In the present study, between 11
and 50% positive staining of cytosolic β-catenin and an
intermediate level of membrane β-catenin was observed more
frequently in pre-malignancy tissues and decreased in tumors of
high CIN stages. Furthermore, β-catenin mRNA and protein were not
differently expressed between CIN 2 and invasive SCC; however,
intermediate-level cytosolic β-catenin accumulated between chronic
cervicitis and invasive SCC. These results were consistent with
that of a previous study, where β-catenin expression was not
significantly different in SCC but was increased in high-grade
squamous intraepithelial lesions (equal to CIN2-CIN3) and SCC,
compared with pre-malignant lesions (14). Thus, discrepancies in β-catenin
protein expression between pre-malignant and cancerous tissues in
previous studies may be due to different definitions of
‘positivity’ in data interpretation. The results of the present
study indicated that increased levels (intermediate to strong) of
β-catenin protein accumulation may be associated with tumorigenesis
in cervical cancer; however, its role in advanced disease
progression remains unknown.
Chibby is an antagonist involved in the
Wnt/β-catenin signaling pathway and is hypothesized to serve a
function in tumorigenesis. Previous studies have found that Chibby
is downregulated in a number of types of cancer including
neuroblastoma (19), lung cancer
(20), ependymoma (21) and colorectal cancer (22,23). In
neuroblastoma, Chibby has been suggested to interact with two tumor
suppressors, neuroblastoma breakpoint family member 1 and clusterin
(19). In a colon cancer line, Chibby
was identified to bind to 14-3-3 protein and enabled β-catenin to
be sequestered for nuclear exportation (23). Additionally, Chibby has been revealed
to regulate β-catenin expression in lung cancer (20). In spite of these previous results, the
associations between Chibby, pre-malignancy and tumorigenesis in
cervical cancer remain unknown. Compared with chronic cervicitis
tissue, nuclear and cytosolic Chibby expression was significantly
decreased in lesions with a high CIN stage (P<0.001). For
nuclear Chibby staining, the expression levels varied with CIN
stage, with tumors of higher CIN stage progressively expressing
decreased nuclear Chibby (P<0.001). For cytosolic Chibby
staining, increased protein expression was determined in CIN 1
lesions compared with later CIN tumor stages. The results of the
present study indicated that the downregulation of Chibby in the
nucleus and cytoplasm is associated with tumorigenesis in cervical
cancer. Furthermore, Chibby expression is negatively associated
with disease progression, suggesting that Chibby is a functional
tumor suppressor. In the present study, it was hypothesized that
Chibby is involved in the Wnt signaling pathway and antagonizes the
β-catenin interaction with TCF/LEF. In addition, Chibby may
coordinate with 14-3-3 to execute nuclear exportation of the
TCF/LEF-β-catenin complex and downregulate downstream gene
expression in the Wnt signaling pathway, as described previously
(17,18). It may be hypothesized that when
nuclear Chibby expression is decreased or absent, accumulation of
the TCF/LEF-β-catenin complex, due to the relatively decreased
Chibby/β-catenin ratio, may subsequently trigger downstream Wnt
signaling pathway gene expression, leading to cell proliferation
and cervical cancer tumorigenesis.
Our previous study demonstrated that the Chibby mRNA
and protein are downregulated in two cervical cancer lines, HeLa
and SiH (Huang Y.L., unpublished work). In this study, a
significant difference in Chibby mRNA expression and the
Chibby/β-catenin ratio between different CIN groups was determined.
In addition, nuclear and cytosolic Chibby expression were
significantly downregulated in high CIN stage tumors, in
particular, when Chibby expression was interpreted by positive
staining area (nucleus, >50% area; cytoplasm, 11–50% area).
According to the results of the present study, Chibby nuclear and
cytoplasmic positive staining area and β-catenin cytoplasmic and
membrane intensity were associated with CIN staging. Therefore, the
criteria 01 was constructed and converted into a dummy variable for
logistical regression, to determine whether it may predict SCC.
Criteria 01 was significantly associated with SCC (P=0.002), with
PPV and NPV of 50.0 and 84.3%, respectively. In terms of clinical
relevance, the negative prediction rate of a biomarker is
relatively important since it may beneficial to rule out false
positive cases during the risk assessments. Furthermore, it is not
affected by disease prevalence as PPV. The results of the present
study indicated that determining the protein expression of Chibby
and β-catenin, on the basis of criteria 01, may serve as biomarker
for predicting invasive SCC of cervical cancer.
Acknowledgements
The authors would like to thank the Department of
Pathology of KAFGH (Kaohsiung, Taiwan) for assisting with the
collection of tissue specimens and accessing the medical reviews,
Dr. Kao Wei-Tsung, a deputy director of Laboratory of Medical
Research, KAFGH, for providing the facilities and laboratory space
to perform the molecular biology experiments, and Gary Mawyer,
Managing Editor of Journal of the Wound, Ostomy and Continence
Nurses Society (WOCN), for language editing. The present study was
supported by Outpatient Department (OPD) research (grant no. 101-2)
from KAFGH.
References
|
1
|
Parkin DM and Bray F: Chapter 2: The
burden of HPV-related cancers. Vaccine. 24 Suppl 3:S3/11–25. 2006.
View Article : Google Scholar
|
|
2
|
Shah KV, Kessis TD, Shah F, Gupta JW,
Shibata D and Jones RW: Human papillomavirus investigation of
patients with cervical intraepithelial neoplasia 3, some of whom
progressed to invasive cancer. Int J Gynecol Pathol. 15:127–130.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Parkin DM: Global cancer statistics in the
year 2000. Lancet Oncol. 2:533–543. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Perez-Plasencia C, Duenas-Gonzalez A and
Alatorre-Tavera B: Second hit in cervical carcinogenesis process:
Involvement of wnt/beta catenin pathway. Int Arch Med. 1:102008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lustig B and Behrens J: The Wnt signaling
pathway and its role in tumor development. J Cancer Res Clin Oncol.
129:199–221. 2003.PubMed/NCBI
|
|
6
|
Kwong KY, Zou Y, Day CP and Hung MC: The
suppression of colon cancer cell growth in nude mice by targeting
beta-catenin/TCF pathway. Oncogene. 21:8340–8346. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chien AJ, Moore EC, Lonsdorf AS,
Kulikauskas RM, Rothberg BG, Berger AJ, Major MB, Hwang ST, Rimm DL
and Moon RT: Activated Wnt/beta-catenin signaling in melanoma is
associated with decreased proliferation in patient tumors and a
murine melanoma model. Proc Natl Acad Sci USA. 106:pp. 1193–1198.
2009; View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Uren A, Fallen S, Yuan H, Usubütün A,
Küçükali T, Schlegel R and Toretsky JA: Activation of the canonical
Wnt pathway during genital keratinocyte transformation: A model for
cervical cancer progression. Cancer Res. 65:6199–6206. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pollack AL, Barth AI, Altschuler Y, Nelson
WJ and Mostov KE: Dynamics of beta-catenin interactions with APC
protein regulate epithelial tubulogenesis. J Cell Biol.
137:1651–1662. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takemaru KI and Moon RT: The
transcriptional coactivator CBP interacts with beta-catenin to
activate gene expression. J Cell Biol. 149:249–254. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Calvisi DF, Ladu S, Factor VM and
Thorgeirsson SS: Activation of beta-catenin provides proliferative
and invasive advantages in c-myc/TGF-alpha hepatocarcinogenesis
promoted by phenobarbital. Carcinogenesis. 25:901–908. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Morin PJ, Sparks AB, Korinek V, Barker N,
Clevers H, Vogelstein B and Kinzler KW: Activation of
beta-catenin-Tcf signaling in colon cancer by mutations in
beta-catenin or APC. Science. 275:1787–1790. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Polakis P: Wnt signaling and cancer. Genes
Dev. 14:1837–1851. 2000.PubMed/NCBI
|
|
14
|
Yang JZ, Zhang XH, Wu WX, Yan X, Liu YL,
Wang JL and Wang FR: Expression of EP-CAM, beta-catenin in the
carcinogenesis of squamous cell carcinoma of uterine cervix.
Zhonghua Zhong Liu Za Zhi. 25:372–375. 2003.(In Chinese).
PubMed/NCBI
|
|
15
|
Saito-Diaz K, Chen TW, Wang X, Thorne CA,
Wallace HA, Page-McCaw A and Lee E: The way Wnt works: Components
and mechanism. Growth Factors. 31:1–31. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zirn B, Wittmann S, Graf N and Gessler M:
Chibby, a novel antagonist of the Wnt pathway, is not involved in
Wilms tumor development. Cancer Lett. 220:115–120. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li FQ, Mofunanya A, Harris K and Takemaru
K: Chibby cooperates with 14-3-3 to regulate beta-catenin
subcellular distribution and signaling activity. J Cell Biol.
181:1141–1154. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li FQ, Mofunanya A, Fischer V, Hall J and
Takemaru K: Nuclear-cytoplasmic shuttling of Chibby controls
beta-catenin signaling. Mol Biol Cell. 21:311–322. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vandepoele K, Staes K, Andries V and van
Roy F: Chibby interacts with NBPF1 and clusterin, two candidate
tumor suppressors linked to neuroblastoma. Exp Cell Res.
316:1225–1233. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu HT, Li QC, Dai SD, Xie XM, Liu DI and
Wang EH: The expression patterns and correlations of chibby,
β-catenin and DNA methyltransferase-1 and their clinicopathological
significance in lung cancers. APMIS. 119:750–758. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Karakoula K, Suarez-Merino B, Ward S,
Phipps KP, Harkness W, Hayward R, Thompson D, Jacques TS, Harding
B, Beck J, et al: Real-time quantitative PCR analysis of pediatric
ependymomas identifies novel candidate genes including TPR at 1q25
and CHIBBY at 22q12-q13. Genes Chromosomes Cancer. 47:1005–1022.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fischer V, Brown-Grant DA and Li FQ:
Chibby suppresses growth of human SW480 colon adenocarcinoma cells
through inhibition of β-catenin signaling. J Mol Signal. 7:62012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schuierer MM, Graf E, Takemaru K,
Dietmaier W and Bosserhoff AK: Reduced expression of β-catenin
inhibitor Chibby in colon carcinoma cell lines. World J
Gastroenterol. 12:1529–1535. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou CZ, Qiu GQ, Zhang F, He L and Peng
ZH: Loss of heterozygosity on chromosome 1 in sporadic colorectal
carcinoma. World J Gastroenterol. 10:1431–1435. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jenkins D: Histopathology and
cytopathology of cervical cancer. Dis Markers. 23:199–212. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kerns BJ, Jordan PA, Moore MB, Humphrey
PA, Berchuck A, Kohler MF, Bast RC Jr, Iglehart JD and Marks JR:
p53 overexpression in formalin-fixed, paraffin-embedded tissue
detected by immunohistochemistry. J Histochem Cytochem.
40:1047–1051. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Giulietti A, Overbergh L, Valckx D,
Decallonne B, Bouillon R and Mathieu C: An overview of real-time
quantitative PCR: Applications to quantify cytokine gene
expression. Methods. 25:386–401. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin HY, Huang CH, Yu TJ, Wu WJ, Yang MC
and Lung FW: p53 codon 72 polymorphism as a progression index for
bladder cancer. Oncol Rep. 27:1193–1199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Barker N and Clevers H: Mining the Wnt
pathway for cancer therapeutics. Nat Rev Drug Discov. 5:997–1014.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
He TC, Sparks AB, Rago C, Hermeking H,
Zawel L, da Costa LT, Morin PJ, Vogelstein B and Kinzler KW:
Identification of c-MYC as a target of the APC pathway. Science.
281:1509–1512. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tetsu O and McCormick F: Beta-catenin
regulates expression of cyclin D1 in colon carcinoma cells. Nature.
398:422–426. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu B, Crampton SP and Hughes CC: Wnt
signaling induces matrix metalloproteinase expression and regulates
T cell transmigration. Immunity. 26:227–239. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang K, Li N, Yeung CH, Cooper TG, Liu XX,
Liu J, Wang WT, Li Y, Shi H and Liu FJ: Comparison of gene
expression of the oncogenic Wnt/β-catenin signaling pathway
components in the mouse and human epididymis. Asian J Androl.
17:1006–1011. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jamieson C, Sharma M and Henderson BR: Wnt
signaling from membrane to nucleus: β-catenin caught in a loop. Int
J Biochem Cell Biol. 44:847–850. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shinohara A, Yokoyama Y, Wan X, Takahashi
Y, Mori Y, Takami T, Shimokawa K and Tamaya T: Cytoplasmic/nuclear
expression without mutation of exon 3 of the beta-catenin gene is
frequent in the development of the neoplasm of the uterine cervix.
Gynecol Oncol. 82:450–455. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pereira-Suarez AL, Meraz MA, Lizano M,
Estrada-Chávez C, Hernández F, Olivera P, Pérez E, Padilla P, Yaniv
M, Thierry F and García-Carrancá A: Frequent alterations of the
beta-catenin protein in cancer of the uterine cervix. Tumour Biol.
23:45–53. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rodríguez-Sastre MA, González-Maya L,
Delgado R, Lizano M, Tsubaki G, Mohar A and García-Carrancá A:
Abnormal distribution of E-cadherin and beta-catenin in different
histologic types of cancer of the uterine cervix. Gynecol Oncol.
97:330–336. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Walther A, Johnstone E, Swanton C, Midgley
R, Tomlinson I and Kerr D: Genetic prognostic and predictive
markers in colorectal cancer. Nat Rev Cancer. 9:489–499. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jin J, Zhan P, Katoh M, Kobayashi SS, Phan
K, Qian H, Li H, Wang X, Wang X and Song Y; written on behalf of
the AME Lung Cancer Collaborative Group, : Prognostic significance
of β-catenin expression in patients with non-small cell lung
cancer: A meta-analysis. Transl Lung Cancer Res. 6:97–108. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang S, Wang Z, Shan J, Yu X, Li L, Lei
R, Lin D, Guan S and Wang X: Nuclear expression and/or reduced
membranous expression of β-catenin correlate with poor prognosis in
colorectal carcinoma: A meta-analysis. Medicine (Baltimore).
95:e55462016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang DP, Li XW and Lang JH: Prognostic
value of β-catenin expression in breast cancer patients: A
meta-analysis. Asian Pac J Cancer Prev. 16:5625–5633. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu W, Tian Y, Wan H, Ma J, Song Y, Wang Y
and Zhang L: Expression of β-catenin and E- and N-cadherin in human
brainstem gliomas and clinicopathological correlations. Int J
Neurosci. 123:318–323. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Imura J, Ichikawa K, Takeda J and Fujimori
T: Beta-catenin expression as a prognostic indicator in cervical
adenocarcinoma. Int J Mol Med. 8:353–358. 2001.PubMed/NCBI
|
|
45
|
Takemaru K, Yamaguchi S, Lee YS, Zhang Y,
Carthew RW and Moon RT: Chibby, a nuclear beta-catenin-associated
antagonist of the Wnt/Wingless pathway. Nature. 422:905–909. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pestereli HE, Erdogan GG, Karavel S and
Simsek T: Cadherin/catenin expression in squamous lesions of
uterine cervix. Turkish J Cancer. 36:64–68. 2006.
|
|
47
|
García-Tamayo J, Molina J and
Blasco-Olaetxea E: Importance of immunohistochemical studies in the
diagnosis and the prognostic evaluation of cervical intraepithelial
neoplasia and invasive squamous cell carcinoma of the uterine
cervix. Review. Invest Clin. 50:241–250. 2009.(In Spanish).
|
|
48
|
Bahrami A, Hasanzadeh M, ShahidSales S,
Yousefi Z, Kadkhodayan S, Farazestanian M, Mashhad M Joudi, Gharib
M, Hassanian S Mahdi and Avan A: Clinical significance and
prognosis value of Wnt signaling pathway in cervical cancer. J Cell
Biochem. 2017.(Epub ahead of print). View Article : Google Scholar
|